Abstract
Developing new methods to detect biomass information on freshwater fish in farm conditions enables the creation of decision bases for precision feeding. In this study, an approach based on Keypoints R-CNN is presented to identify species and measure length automatically using an underwater stereo vision system. To enhance the model’s robustness, stochastic enhancement is performed on image datasets. For further promotion of the features extraction capability of the backbone network, an attention module is integrated into the ResNeXt50 network. Concurrently, the feature pyramid network (FPN) is replaced by an improved path aggregation network (I-PANet) to achieve a greater fusion of effective feature maps. Compared to the original model, the mAP of the improved one in object and key point detection tasks increases by 4.55% and 2.38%, respectively, with a small increase in the number of model parameters. In addition, a new algorithm is introduced for matching the detection results of neural networks. On the foundation of the above contents, coordinates of head and tail points in stereo images as well as fish species can be obtained rapidly and accurately. A 3D reconstruction of the fish head and tail points is performed utilizing the calibration parameters and projection matrix of the stereo camera. The estimated length of the fish is acquired by calculating the Euclidean distance between two points. Finally, the precision of the proposed approach proved to be acceptable for five kinds of common freshwater fish. The accuracy of species identification exceeds 94%, and the relative errors of length measurement are less than 10%. In summary, this method can be utilized to help aquaculture farmers efficiently collect real-time information about fish length.
1. Introduction
In recent years, fish species with a pleasant taste and high content of animal protein, have been increasingly popular in human daily consumption [1,2]. In aquaculture, fish length and species provide important biomass information; these are not only important indicators of product classification [3] but also serve as the foundation for making intelligent feeding decisions [4]. Therefore, for optimal feed utilization and increased breeding income, it is crucial to estimate the length of the freshwater fish body at various growth stages. Manual measurement has been used traditionally to determine the fish length and species and is still widely employed today. This method, however, falls short of current aquaculture requirements due to defects like low measurement efficiency and significant errors.
Since image-based measurement approaches were first proposed, it has attracted lots of attention from scholars as a result of being a digital and contactless nondestructive testing method [5,6]. Numerous studies have confirmed that these approaches are applicable to many tasks: stock behavior diagnosing [7], fish population counting [8,9], fish species recognition or size measuring [10,11,12], and quality estimation of fishes [13,14]. The methods could be divided into two categories, semi-automatic and automatic, depending on the level of automation of the model.
Research on semi-automatic methods mainly appeared in the early stages in the field of fish length measurement. Harvey et al. developed a procedure in the foundation of a stereo-vision system to measure the body length of tuna [15]. Hsieh et al. reported a semi-automated method using a calibration plate as a standard for dimensional correction by which a lack of scale in monocular images can be made up [16]. Shafait et al. presented a semi-automatic approach, which is taking stereo photos automatically and labeling points manually, to measure the body length of tuna, which proved to be more efficient than manual operation [17]. In fish species detection, White et al. constructed a discrimination model for seven species of fish through the linear combination of 114 color and 10 shape variables [18]. Alsmadi et al. presented a neural network model to recognize fish species, which takes the distance and angle of fish feature points as input [19]. Although the semi-automatic methods are better than manual operations, it is still difficult to meet the demands in high-density fish farming mode. This is because time-consuming manual operations, such as capturing image coordinates of head and tail points or extracting color and texture features, are still necessary. Moreover, in order to obtain better image characteristics, there are certain requirements for image acquisition. Thus, these methods are more suitable for post-process rather than real-time detection in cultural conditions.
With the rapid development of computer performance, prediction models based on CNN (convolutional neural network) have been gradually extended from classification tasks to posture (key points) detection and instance segmentation, forming a new research direction [20,21,22]. Tseng et al. designed an algorithm for automatic fish length measurement and reached a mean relative error of not more than 5%. This consisted of a CNN classifier and image processing part. Among them, the regions of the head, tail fork, and calibration plate were detected by the neural network, while the coordinates of the snout and the middle point of the tail fork were determined by image processing [23]. Yu et al. proposed a measuring method based on Mask R-CNN. This method estimated the fish length through the segmented morphological features extracted by the network. In this way, its max relative error is less than 5% in complex photographing background [24]. Another method based on a 3D points cloud was raised to measure fish length. The area of each fish in the images was segmented by Mask R-CNN, before matching the depth map and acquiring 3D points information. The length of the fish body can be calculated after 3D reconstruction. The advantage of this method is that measurement error is not related to the posture of the fish [25]. In fish species recognition, Qiu et al. presented a CNN model using a squeeze-and-excitation structure to improve bilinear networks. The improved network achieved better performance with a 2–3% improvement in low-quality and small-scale datasets [26]. To face the challenge of imbalance in sample numbers in categories, Xu et al. presented a new loss function using class-weighting factors to re-balance the original focal loss [27]. Methods based on CNN allow the models to extract from images automatically, as well as achieve length measurement and species identification simultaneously, which is beneficial to the implementation of the automatic detection method.
