Identification and Characterization of PIWI-Interacting RNAs in Spinyhead Croakers (Collichthys lucidus) by Small RNA Sequencing
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Small RNA Library Preparation and Sequencing
2.3. piRNA Identification
2.4. Differential Expression Analysis, Target Gene Prediction, and Functional Analysis
2.5. piRNA Cluster Identification
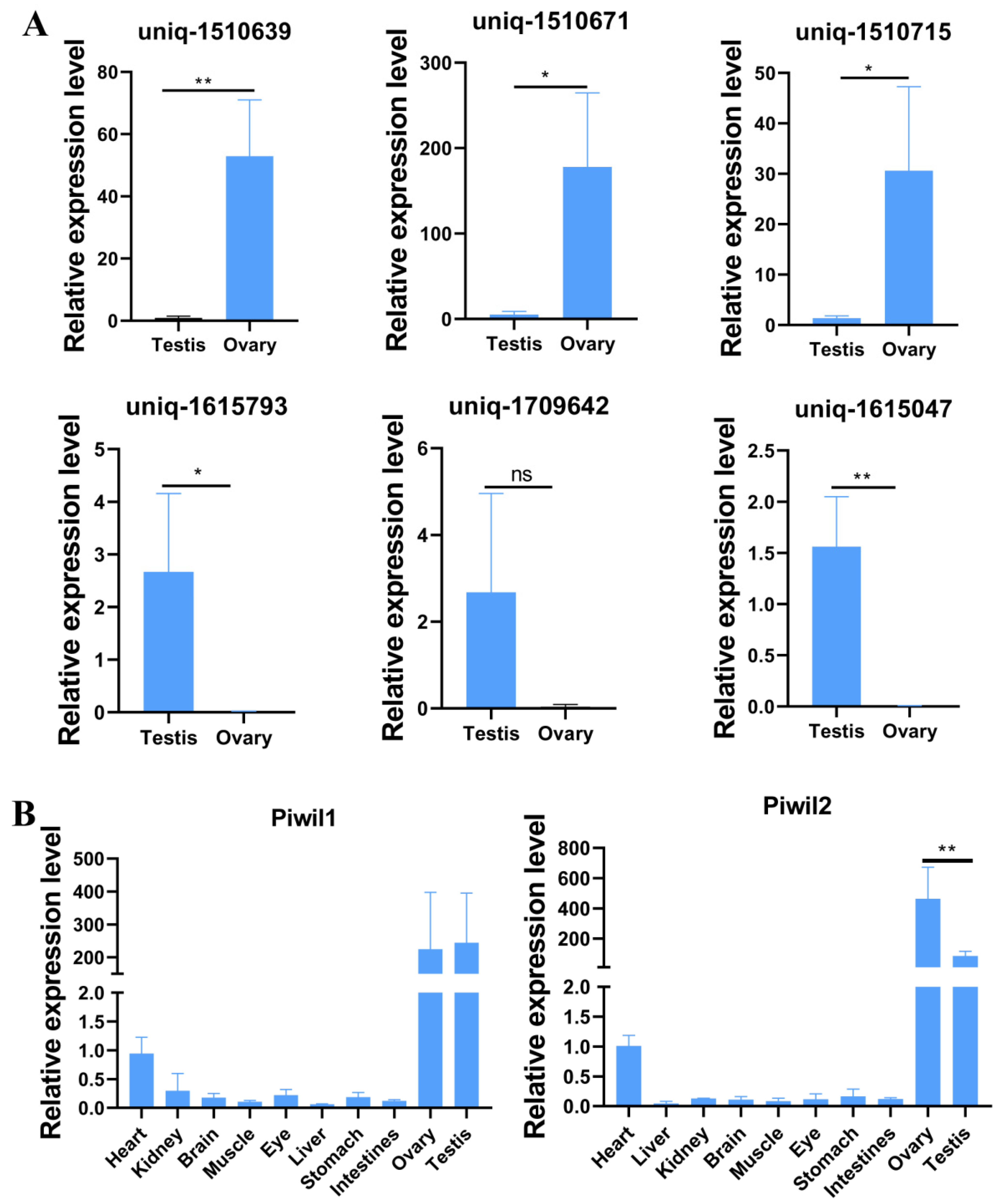
2.6. Validation of Differentially Expressed piRNAs by Real-Time PCR
3. Results
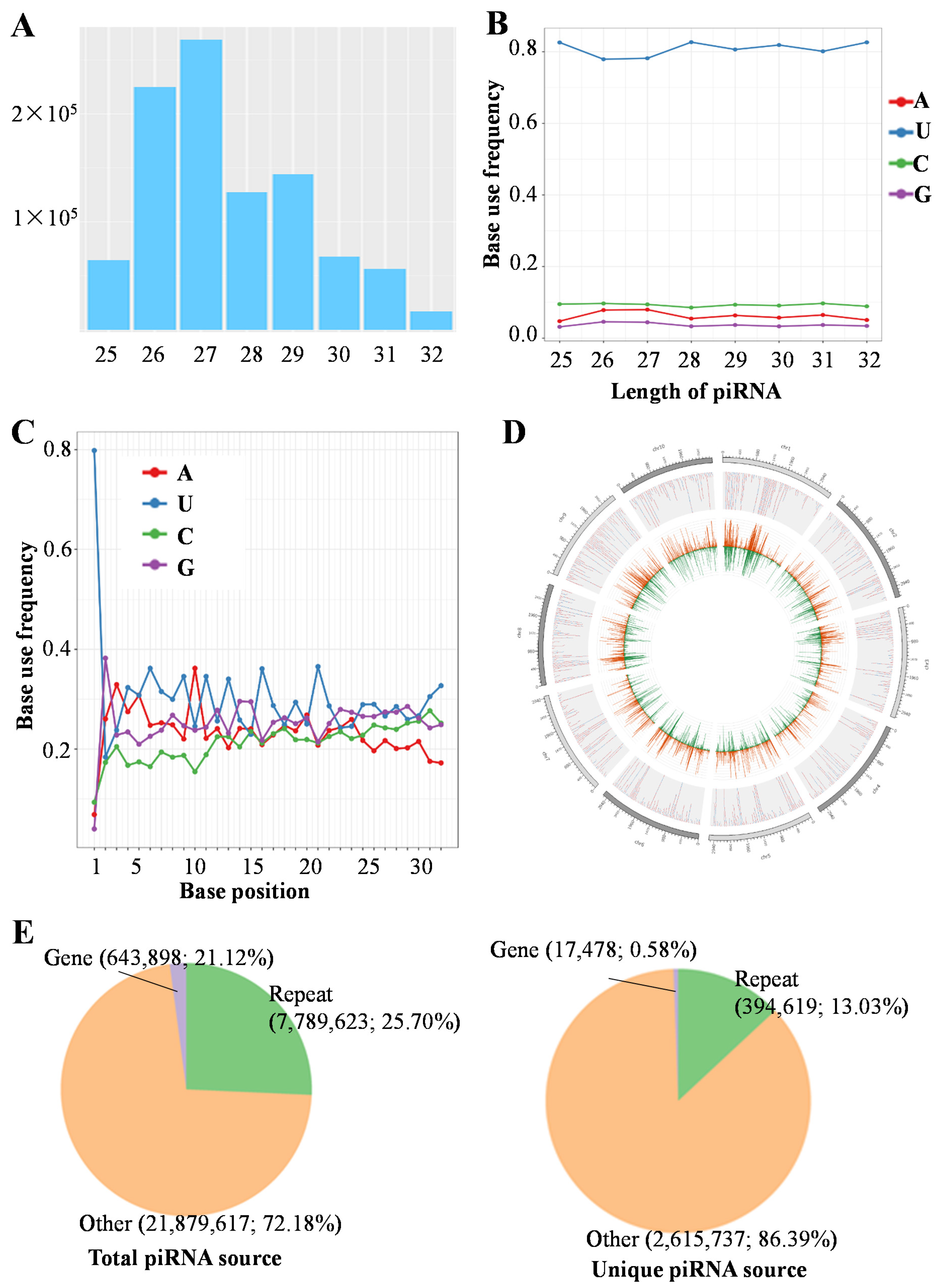
3.1. Characteristics of Gonad piRNAs in C. lucidus
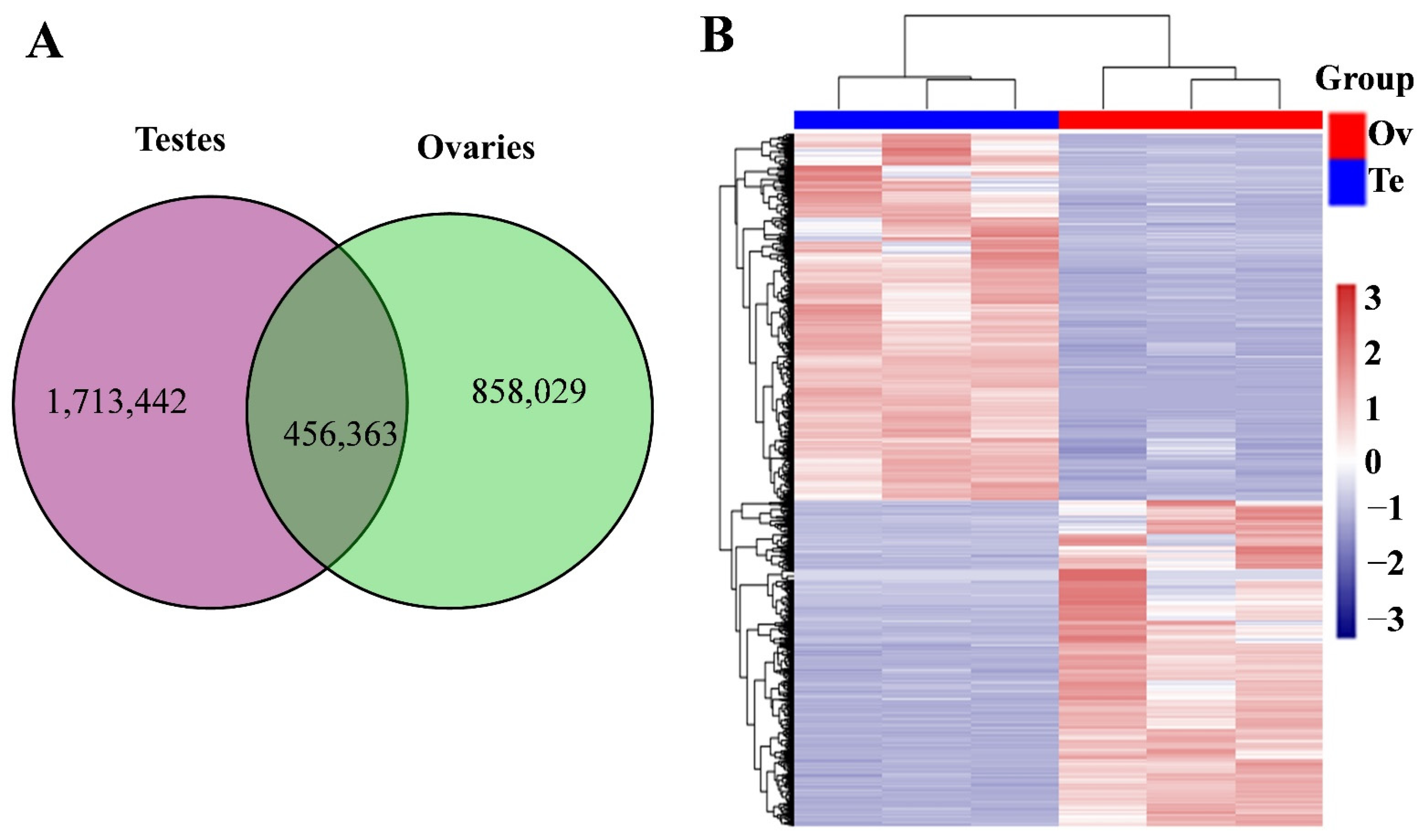
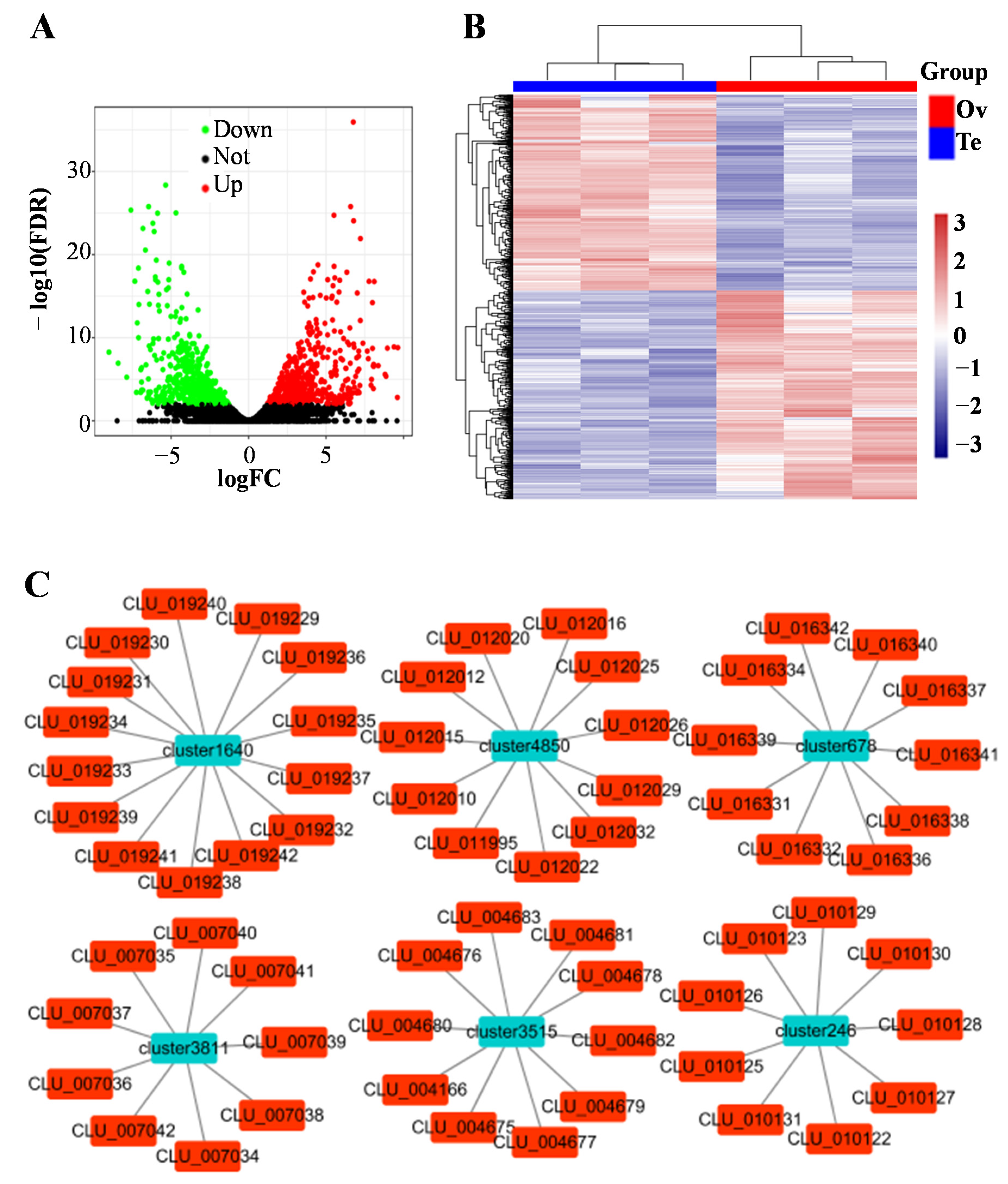
3.2. Differentially Expressed piRNAs
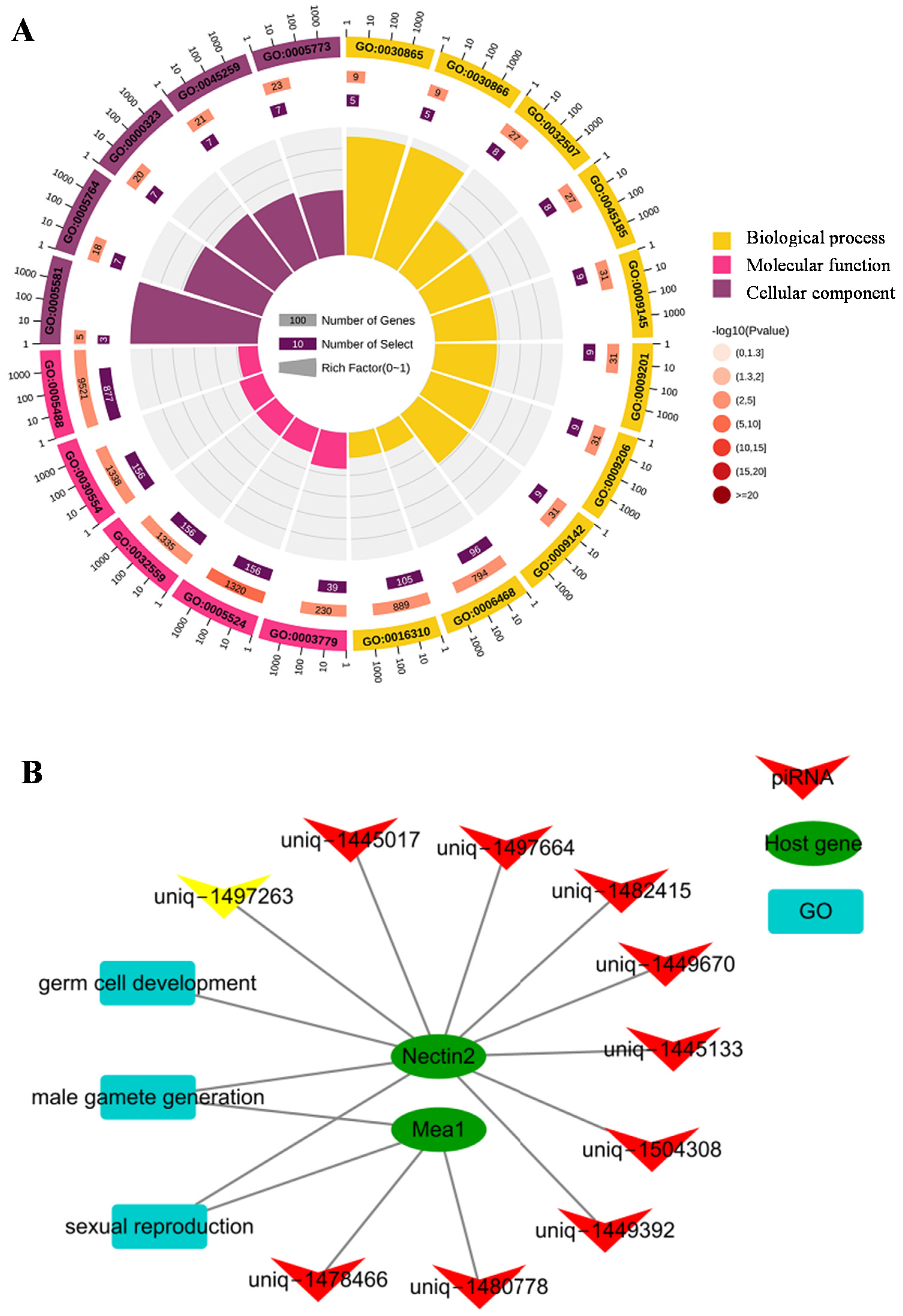
3.3. Putative Functions and Pathways of Differentially Expressed piRNAs
3.4. Candidate Host Genes of piRNAs
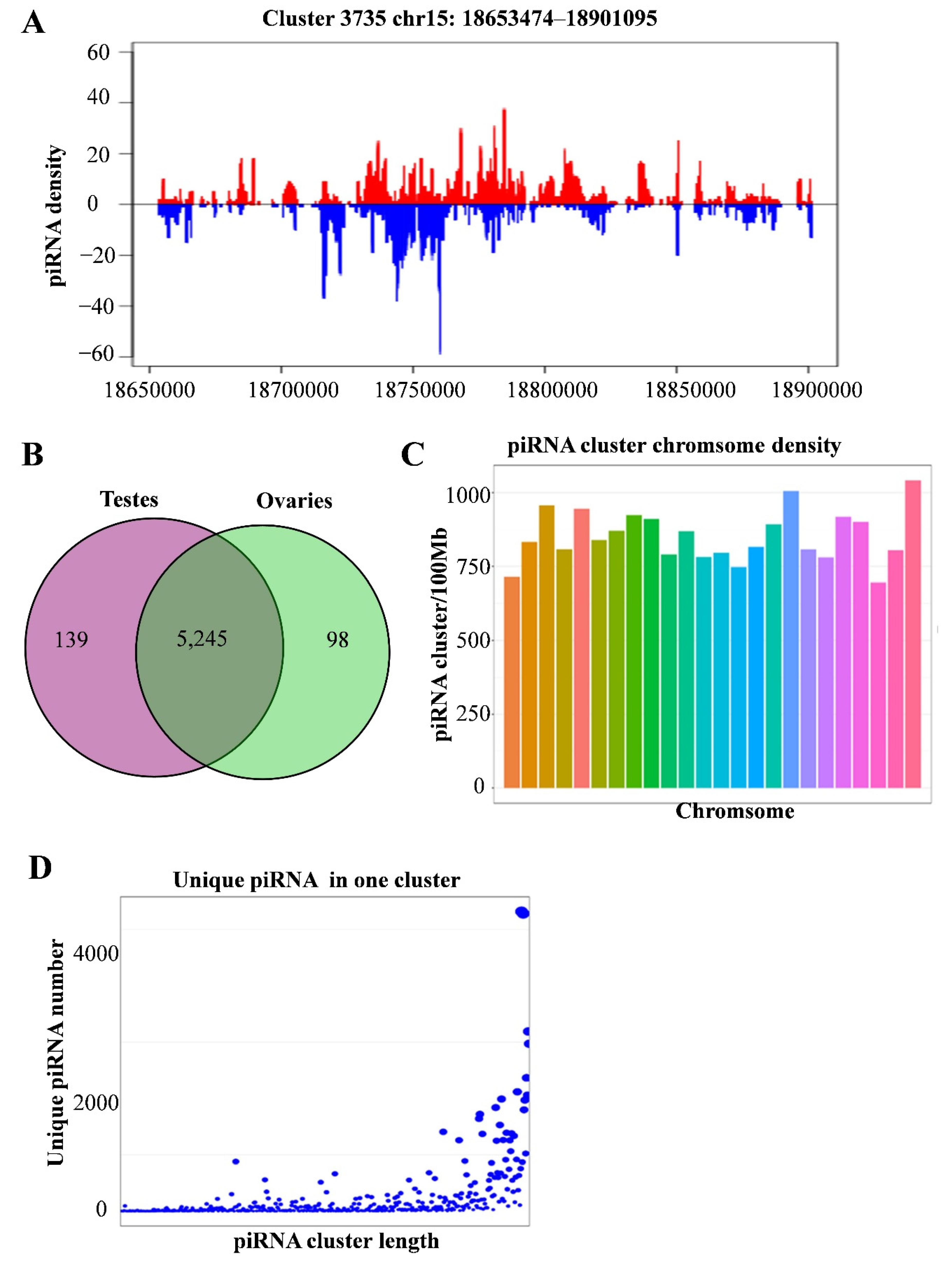
3.5. Identification of piRNA Clusters
3.6. Differential Expression Analysis of piRNA Clusters
3.7. Validation of Differentially Expressed piRNAs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Cheng, J.; Ma, G.-Q.; Miao, Z.-Q.; Shui, B.-N.; Gao, T.-X. Complete mitochondrial genome sequence of the spinyhead croaker Collichthys lucidus (Perciformes, Sciaenidae) with phylogenetic considerations. Mol. Biol. Rep. 2012, 39, 4249–4259. [Google Scholar] [CrossRef]
- Sang, C.; Lin, Y.; Jiang, K.; Zhang, F.; Ma, C.; Ma, L.; Song, W. Molecular cloning and expression analysis of MyD88 in spiny head croaker, Collichthys lucidus. Genet. Mol. Res. 2015, 14, 4666–4676. [Google Scholar] [CrossRef]
- Sang, C.; Lin, Y.; Jiang, K.; Zhang, F.; Song, W. Molecular cloning and mRNA expression of a hepcidin gene from the spinyhead croaker, Collichthys lucidus. Genet. Mol. Res. 2015, 14, 16050–16059. [Google Scholar] [CrossRef] [PubMed]
