Abstract
Exposure to acidic and alkaline pHs results in an ionic imbalance. Cellular responses involved in osmoregulation in silver catfish exposed to different pHs (5.5, 7.5, and 9.0) for 24 h were evaluated. The gills and kidney were collected to measure Na+/K+-ATPase (NKA) and H+-ATPase (V-ATPase) activities and to evaluate the expression of ion transporter-related genes: NKA (atp1a1), H+-ATPases (atp6v0a1b, atp6v0a2a, atp6v0a2b), Na+/H+ antiporter (slc9a3), K+/Cl− symporters (slc12a4, slc12a6, slc12a7a, slc12a7b), Na+/K+/2Cl− symporter (slc12a2), and ammonium transporter Rh type b (rhbg). The gills presented greater responses to pH changes than the kidney. The pH alterations changed the atp1a1 gene expression and NKA activity, whereas the H+-ATPase activity increased in the gills in alkaline water, probably to maintain ionic balance. The slc9a3 and slc12a2 genes play more prominent roles in the ion uptake at acidic pH than H+-ATPase. The slc12a7a was the only isoform of this transporter affected by pH. The rhbg is apparently related to ammonia excretion through the gills and kidney (minor scale). Exposure to alkaline pH seems to be battled by impairment of NKA and H+-ATPase activities in the gills, whereas the expression of some ion transporters in silver catfish changes during both acidic and alkaline pHs.
1. Introduction
In the aquatic environment, fish may encounter severe and challenging ionic/osmotic gradients to maintain internal homeostasis. The low concentration of ions in freshwater (FW) causes fish to lose ions and gain water by osmosis [1,2,3]. The problem of osmotic water gain is solved by low water ingestion [4], decreasing the water permeability of the integument [5] and producing large amounts of diluted urine [6]. The net ion loss is mainly solved by the combined response of a reduction in diffusive loss [5] and an increased ability to absorb ions to maintain liquid ionic status [7]. To maintain this internal balance, fish have developed sophisticated mechanisms that are achieved by osmoregulatory organs [8]. Fish gills are the main organ for gas exchange, ion regulation, acid-base balance, and nitrogen waste excretion [3,7,9]. The gills are the main target of acidic or alkaline water because they are in direct contact with the environment [10]. The kidney, together with the gills, is essential for osmoregulatory processes in freshwater teleosts [11]. The kidney is the primary organ for the elimination of water, being particularly important for freshwater species due to the efficient ion reabsorption mechanisms to minimize ion loss [12]. Considering the importance of these two organs in the osmoregulatory process, the gills and kidney were the focus of this study.
At the cellular level, ion transport is accomplished through mitochondria-rich cells, the ionocytes, which express specific ion transporters (or enzymes) and, therefore, are responsible for ion transport (primarily Na+, Cl−, and Ca2+ uptake and H+, HCO3− uptake/secretion) and NH3 excretion in FW [1,2,7,13]. Different transmembrane proteins have been described as being involved in ion exchange (ion channels, co-carriers). The specificity, location (apical/basolateral), and relative abundance and expression of these proteins in ionocytes result in the uptake or excretion of ions [14].
The pH of water is a very important parameter to be considered because it has a direct effect on homeostasis, metabolism, physiological processes, and fish survival [15]. The pH of water is usually regulated by the carbonate–bicarbonate system, generally remaining between 6.0 and 8.0 in freshwater [16]. Oscillations over this range may be due to the abundance of phytoplankton or the presence of high concentrations of dissolved HCO3− and CO32− salts (alkaline pH) or even by an excess of mineral and organic acids (acidic pH) [17]. When exposed to acidic or alkaline waters, the normal functions of the gills tend to decrease [15].
In fish exposed to low pH, acid loading through the gills is a source of acid-based disturbance, with increased H+ and NH4+ excretion in the urine to compensate for this problem [18]. In addition, it increases the ion loss [19,20], which decreases plasma ion levels and the pH [18,21]. Very acidic pH levels impair branchial protein junctions, which increases paracellular ion loss [22,23]. A decrease in Na+ uptake also occurs in fish exposed to acidic waters due to the inhibition of Na+/H+ or Na+/NH4+ and H+-ATPase (V-ATPase) apical transporters [23].