To summarize, as a kind of supervised learning model, the neural network is able to meet different types of requirements. However, challenges in underwater vision systems based on this model still remain. For example, acquiring images underwater may affect the quality of images, which is not conducive to image processing later. In addition, underwater calibration operations cannot fully compensate for the errors caused by refraction. Designed to seek an automatic algorithm that is appropriate to industrial farming models, this study develops a method for category recognition and length estimation of fish underwater concurrently. This problem has been concerning for many years and has yet to be solved by existing studies.
The rest of this paper is as follows. Section 2 describes the experimental facilities and dataset in this research, as well as the specifics of the Keypoints R-CNN model along with stereo matching and length measuring methods. Section 3 illustrates the results and analysis of the experiment. Section 4 discusses the main findings and limitations of this paper. Lastly, Section 5 concludes the ratiocination inferred from the results in this research and looks ahead to further work.
2. Materials and Methods
2.1. Experimental Materials and Facility
Experimental images were collected by the self-built underwater image acquisition platform in the electromechanical engineering training center of Huazhong Agricultural University (Wuhan, China). The platform, as Figure 1 shows, consists of a culture barrel (diameter 2 m, height 1 m), a waterproof case made of PMMA, a stereo camera (CAM-AR0135-3T16, f3.6, USB3.0), and an underwater light source (20.5 w). The camera was set in auto-shooting mode, and takes 1 photo every 15 s, for a total of 24 h.

Figure 1.
Schematic diagram of the image acquisition platform, (a) Structure of the platform, (b) Photo of the actual device.
In this study, five species of common freshwater fish, namely, largemouth bass, crucian carp, grass carp, snakehead, and catfish, were chosen to be the research objects. There were two circumstances when capturing photos. The culture barrel in scenario one contained multiple fish of the same species, whereas, in scenario two, fish from all five species were cultivated there.
2.2. Dataset and Annotations
In order to make the target features in different images more diverse, only one picture from the set acquired by the left and right cameras is selected to form the dataset. Then, in a total of 5760 pictures, those in which there are no fish in the field of view, or those in which all fish bodies do not meet the requirements of clear visibility due to occlusion and inclination, were removed. Finally, 3300 original images with a resolution of 640 × 480 were selected as the raw dataset. Raw images were labeled with labelme annotation software, in the light of human key point annotations in the coco dataset format.
Images offline enhancement in the training set was carried out for the original dataset using Python programming language and Opencv toolkit, in which random two attributes from saturation, brightness, contrast, and sharpness were adjusted stochastically. The dataset is then divided into training sets, validation sets, and test sets according to the ratio of 8:2:1. The body length ranges of the freshwater fishes and the numbers of each set in this study are described in Table 1.

Table 1.
Detailed information about the dataset.
2.3. Design of Improved Keypoints R-CNN Network
We selected Keypoints R-CNN, an improved two-stage model for key points detection based on mask R-CNN, as the template of the network model in this research. In order to further enhance the model performance, the backbone and feature pyramid networks were mainly adjusted in this research. The structure of the improved network was displayed in Figure 2.
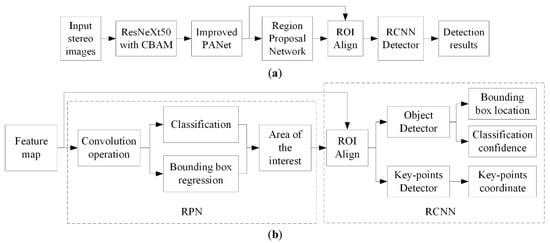
Figure 2.
Schematic diagram of the improved Keypoints RCNN model, (a) General structure of the model, (b) Main process of the RPN and RCNN part.
2.3.1. ResNeXt with CBAM
Compared with ResNet in a traditional Keypoints R-CNN network, ResNeXt is a new feature extraction network that combines both the advantages of residual structure and an inception block. The residual unit can avoid defects of feature degradation and vanishing gradients due to the increasing network depth [28]. Owing to bottleneck structure and group convolution from the inception block, the computation of the model can be effectively reduced. The main structure of the ResNeXt50 network was presented in Table 2.

Table 2.
Architecture of the main calculations in ResNeXt50.