- Cai, M.; Zou, Y.; Xiao, S.; Li, W.; Han, Z.; Han, F.; Xiao, J.; Liu, F.; Wang, Z. Chromosome assembly of Collichthys lucidus, a fish of Sciaenidae with a multiple sex chromosome system. Sci. Data 2019, 6, 132. [Google Scholar] [CrossRef] [PubMed]
- Song, W.; Zhang, Y.; Zhang, X.; Gui, J. De novo transcriptome assembly of four organs of Collichthys lucidus and identification of genes involved in sex determination and reproduction. PLoS ONE 2020, 15, e0230580. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Song, W.; Zhu, Y.F.; Zhu, T.Y.; Ma, L.B.; Li, M.Y. Evolutionarily conserved vasa identifies embryonic and gonadal germ cells in spinyhead croaker Collichthys lucidus. J. Fish Biol. 2019, 94, 772–780. [Google Scholar] [CrossRef]
- Ozata, D.M.; Gainetdinov, I.; Zoch, A.; O’Carroll, D.; Zamore, P.D. PIWI-interacting RNAs: Small RNAs with big functions. Nat. Rev. Genet. 2018, 20, 89–108. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Lin, H. Roles of piRNAs in transposon and pseudogene regulation of germline mRNAs and lncRNAs. Genome Biol. 2021, 22, 27. [Google Scholar] [CrossRef]
- Brennecke, J.; Aravin, A.A.; Stark, A.; Dus, M.; Kellis, M.; Sachidanandam, R.; Hannon, G.J. Discrete Small RNA-Generating Loci as Master Regulators of Transposon Activity in Drosophila. Cell 2007, 128, 1089–1103. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Z.-H.; Zhao, Y.-M. The regulatory functions of piRNA/PIWI in spermatogenesis. Hereditas 2017, 39, 683–691. [Google Scholar] [PubMed]
- Zhang, P.; Kang, J.Y.; Gou, L.T.; Wang, J.; Xue, Y.; Skogerboe, G.; Dai, P.; Huang, D.W.; Chen, R.; Fu, X.D.; et al. MIWI and piRNA-mediated cleavage of messenger RNAs in mouse testes. Cell Res. 2015, 25, 193–207. [Google Scholar] [CrossRef] [PubMed]