In alkaline waters, the main problems are related to the inhibition of ammonia excretion [18] and increased excretion of CO2 [24]. Specimens of Amur ide (Leuciscus waleckii) that live in alkaline waters (pH 9.6) showed higher expression of some slc12 genes (Na+/K+/2Cl−, K+/Cl−, and Na+/Cl− symporters), which may facilitate Cl− uptake [25,26]. In FW fish, ammonia excretion is maintained through a Na+/NH4+ exchange complex, which consists of several membrane transporters (Rhbg, H+-ATPase, Na+/H+) that together create an acid capture mechanism in the gill boundary layer [27]. At neutral pH, ammonia leaves the gills by diffusion in the form of NH3 (non-ionized), which is converted to NH4+ (ionized) in water, maintaining a favorable gradient for NH3 diffusion [16]. Thus, any disruption in the acidified gill microenvironment may cause impairment of the excretion mechanism and result in an internal accumulation of ammonia [28]. These physiological effects, combined with ion loss, can be considered the main mechanism associated with mortality in fish exposed to high environmental pH [29].
The silver catfish inhabits different FW environments from Central and South America [30] and has economic and ecological importance [20]. This species can survive acute changes in the 4.0 to 9.0 pH range for 96 h without significant mortality [31], but plasma and urinary Na+ and Cl− levels were altered after 24 h exposure to different water pH [18]. Studies on physiology under various water pH levels are still necessary because stressful conditions of water pH may negatively affect fish performance and survival [32,33,34].
Considering that cellular responses to acute pH changes in this species remain uncertain and the lack of studies analyzing the activity of enzymes and expression of genes involved in FW fish osmoregulation, the aim of this study was to evaluate the activity of the enzymes involved in ion and ammonia exchange, such as the sodium potassium pump (Na+/K+-ATPase, NKA), the proton pump (H+-ATPase), and the expression of ion transporter-related genes: NKA (atp1a1), H+-ATPases (atp6v0a1b, atp6v0a2a, atp6v0a2b), Na+/H+ antiporter (slc9a3), K+/Cl− symporters (slc12a4, slc12a6, slc12a7a, slc12a7b), Na+/K+/2Cl− symporter (slc12a2), and ammonium transporter Rh type b (rhbg), in the gill and kidney of silver catfish exposed to acute pH changes.
2. Materials and Methods
2.1. Experimental Animals and Handling Conditions
Silver catfish juveniles (7.32 ± 0.40 g and 11.35 ± 1.30 cm; mean ± SEM) (n = 36) were obtained from a fish farm at Santa Maria, southern Brazil. These juveniles were acclimated for one week in a continuously aerated 250 L tank (two air pumps of 12 W each) at pH levels of 7.0–7.5. The animals were fed once daily to satiety with a commercial feed (32% crude protein).
2.2. Experimental Conditions
After acclimation, juveniles were placed in three recirculation systems with biological filters containing four 40 L tanks each (three fish in each replicate, four replicates). Temperature and dissolved oxygen levels were determined with the oxygen meter Y5512 (YSI Inc., Yellow Springs, OH, USA). Total ammonia (Labcon Test) and water hardness (EDTA titrimetric method) levels were verified. In each system, the pH was adjusted to the following values: 5.5, 7.5, and 9.0. This acidic to alkaline pH range was chosen because it reduces silver catfish growth but does not cause mortality [35]. The pH adjustment and control were performed every hour with a DMPH-2 pH meter (Digimed, São Paulo, SP, Brazil). The water was acidified with 1 N H2SO4 and alkalinized with 1 N NaOH. The fish were transferred directly from the acclimation tanks (pH 7.0–7.5) to the experimental pH and collected after 24 h of exposure.
2.3. Sample Collection and Analysis
Fish from each tank (n = 12 per treatment) were sampled and collected after 24 h of exposure to the experimental pH, anesthetized with 50 μL/L eugenol [36], and then euthanized by spinal cord section to remove gills and caudal kidneys. A small portion of these organs was stored in 2 mL RNase-free tubes containing TRIzol® Reagent (Invitrogen, Waltham, MA, USA) for analysis of gene expression. The remaining collected tissues were immersed in liquid nitrogen and subsequently stored at −80 °C until the enzyme analysis could be performed.