To further enhance the feature extraction capability of the model for underwater images, we added CBAM(Convolutional Block Attention Module) to the basic ResNeXt model [29]. CBAM is a lightweight attention module in CNN, which combines both channel and spatial attention mechanisms. This module was placed in each valid feature layer output node of the model. After being weighted through this module, the image features extracted by the convolution operation will be retained if they contribute more to the recognition result and suppressed if they contribute less. The operation procedures of the CBAM module are shown in Figure 3.
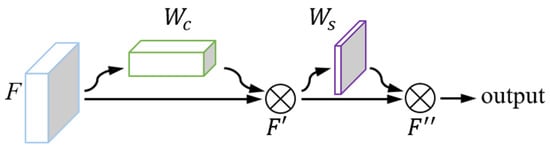
Figure 3.
Structure schematic of the CBAM module. Note: is the channel attention weight, is the spatial attention weight, F is the original feature map, F′ is the channel weighted feature map, and F″ is the spatial weighted feature map.
2.3.2. Improved PANet
It has been proven that FPN (Feature Pyramid Network) can achieve a higher detection accuracy by fusing features from feature maps on different scales [30]. However, there are still restrictions in the aforementioned network. The model is unsuitable for large target detection missions because single-direction integrations from the top down still do not enable top features to acquire geometric characteristics from the low level. Additionally, the original FPN’s Nearest-Neighbor Interpolation Algorithm is prone to produce significant errors during continuous upsampling.
To address the shortcomings mentioned in FPN, we proposed an improved double direction features fusion network I-PANet (Improved-Path Aggregation Network) [31]. The structure is shown in Figure 4. In the beginning, bi-cubic interpolation, which is advantageous for information around the sampled points being fully exploited, was performed to upsample the upper features for the first features fusion. Then, outputs from the initial fusion would be further refined using a convolution block with an inverted bottleneck structure. This block is composed of various basic computation units including convolution, batch normalization, and activation function. The first and last convolutional operations were used to alter the dimension of the input tensor, and the second one with a bigger number of channels was designed to extract features. Lastly, revised feature maps were downsampled to accomplish the second fusion. In addition, the features of layer P6 are produced by downsampling from P5.
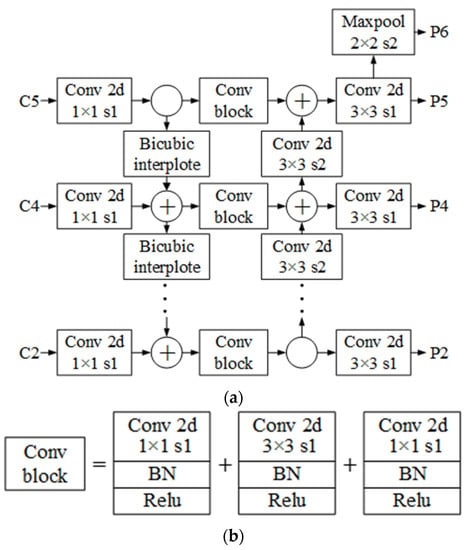
Figure 4.
Schematic diagram of the I-PANet, (a) Structure of the I-PANet, (b) Structure of the “Conv block”.
2.3.3. Training Procedures
The training strategy is a significant aspect that determines the convergence and detection accuracy of the model. In this study, the initial learning rate was adjusted to discover the model with the best performance. During training, the learning rate was dynamically decayed in each epoch according to a cosine function whose half period was the number of epochs. The approach of transfer learning was utilized. A warm-up strategy was also added to the first epoch to promote better convergence of the model. Furthermore, all images were resized to 384 × 384 pixels before input into the network with the mini-batch size being 8. The number of total iterations was set to 27,000 and SGD (Stochastic Gradient Descent) was selected as the model optimizer with a momentum of 0.9. The model was established based on Python 3.7 and PyTorch 1.10. The configuration of the hardware and compiler environment in this study was shown in Table 3.

Table 3.
Configuration of the computer in this study.
2.4. Stereo Matching and Fish Length Measurement
Traditional stereo-matching methods that take aggregation cost as an evaluation metric typically have to search the entire image with a fixed-size window, leading to intensive computation. We proposed a new matching method that matches the key points detection results from left and right images. It refers to the illumination of IoU (Intersection over Union) and OKS (Object Keypoint Similarity). Considering the existence of parallax in the left and right images, in this study, the image width of 0.05 times was chosen as the compensating offset for the key points horizontal coordinate in the right images. After the compensation process, corresponding detection results in the left and right images had closer coordinates, the combination of which possesses the larger matching similarity index. The index was defined in the following equation:
where (Matching Similarity) indicated the index of matching similarity, and represent the key points coordinate vector of No. detection target in the left image and No. in right, respectively; is the IoU of bounding boxes in No. and No. detection target; is the scaling factor, taken as 0.1.