- Thomson, T.; Lin, H. The Biogenesis and Function of PIWI Proteins and piRNAs: Progress and Prospect. Annu. Rev. Cell Dev. Biol. 2009, 25, 355–376. [Google Scholar] [CrossRef]
- Barucci, G.; Cornes, E.; Singh, M.; Li, B.; Ugolini, M.; Samolygo, A.; Didier, C.; Dingli, F.; Loew, D.; Quarato, P.; et al. Small-RNA-mediated transgenerational silencing of histone genes impairs fertility in piRNA mutants. Nat. Cell Biol. 2020, 22, 235–245. [Google Scholar] [CrossRef] [PubMed]
- Juliano, C.; Wang, J.; Lin, H. Uniting Germline and Stem Cells: The Function of Piwi Proteins and the piRNA Pathway in Diverse Organisms. Annu. Rev. Genet. 2011, 45, 447–469. [Google Scholar] [CrossRef] [PubMed]
- Tao, W.; Sun, L.; Chen, J.; Shi, H.; Wang, D. Genomic identification, rapid evolution, and expression of Argonaute genes in the tilapia, Oreochromis niloticus. Dev. Genes Evol. 2016, 226, 339–348. [Google Scholar] [CrossRef] [PubMed]
- Houwing, S.; Kamminga, L.M.; Berezikov, E.; Cronembold, D.; Girard, A.; van den Elst, H.; Filippov, D.V.; Blaser, H.; Raz, E.; Moens, C.B.; et al. A Role for Piwi and piRNAs in Germ Cell Maintenance and Transposon Silencing in Zebrafish. Cell 2007, 129, 69–82. [Google Scholar] [CrossRef]
- Song, H.; Xing, C.; Lu, W.; Liu, Z.; Wang, X.; Cheng, J.; Zhang, Q. Rapid evolution of piRNA pathway and its transposon targets in Japanese flounder (Paralichthys olivaceus). Comp. Biochem. Physiol. Part D: Genom. Proteom. 2019, 31, 100609. [Google Scholar] [CrossRef]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1989. [Google Scholar]