2.4. Expression of Genes Related to Ion and Ammonia Transport
In order to determine the sequence of the genes of NKA (sodium potassium-transporting ATPase subunit alpha-1, atp1a1), H+-ATPases (v-type proton ATPase subunit a, atp6v0a1b, atp6v0a2a, atp6v0a2b), Na+/H+ antiporter (sodium/hydrogen exchanger, slc9a3), K+/Cl− symporters (slc12a4, slc12a6, slc12a7a, slc12a7b), Na+/K+/2Cl− symporter (solute carrier family 12 member 2, slc12a2), and ammonium transporter Rh type B (rhbg) in silver catfish, in brief, samples of the head and caudal kidney, brain including the pituitary gland, and liver were mass sequenced as previously described [37]. Subsequently, other alternatives, such as the alignment of the readings against related species, were performed. Primers were then designed using the Primer Express software 3.0 (Applied Biosystems, San Francisco, CA, USA) and validated by standard curves. The genes actin beta (actb) and eukaryotic elongation factor 1-alpha (eef1a), described in [37,38], respectively, served as internal reference genes (Table 1). All of the gene names followed the ZFIN Zebrafish Nomenclature Conventions and were named after the mammalian orthologues. These genes were chosen because their complete sequences were available in the mass sequence of silver catfish and were expressed in other freshwater-adapted teleosts [1,3,8,11,39,40,41,42,43,44,45,46,47,48]. In addition, previous analysis with the respective primers tested indicated the expression of these genes in the gills and kidney of silver catfish.

Table 1.
Primer oligonucleotide sequences used for RT-PCR analysis.
Total RNA from the gills and kidney was extracted using TRIzol®® Reagent (Thermo Fisher Scientific, São Paulo, SP, Brazil), as indicated in the manufacturer’s instructions. Quantification and determination of RNA purity was performed as described previously [37]. Total RNA (500 ng) was treated with 0.1 U DNase Amplification Grade (Invitrogen) for 15 min at 27 °C, followed by DNase inactivation with 1 µL of EDTA at 65 °C for 10 min. Then, the complementary DNA (cDNA) was synthetized by reverse transcription reaction using an iScript™ cDNA Synthesis Kit (BioRad, São Paulo, SP, Brazil), according to the manufacturer’s instructions. The reverse-transcription was performed using a SimpliAmp™ Thermal Cycler (Applied Biosystems™, São Paulo, SP, Brazil), and the program consisted of 5 min at 25 °C, 30 min at 42 °C, and 5 min at 85 °C, finishing with a hold at 4 °C. Real-time quantitative PCR (qPCR) reactions were performed with 2 µL cDNA (5 ng of cDNA in a final reaction volume of 10 µL), 1 µL of specific forward and reverse primers for each gene at a final concentration of 200 nM, 5 µL of SYBR Green (GoTaq® qPCR Master Mix, Promega, São Paulo, SP, Brazil), and 1 µL of nuclease-free water. The qPCR reactions were conducted in a CFX384 thermocycler (BioRad), and the thermal profile was as follows: initial denaturation at 95 °C for 3 min followed by 40 cycles of denaturation at 95 °C for 10 s and annealing/extension at 60 °C for 1 min, melting curve from 65 to 95 °C increasing 0.5 °C every 5 s. Melting curve analyses were performed to verify that a single product was amplified and to check the absence of primer-dimer artifacts. Samples were run in duplicate and relative gene quantification was performed using the 2−ΔΔCq method [49], corrected for efficiencies [50] and by geometric averaging of two internal reference genes [51].
The primer oligonucleotide sequences used for RT-PCR analyses are shown in Table 1. E (%) is the reaction efficiency and R2 the coefficient of determination for each pair of oligonucleotides, both in gills and kidney. The genes actb and eef1a served as internal reference genes.
2.5. Determination of NKA and H+-ATPase Activities
The branchial and kidney tissues were homogenized (1:10 w/v) in a homogenization buffer (150 mM sucrose, 50 mM imidazole, and 10 mM EDTA, pH 7.5) and centrifuged at 1000× g for 10 min at 4 °C, and the supernatant was stored at −80 °C until use. Branchial and renal NKA and H+-ATPase activities were determined in microplates, as described by [52]. Briefly, 5 µL of homogenate and 200 µL of reaction solution (30 mM imidazole, 45 mM NaCl, 15 mM KCl, 3 mM MgCl2, 0.4 mM KCN 1 mM ATP, 0.2 mM NADH, 0.1 mM fructose 1,6 diphosphate, and 2 mM phosphoenolpyruvate) were added to each sample. Ouabain (2 mM) was used as an inhibitor. H+-ATPase (HA) activity was measured in the same manner as the NKA, using Bafilomycin A1 as a specific inhibitor of the V-type H+-ATPase [53] in a final concentration of 100 nM. Results were expressed as mmol ADP released/min/mg of protein.