An algorithm that determines the body length of the fish automatically using underwater stereo images was devised. Firstly, the detection results acquired by the Keypoints RCNN network from distortion-corrected stereo images were matched using the method suggested in this study. Secondly, 3D reconstruction of the key points was conducted utilizing the projection matrix and stereo camera parameters. Finally, the fish length was acquired by computing the Euclidean distance between the two key points.
2.5. Performance Evaluation Metrics
In this study, the performance of the model was evaluated by mAP (mean Average Precision), mAR (mean Average Recall), and model size. The performance of the species recognition experiments was evaluated by precision, recall and F1-score. The accuracy of submarine length measurement was evaluated using relative measurement error. Those metrics are described in Equations (2)–(7).
where is the integral of the precision-recall curve over the domain for the class target, is the integral of the recall-IoU curve from 0.5 to 1 for the class target, and is the total number of classes. TP, FP, and FN are the numbers of four types of detection results, true positives, false positives, and false negatives, respectively. represents the relative error between the results from the algorithm and the real value of measurement, with and indicates the calculated length and the real length, respectively.
3. Results
In this section, different hyper-parameters, such as the initial learning rate and its decay factor, were set to test model performance. A total of four combinations between the backbone network and FPN were compared using the evaluator in Section 2.5. In addition, the detection results from the left and right images were matched by applying the approach in Section 2.4. The results of the comparison were shown in Table 4, Table 5 and Table 6.

Table 4.
Model performance with different initial learning rate.

Table 5.
Model performance with different learning rate decay factor.

Table 6.
Model performance with different combinations of backbone network and FPN.
3.1. Model Performance Evaluation
Table 4 described the model performance under the initial learning rates of 0.01, 0.005, and 0.001, with the decay factor setting of 0.5. Table 5 displayed the performance under the learning rate decay factor of 0.33, 0.5, and 0.66, accordingly, when the initial learning rate was 0.005. Note that the model with optimal performance was obtained when the initial learning rate was 0.005 with a decay factor of 0.5. The results indicated that a large initial learning rate and small decay factor tended to cause oscillations that occurred during the iterative process, while the opposite settings commonly led to the model falling into a local optimal solution early.
To test the contribution of the backbone network and the feature pyramid network to the model’s performance, the original and improved networks were combined in a different order. We reported the results of the evaluation of the models in different structures in Table 6. The model size was one of the key indicators of computational performance. There was no evident difference in the number of parameters with the model of diverse structures. Additionally, the results pointed out that, out of the four different model structures, RexNeXt with BIC-PANet possessed the optimal comprehensive score under the other four evaluation indexes. The mAP and mAR values of the bounding box in this combination were 0.873 and 0.807, respectively, increasing by 4.5% and 3.7% compared to basic ResNet with FPN. Concurrently, the mAP and mAR values of the key points were only lower than the third combination with the discrepancies being almost ignorable. It demonstrated that ResNeXt and BIC-PANet have a better capability of extracting features from images.
The training process of the model with optimal structure and hyper-parameters was shown in Figure 5. The three curves represented the change in loss and mAP values during the learning process. Loss was the indicator of convergence in the training phase, and mAP responded to the generalization capacity. Note that the loss and map values tended to stabilize after about 40,000 iterations, which demonstrates that the model had nearly converged at this time. The mAP curves did not display a diminishing trend after 40,000 iterations, indicating that the model was not overfitted.
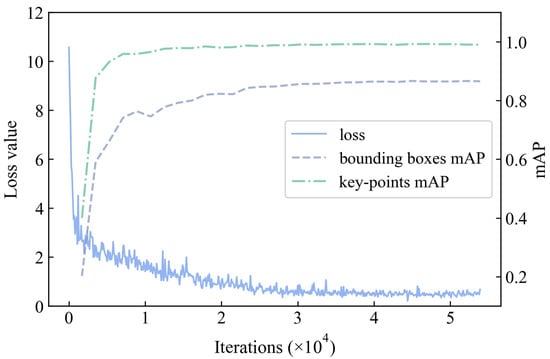
Figure 5.
Training process of ResNeXt with I-PANet.
3.2. Fish Species Recognition Experiments
To further test the detection effect of the improved Keypoints R-CNN network, the proposed model was utilized to identify the species of fish in the test set. Table 7 presented the evaluation indexes of the recognition experiment for five freshwater fishes. Figure 6 shows the confusion matrix with a confidence threshold of 0.75.

Table 7.
Recognition results for different species of freshwater fish.