- Zhou, Y.; Sun, W.; Cai, H.; Bao, H.; Zhang, Y.; Qian, G.; Ge, C. The Role of Anti-Mullerian Hormone in Testis Differentiation Reveals the Significance of the TGF-beta Pathway in Reptilian Sex Determination. Genetics 2019, 213, 1317–1327. [Google Scholar] [CrossRef]
- Xue, R.; Lin, W.; Sun, J.; Watanabe, M.; Xu, A.; Araki, M.; Nasu, Y.; Tang, Z.; Huang, P. The role of Wnt signaling in male reproductive physiology and pathology. Mol. Hum. Reprod. 2021, 27, gaaa085. [Google Scholar] [CrossRef]
- Yang, F.; Whelan, E.C.; Guan, X.; Deng, B.; Wang, S.; Sun, J.; Avarbock, M.R.; Wu, X.; Brinster, R.L. FGF9 promotes mouse spermatogonial stem cell proliferation mediated by p38 MAPK signalling. Cell Prolif. 2021, 54, e12933. [Google Scholar] [CrossRef]
- La, H.M.; Chan, A.-L.; Legrand, J.; Rossello, F.J.; Gangemi, C.G.; Papa, A.; Cheng, Q.; Morand, E.; Hobbs, R.M. GILZ-dependent modulation of mTORC1 regulates spermatogonial maintenance. Development 2018, 145, dev165324. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Zhao, W.; Liu, J.; Wang, Y.; Yuan, C.; Zhang, F.; Jin, G.; Qin, Q. Effects of HIF-1α on Spermatogenesis of Varicocele Rats by Regulating VEGF/PI3K/Akt Signaling Pathway. Reprod. Sci. 2020, 28, 1161–1174. [Google Scholar] [CrossRef] [PubMed]
- Dai, P.; Wang, X.; Liu, M.-F. A dual role of the PIWI/piRNA machinery in regulating mRNAs during mouse spermiogenesis. Sci. China Life Sci. 2020, 63, 447–449. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.; Seth, M.; Tu, S.; Shen, E.-Z.; Li, Q.; Shirayama, M.; Weng, Z.; Mello, C.C. A Sex Chromosome piRNA Promotes Robust Dosage Compensation and Sex Determination in C. elegans. Dev. Cell 2018, 44, 762–770.e3. [Google Scholar] [CrossRef]