2.6. Phylogenetic Trees
Phylogenetic trees for the deduced protein sequences from genes atp1a1, atp6v0a1b, atpv0a2a, atpv0a2b, slc9a3, slc12a4, slc12a6, slc12a7a, slc12a7b, slc12a2, and rhbg were performed using NGPhylogeny.fr, with default parameters (PhyML), visualizing the tree with the interactive Tree of Life (iTOL; https://itol.embl.de (accessed on 1 August 2022)). The accession number of the Genbank and NCBI reference sequences are in Supplementary Figures S1–S8.
2.7. Statistical Analysis
A Kolmogorov–Smirnov test was conducted to evaluate the homogeneity of the variances. The effect of tanks (replicates) within each treatment was analyzed using one-way analysis of variance (ANOVA), and no significant difference was found, so data from all replicates were pooled. The data were compared using one-way ANOVA followed by Tukey’s test. All analyses were performed using the GraphPad Prism 5.0® software. The minimum level of significance was p < 0.05. The results for enzyme activities were expressed as the mean ± standard error of the mean (SEM), whereas for gene expression data were expressed according to BioRad CFX Maestro 2.3 software (Bio-Rad Laboratories, Hercules, CA, USA), using SEM in the error bars.
3. Results
Water physicochemical parameters did not differ significantly between treatments, except pH (Table 2). No fish died during the whole experimental period.

Table 2.
Water parameters measured during the experimental period.
3.1. Phylogenetic Comparisons of the Sequences
The deduced protein sequences for atp1a1, atp6v0a1b, atp6v0a2a, and atp6v0a2b in silver catfish showed over 90% identity with the same proteins and isoforms of other Siluriformes. The Atp1a1 protein has an 88.33% identity with zebrafish, Danio rerio. The Atp6v0a1b protein also has an identity with zebrafish of 92.29% and 91.15% with rainbow trout, Oncorhynchus mykiss. The Atpv0a2b protein showed 82.73% identity with rainbow trout and 82.45% with zebrafish. The Atpv0a2a protein also showed 82.22% identity with rainbow trout and 82.22% with zebrafish (Supplementary Figures S1 and S2). The protein sequences for Slc9a3 showed 83–87% identity with other Siluriformes and 67.74% with zebrafish (Supplementary Figure S3). The protein sequence for Slc12a4 showed 88–97% identity with other Siluriformes: 93.0% with zebrafish, and 90.59% with rainbow trout (Supplementary Figure S4). The protein sequence for Slc12a6 showed 88–97% identity with other Siluriformes (Supplementary Figure S5). The protein sequence for Slc12a7a showed 77.50% identity with rainbow trout and 74.36% with zebrafish, and Slc12a7b showed 85.66% identity with zebrafish (Supplementary Figure S6). The protein sequence for Sc21a2 showed 84.44% identity with zebrafish and 81.63% with rainbow trout (Supplementary Figure S7). The protein sequence for Rhbg showed 85–89% identity with other Siluriformes, 80.26% with rainbow trout, and 79.91% with zebrafish (Supplementary Figure S8).
3.2. Gene Expression
Three different H+-ATPase isoforms were expressed in the gills and kidney of silver catfish: atp6v0a1b, atp6v0a2a, and atp6v0a2b. The treatments did not significantly affect their expression in the gills and kidney (Figure 1a–c and Figure 2a–c). Silver catfish that were exposed to pH levels of 5.5 and 9.0 showed significantly higher atp1a1 expression in gills when compared to those kept at a pH of 7.5 (Figure 1d). The contrary occurred in the kidney, where there was lower expression in fish exposed to pH levels of 5.5 and 9.0, when compared to a pH of 7.5 (Figure 2d).
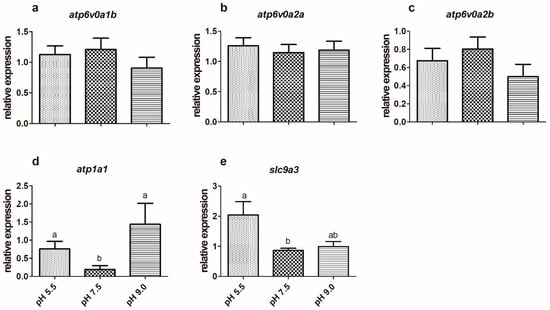
Figure 1.