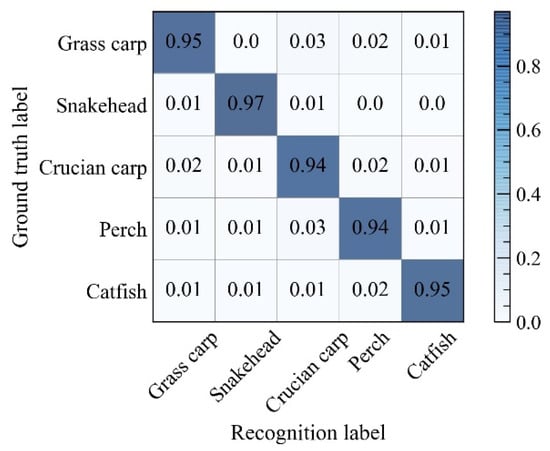
Figure 6.
Confusion matrix of recognition results.
As can be seen in Table 7, the values of precisions for all species exceeded 94%. Among them, the detection precision of snakehead was 97.12%, which was higher than the remaining four species of fish. Results of the crucian carp and the perch, with precisions of 94.02% and 94.48% severally, were at a relatively low level. Concurrently, the recall and F1-score of these five species were not less than 93%. These results implied that the model retains a high classification accuracy with small scales of missing detections, at the current threshold.
3.3. Fish Length Measurement Experiments
3.3.1. Stereo Matching
Figure 7 showed the detection results presented in the original images, as well as the matching similarity matrix of detection targets from a pair of left and right images. According to the matrix, the related targets with better overlap after compensation possessed a higher similarity index. The order numbers of the detection results were generated after being sorted by confidence level from highest to lowest. So, the matching relationship could be obtained by indexing the maximum value by rows or columns. Owing to the influence of parallax, part of the fish might not be intact in the left or right images. Hence, the number of detection results from a pair of images might not be equal. It was necessary to select the one with a smaller number from rows or columns as the index direction. Since the matching process was a one-to-one correspondence, the maximum value should not appear in the same row or column.
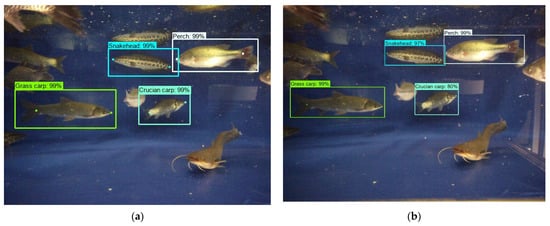
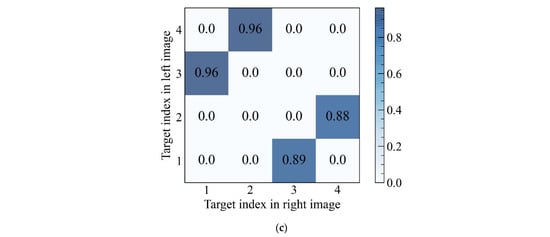
Figure 7.
Detection and stereo matching results of the left and right image, (a) the left image, (b) the right image, (c) the MS matrix.
3.3.2. Fish Length Measurements
The left subplot of Figure 8 showed the length measurement results presented in the left image. Additionally, the right subplot displayed the relative errors of measurements in a box plot. Each rectangle box in this graph reflected the 25th to 75th percentiles of measuring results for a particular species of fish. The median of the measurement error was shown by the red line in the center of the box. These five fish species had mean measurement errors of 5.08 ± 2.45% (mean ± standard deviation), 4.28 ± 1.95%, 4.29 ± 2.32%, 4.52 ± 2.52%, and 5.58 ± 2.91%. Among them, the snakehead had a higher measurement precision than the other four fish species, while the catfish had one of the lowest.
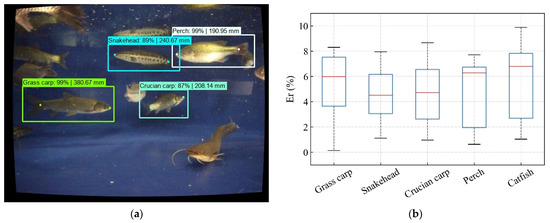
Figure 8.
The results of the fish length measurement experiment, (a) result mark on the original image, (b) relative errors of the length measuring.