- Kiuchi, T.; Koga, H.; Kawamoto, M.; Shoji, K.; Sakai, H.; Arai, Y.; Ishihara, G.; Kawaoka, S.; Sugano, S.; Shimada, T.; et al. A single female-specific piRNA is the primary determiner of sex in the silkworm. Nature 2014, 509, 633–636. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; He, X.; Zhao, Y.; Bai, D.; Bou, G.; Zhang, X.; Su, S.; Dao, L.; Liu, R.; Wang, Y.; et al. Identification of piRNAs and piRNA clusters in the testes of the Mongolian horse. Sci. Rep. 2019, 9, 5022. [Google Scholar] [CrossRef]
- Yang, Q.; Hua, J.; Wang, L.; Xu, B.; Zhang, H.; Ye, N.; Zhang, Z.; Yu, D.; Cooke, H.J.; Zhang, Y.; et al. MicroRNA and piRNA Profiles in Normal Human Testis Detected by Next Generation Sequencing. PLoS ONE 2013, 8, e66809. [Google Scholar] [CrossRef]
- Lau, N.C.; Seto, A.G.; Kim, J.; Kuramochi-Miyagawa, S.; Nakano, T.; Bartel, D.P.; Kingston, R.E. Characterization of the piRNA Complex from Rat Testes. Science 2006, 313, 363–367. [Google Scholar] [CrossRef]
- Han, B.W.; Wang, W.; Li, C.; Weng, Z.; Zamore, P.D. piRNA-guided transposon cleavage initiates Zucchini-dependent, phased piRNA production. Science 2015, 348, 817–821. [Google Scholar] [CrossRef]
- Fofanov, M.; Prokopov, D.; Kuhl, H.; Schartl, M.; Trifonov, V. Evolution of MicroRNA Biogenesis Genes in the Sterlet (Acipenser ruthenus) and Other Polyploid Vertebrates. Int. J. Mol. Sci. 2020, 21, 9562. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, X.; Kang, L. A k-mer scheme to predict piRNAs and characterize locust piRNAs. Bioinformatics 2011, 27, 771–776. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Ge, Y.; Cheng, D.; Nie, Z.; Lv, Z. Detection of piRNAs in whitespotted bamboo shark liver. Gene 2016, 590, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.-L.; Wang, Z.-P.; Wang, J.-Q.; Li, M.-Y.; Chen, X.-W. Identification of candidate piRNAs in the gonads of Paralichthys olivaceus (Japanese flounder). Zool. Res. 2016, 37, 301–306. [Google Scholar] [CrossRef] [PubMed]
- Waiho, K.; Fazhan, H.; Zhang, Y.; Li, S.; Zhang, Y.; Zheng, H.; Ikhwanuddin, M.; Ma, H. Comparative profiling of ovarian and testicular piRNAs in the mud crab Scylla paramamosain. Genomics 2020, 112, 323–331. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhong, H.; Xiao, J.; Yan, J.; Luo, Y.; Gan, X.; Yu, F. Identification and comparative analysis of piRNAs in ovary and testis of Nile tilapia (Oreochromis niloticus). Genes Genom. 2016, 38, 519–527. [Google Scholar] [CrossRef]
- Chen, R.; Du, J.; Ma, L.; Wang, L.-Q.; Xie, S.-S.; Yang, C.-M.; Lan, X.-Y.; Pan, C.-Y.; Dong, W.-Z. Comparative microRNAome analysis of the testis and ovary of the Chinese giant salamander. Reproduction 2017, 154, 269–279. [Google Scholar] [CrossRef]
- Zhang, X.; Lui, W.-Y. Dysregulation of nectin-2 in the testicular cells: An explanation of cadmium-induced male infertility. Biochim. Biophys. Acta 2014, 1839, 873–884. [Google Scholar] [CrossRef]
- Bronson, R.; Mikhailik, A.; Schwedes, J.; Gnatenko, D.; Hatchwell, E. Detection of candidate nectin gene mutations in infertile men with severe teratospermia. J. Assist. Reprod. Genet. 2017, 34, 1295–1302. [Google Scholar] [CrossRef]
- Tang, R.; Liu, X.; Peng, Y.; Ju, W.; Hao, W.; Peng, X.; Chen, R. Nectin-like molecule 2, a necessary sexual maturation regulator, participates in congenital hypogonadotropic hypogonadism. Gene 2020, 754, 144885. [Google Scholar] [CrossRef]
- Lau, Y.F.; Chan, K.M.; Sparkes, R. Male-enhanced antigen gene is phylogenetically conserved and expressed at late stages of spermatogenesis. Proc. Natl. Acad. Sci. USA 1989, 86, 8462–8466. [Google Scholar] [CrossRef]