Presumably, the spots on the carapace of the blackfish made it distinctive compared to the other four species. Therefore, it was easier to extract its visual characters from the image. However, the catfish were black and scaleless, which made it difficult for them to reflect light into the camera. Hence, the fish body was visually closer to the background shadows and difficult to be detected precisely. In the detection results for the perch and grass carp, inaccuracy was typically either less than 2% or greater than 6%. It might be connected to the particular posture of the fish. The bent body would render the Euclidian distance between the head and tail smaller than the actual body length when the fish was swimming. In this case, the measurement error would increase remarkably. There was no significant effect (p > 0.05) due to fish species on measurement accuracy after statistical analysis, which revealed that this method has good robustness. Overall, the upper limit of the relative errors did not exceed 10%. The accuracy of this procedure was adequate in general.
4. Discussion
4.1. Precision of Fish Species Recognition
According to the results in Section 3.2, it could be inferred that the effects on the detection of the snakehead are better as a result of its great difference in color and shape compared with other kinds of fish. Furthermore, the appearance features of the crucian carp were similar to that of the perch. So, false detections would occur when fish are far away from the imaging, due to the lack of morphology details. Additionally, the major difference between the crucian carp and the grass carp was body length. Owing to the absence of scale information, there was a pattern of large, near, and small distances in the planar images. So that the grass carp or crucian carp might be recognized as each other mistakenly when they were at different distances from the camera or partially shaded.
4.2. Precision of Fish Body Length Estimation
There were three primary reasons for the large measurement error according to the experiment results and our speculation. Firstly, bent fish accounted for a large proportion of the total number of samples, which brought about a rise in average measuring errors. Secondly, the accuracy of calibration results in underwater stereo systems was worse than that in a single media condition, due to image distortion brought on by media change [32,33,34]. Moreover, the optical performance and assembly accuracy of the USB camera amplified the negative effects of distortion on the underwater images. The superposition of these two occurrences created a major 3D reconstruction error, which finally impaired the precision of the fish length estimation results. Thirdly, common freshwater fish were substantially smaller in length than marine fish, which led to a greater sensitivity to error fluctuations. For example, the snout fork length of the tuna was commonly over 1200 mm [17,35], but none of the five freshwater fish samples selected for this study had a body length of more than 450 mm. This implied that the relative error of freshwater fish would be several times higher than that of marine fish when the absolute error was close.
5. Conclusions
A new algorithm based on improved Keypoints R-CNN was developed for freshwater fish species recognition and length measurement in underwater conditions. By adding the attention module and replacing I-PANet, the new detection model has achieved better performance than the original one. The mAP of object detection and key points recognition tasks reach 0.873 and 0.990, respectively. It has been confirmed that the species identification accuracy of all five kinds of freshwater fish mentioned in this study reached over 94% through experiments. Through the application of the proposed stereo-matching method, the coordinates of key points from detection results were matched quickly and accurately. Additionally, the proposed fish length measuring method was verified and proved to possess acceptable detection precision with relative errors of less than 10%.
In future work, we are going to acquire images from different angles in order to reduce the effect of fish posture on body length measuring. Moreover, we want to explore some other methods concerning the geometric features that can be used to describe the size of the fish so that a more accurate model for fish weight estimation can be constructed. Finally, we hope to establish a fish biomass detection system on commercial farms.
Author Contributions
Data curation, Y.D. and M.T.; formal analysis, D.Z.; funding acquisition, M.Z.; investigation, Y.L.; methodology, Y.D. and M.T.; project administration, Y.D. and H.T.; resources, H.T.; software, Y.D and M.T.; supervision, H.T.; visualization, Y.D.; writing—original draft preparation, Y.D. and D.Z.; writing—review and editing, H.T. All authors have read and agreed to the published version of the manuscript.
Funding
This paper was supported by the Fundamental Research Funds for the Central Universities (2662020GXPY003, 107/11041910104).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Conflicts of Interest
The authors declare that there is no known conflict of interest or personal relationships that could have appeared to influence the work reported in this paper.