- Ohinata, Y.; Sutou, S.; Kondo, M.; Takahashi, T.; Mitsui, Y. Male-Enhanced Antigen-1 Gene Flanked by Two Overlapping Genes Is Expressed in Late Spermatogenesis1. Biol. Reprod. 2002, 67, 1824–1831. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhang, Y.; Xu, X.; Li, J.; Yuan, F.; Bo, S.; Qiao, J.; Xia, G.; Su, Y.; Zhang, M. Transforming growth factor-beta is involved in maintaining oocyte meiotic arrest by promoting natriuretic peptide type C expression in mouse granulosa cells. Cell Death Dis. 2019, 10, 558. [Google Scholar] [CrossRef] [PubMed]
- Moraveji, S.-F.; Esfandiari, F.; Taleahmad, S.; Nikeghbalian, S.; Sayahpour, F.-A.; Masoudi, N.-S.; Shahverdi, A.; Baharvand, H. Suppression of transforming growth factor-beta signaling enhances spermatogonial proliferation and spermatogenesis recovery following chemotherapy. Hum. Reprod. 2019, 34, 2430–2442. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.-L.; Batzel, P.; Titus, T.; Sydes, J.; Desvignes, T.; BreMiller, R.; Draper, B.; Postlethwait, J.H. A Hormone That Lost Its Receptor: Anti-Müllerian Hormone (AMH) in Zebrafish Gonad Development and Sex Determination. Genetics 2019, 213, 529–553. [Google Scholar] [CrossRef]
- Deepa, S.; Senthilkumaran, B. Interactive role of Wnt signaling and Zn in regulating testicular function of the common carp, Cyprinus carpio. Theriogenology 2020, 161, 161–175. [Google Scholar] [CrossRef]
- Cao, Z.; Mao, X.; Luo, L. Germline Stem Cells Drive Ovary Regeneration in Zebrafish. Cell Rep. 2019, 26, 1709–1717.e3. [Google Scholar] [CrossRef]