References
- Taheri-Garavand, A.; Fatahi, S.; Banan, A.; Makino, Y. Real-Time Nondestructive Monitoring of Common Carp Fish Freshness Using Robust Vision-Based Intelligent Modeling Approaches. Comput. Electron. Agric. 2019, 159, 16–27. [Google Scholar] [CrossRef]
- Usydus, Z.; Szlinder-Richert, J. Functional Properties of Fish and Fish Products: A Review. Int. J. Food Prop. 2012, 15, 823–846. [Google Scholar] [CrossRef]
- Banan, A.; Nasiri, A.; Taheri-Garavand, A. Deep Learning-Based Appearance Features Extraction for Automated Carp Species Identification. Aquac. Eng. 2020, 89, 102053. [Google Scholar] [CrossRef]
- An, D.; Hao, J.; Wei, Y.; Wang, Y.; Yu, X. Application of Computer Vision in Fish Intelligent Feeding System—A Review. Aquac. Res. 2021, 52, 423–437. [Google Scholar] [CrossRef]
- Hao, M.; Yu, H.; Li, D. The Measurement of Fish Size by Machine Vision—A Review. In Computer and Computing Technologies in Agriculture IX; Li, D., Li, Z., Eds.; Springer: Cham, Switzerland, 2016; Volume 479, pp. 15–32. ISBN 978-3-319-48353-5. [Google Scholar]
- Domasevich, M.A.; Hasegawa, H.; Yamazaki, T. Quality Evaluation of Kohaku Koi (Cyprinus Rubrofuscus) Using Image Analysis. Fishes 2022, 7, 158. [Google Scholar] [CrossRef]
- Iqbal, U.; Li, D.; Akhter, M. Intelligent Diagnosis of Fish Behavior Using Deep Learning Method. Fishes 2022, 7, 201. [Google Scholar] [CrossRef]
- Labuguen, R.T.; Volante, E.J.P.; Causo, A.; Bayot, R.; Peren, G.; Macaraig, R.M.; Libatique, N.J.C.; Tangonan, G.L. Automated Fish Fry Counting and Schooling Behavior Analysis Using Computer Vision. In Proceedings of the 2012 IEEE 8th International Colloquium on Signal Processing and its Applications, Malacca, Malaysia, 23–15 March 2012; pp. 255–260. [Google Scholar]
- Zhang, S.; Yang, X.; Wang, Y.; Zhao, Z.; Liu, J.; Liu, Y.; Sun, C.; Zhou, C. Automatic Fish Population Counting by Machine Vision and a Hybrid Deep Neural Network Model. Animals 2020, 10, 364. [Google Scholar] [CrossRef]
- Hu, J.; Li, D.; Duan, Q.; Han, Y.; Chen, G.; Si, X. Fish Species Classification by Color, Texture and Multi-Class Support Vector Machine Using Computer Vision. Comput. Electron. Agric. 2012, 88, 133–140. [Google Scholar] [CrossRef]
- Rosen, S.; Jörgensen, T.; Hammersland-White, D.; Holst, J.C. DeepVision: A Stereo Camera System Provides Highly Accurate Counts and Lengths of Fish Passing inside a Trawl. Can. J. Fish Aquat. Sci. 2013, 70, 1456–1467. [Google Scholar] [CrossRef]
- Li, D.; Su, H.; Jiang, K.; Liu, D.; Duan, X. Fish Face Identification Based on Rotated Object Detection: Dataset and Exploration. Fishes 2022, 7, 219. [Google Scholar] [CrossRef]
- Fan, L.; Liu, Y. Automate Fry Counting Using Computer Vision and Multi-Class Least Squares Support Vector Machine. Aquaculture 2013, 380, 91–98. [Google Scholar] [CrossRef]
- He, H.-J.; Wu, D.; Sun, D.-W. Nondestructive Spectroscopic and Imaging Techniques for Quality Evaluation and Assessment of Fish and Fish Products. Crit. Rev. Food Sci. Nutr. 2015, 55, 864–886. [Google Scholar] [CrossRef] [PubMed]
- Harvey, E.; Cappo, M.; Shortis, M.; Robson, S.; Buchanan, J.; Speare, P. The Accuracy and Precision of Underwater Measurements of Length and Maximum Body Depth of Southern Bluefin Tuna (Thunnus Maccoyii) with a Stereo–Video Camera System. Fish. Res. 2003, 63, 315–326. [Google Scholar] [CrossRef]
- Hsieh, C.-L.; Chang, H.-Y.; Chen, F.-H.; Liou, J.-H.; Chang, S.-K.; Lin, T.-T. A Simple and Effective Digital Imaging Approach for Tuna Fish Length Measurement Compatible with Fishing Operations. Comput. Electron. Agric. 2011, 75, 44–51. [Google Scholar] [CrossRef]
- Shafait, F.; Harvey, E.S.; Shortis, M.R.; Mian, A.; Ravanbakhsh, M.; Seager, J.W.; Culverhouse, P.F.; Cline, D.E.; Edgington, D.R. Towards Automating Underwater Measurement of Fish Length: A Comparison of Semi-Automatic and Manual Stereo–Video Measurements. ICES J. Mar. Sci. 2017, 74, 1690–1701. [Google Scholar] [CrossRef]
- White, D.J.; Svellingen, C.; Strachan, N.J.C. Automated Measurement of Species and Length of Fish by Computer Vision. Fish. Res. 2006, 80, 203–210. [Google Scholar] [CrossRef]
- Alsmadi, M.K.; Omar, K.B.; Noah, S.A.; Almarashdeh, I. Fish Recognition Based on Robust Features Extraction from Size and Shape Measurements Using Neural Network. J. Comput. Sci. 2010, 6, 1088. [Google Scholar] [CrossRef]
- Cai, K.; Miao, X.; Wang, W.; Pang, H.; Liu, Y.; Song, J. A Modified YOLOv3 Model for Fish Detection Based on MobileNetv1 as Backbone. Aquac. Eng. 2020, 91, 102117. [Google Scholar] [CrossRef]
- Kakehi, S.; Sekiuchi, T.; Ito, H.; Ueno, S.; Takeuchi, Y.; Suzuki, K.; Togawa, M. Identification and Counting of Pacific Oyster Crassostrea Gigas Larvae by Object Detection Using Deep Learning. Aquac. Eng. 2021, 95, 102197. [Google Scholar] [CrossRef]
- Tang, C.; Zhang, G.; Hu, H.; Wei, P.; Duan, Z.; Qian, Y. An Improved YOLOv3 Algorithm to Detect Molting in Swimming Crabs against a Complex Background. Aquac. Eng. 2020, 91, 102115. [Google Scholar] [CrossRef]
- Tseng, C.-H.; Hsieh, C.-L.; Kuo, Y.-F. Automatic Measurement of the Body Length of Harvested Fish Using Convolutional Neural Networks. Biosyst. Eng. 2020, 189, 36–47. [Google Scholar] [CrossRef]
- Yu, C.; Fan, X.; Hu, Z.; Xia, X.; Zhao, Y.; Li, R.; Bai, Y. Segmentation and Measurement Scheme for Fish Morphological Features Based on Mask R-CNN. Inf. Process. Agric. 2020, 7, 523–534. [Google Scholar] [CrossRef]
- Huang, K.; Li, Y.; Suo, F.; Xiang, J. Stereo Vison and Mask-RCNN Segmentation Based 3D Points Cloud Matching for Fish Dimension Measurement. In Proceedings of the 2020 39th Chinese Control Conference (CCC), Shenyang, China, 27–29 July 2020; pp. 6345–6350. [Google Scholar]
- Qiu, C.; Zhang, S.; Wang, C.; Yu, Z.; Zheng, H.; Zheng, B. Improving Transfer Learning and Squeeze-and-Excitation Networks for Small-Scale Fine-Grained Fish Image Classification. IEEE Access 2018, 6, 78503–78512. [Google Scholar] [CrossRef]
- Xu, X.; Li, W.; Duan, Q. Transfer Learning and SE-ResNet152 Networks-Based for Small-Scale Unbalanced Fish Species Identification. Comput. Electron. Agric. 2021, 180, 105878. [Google Scholar] [CrossRef]
- Xie, S.; Girshick, R.; Dollár, P.; Tu, Z.; He, K. Aggregated Residual Transformations for Deep Neural Networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 5987–5995. [Google Scholar]
- Woo, S.; Park, J.; Lee, J.-Y.; Kweon, I.S. Cbam: Convolutional Block Attention Module. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 3–19. [Google Scholar]
- Lin, T.Y.; Dollár, P.; Girshick, R.; He, K.; Hariharan, B.; Belongie, S. Feature pyramid networks for object detection. In Proceedings of the 30th IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 2117–2125. [Google Scholar]
- Liu, S.; Qi, L.; Qin, H.; Shi, J.; Jia, J. Path Aggregation Network for Instance Segmentation. In Proceedings of the 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Salt Lake City, UT, USA, 18–23 June 2018. [Google Scholar]
- Halder, K.K.; Paul, M.; Tahtali, M.; Anavatti, S.G.; Murshed, M. Correction of Geometrically Distorted Underwater Images Using Shift Map Analysis. JOSA A 2017, 34, 666–673. [Google Scholar] [CrossRef] [PubMed]
- Łuczyński, T.; Pfingsthorn, M.; Birk, A. The Pinax-Model for Accurate and Efficient Refraction Correction of Underwater Cameras in Flat-Pane Housings. Ocean. Eng. 2017, 133, 9–22. [Google Scholar] [CrossRef]
- Zhang, C.; Zhang, X.; Tu, D.; Jin, P. On-Site Calibration of Underwater Stereo Vision Based on Light Field. Opt. Lasers Eng. 2019, 121, 252–260. [Google Scholar] [CrossRef]
- Muñoz-Benavent, P.; Andreu-García, G.; Valiente-González, J.M.; Atienza-Vanacloig, V.; Puig-Pons, V.; Espinosa, V. Enhanced Fish Bending Model for Automatic Tuna Sizing Using Computer Vision. Comput. Electron. Agric. 2018, 150, 52–61. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).