- Nicol, B.; Guiguen, Y. Expression profiling of Wnt signaling genes during gonadal differentiation and gametogenesis in rainbow trout. Sex. Dev. 2011, 5, 318–329. [Google Scholar] [CrossRef]
- Zhao, L.; Yao, C.; Xing, X.; Jing, T.; Li, P.; Zhu, Z.; Yang, C.; Zhai, J.; Tian, R.; Chen, H.; et al. Single-cell analysis of developing and azoospermia human testicles reveals central role of Sertoli cells. Nat. Commun. 2020, 11, 5683. [Google Scholar] [CrossRef]
- Lobo, I.K.C.; Do Nascimento, A.R.; Yamagishi, M.E.B.; Guiguen, Y.; da Silva, G.F.; Severac, D.; Amaral, A.D.C.; Reis, V.R.; de Almeida, F.L. Transcriptome of tambaqui Colossoma macropomum during gonad differentiation: Different molecular signals leading to sex identity. Genomics 2020, 112, 2478–2488. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ji, Q.; Xie, Z.; Gan, W.; Wang, L.; Song, W. Identification and Characterization of PIWI-Interacting RNAs in Spinyhead Croakers (Collichthys lucidus) by Small RNA Sequencing. Fishes 2022, 7, 297. https://doi.org/10.3390/fishes7050297
Ji Q, Xie Z, Gan W, Wang L, Song W. Identification and Characterization of PIWI-Interacting RNAs in Spinyhead Croakers (Collichthys lucidus) by Small RNA Sequencing. Fishes. 2022; 7(5):297. https://doi.org/10.3390/fishes7050297
Chicago/Turabian StyleJi, Qun, Zhengli Xie, Wu Gan, Lumin Wang, and Wei Song. 2022. "Identification and Characterization of PIWI-Interacting RNAs in Spinyhead Croakers (Collichthys lucidus) by Small RNA Sequencing" Fishes 7, no. 5: 297. https://doi.org/10.3390/fishes7050297
APA StyleJi, Q., Xie, Z., Gan, W., Wang, L., & Song, W. (2022). Identification and Characterization of PIWI-Interacting RNAs in Spinyhead Croakers (Collichthys lucidus) by Small RNA Sequencing. Fishes, 7(5), 297. https://doi.org/10.3390/fishes7050297