Expression of H+-ATPase isoforms (a—atp6v0a1b, b—atp6v0a2a, c—atp6v0a2b), d—Na+/K+-ATPase (atp1a1), and e—Na+/H+ antiporter (slc9a3) in the gills of silver catfish (Rhamdia quelen, n = 12) exposed to different water pHs. Different letters indicate significant differences between treatments. One-way ANOVA and Tukey’s test were used to determine statistical significance (p < 0.05).
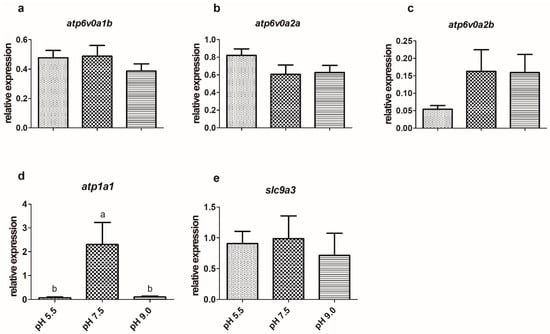
Figure 2.
Expression of H+-ATPase (a—atp6v0a1b, b—atp6v0a2b, c—atp6v0a2a), d—Na+/K+-ATPase (atp1a), and e—Na+/H+ antiporter (slc9a3) in the kidney of silver catfish (Rhamdia quelen, n = 12) exposed to different water pHs. Different letters indicate significant differences between treatments. One-way ANOVA and Tukey’s test were used to determine statistical significance (p < 0.05).
The expression of slc9a3 in the gills of silver catfish exposed to a pH of 5.5 was higher than those exposed to a pH of 7.5, but there was no difference in relation to a pH of 9.0 (Figure 1e), as well as in the expression of this gene in the kidney among the three experimental conditions (Figure 2e). The treatments did not significantly affect the expression of the slc12a6 and slc12a7b genes (Figure 3a,c; Figure 4a,c) in the gills and kidney. Fish exposed to acidic pH levels (5.5) presented higher slc12a7a expression in the gills than fish maintained at a pH of 9.0, but those kept at a pH of 7.5 did not show significant differences from those exposed to acidic and alkaline pH levels (Figure 3b). There was no significant difference in the expression of this gene in the kidney (Figure 4b).
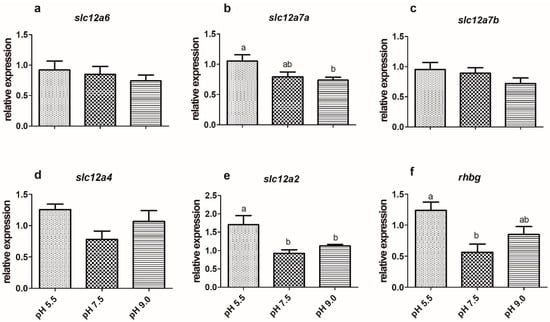
Figure 3.
Expression of K+/Cl− cotransporters (a—slc12a6, b—slc12a7a, c—slc12a7b, d—slc12a4), e—Na+/K+/2Cl– cotransporter (slc12a2), and f—ammonium transporter Rh type C-like, glycoprotein b (rhbg) in the gills of silver catfish (Rhamdia quelen, n = 12) exposed to different water pHs. Different letters indicate significant differences between treatments. One-way ANOVA and Tukey’s test were used to determine statistical significance (p < 0.05).
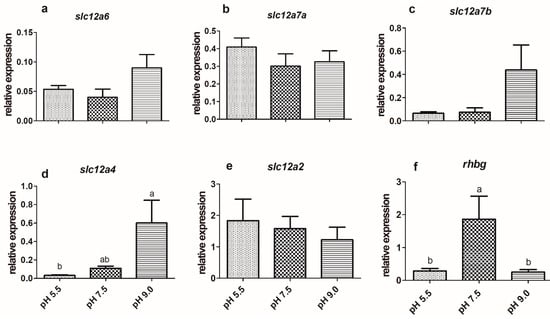
Figure 4.
Expression of K+/Cl− cotransporters (a—slc12a6, b—slc12a7a, c—slc12a7b, d—slc12a4), e—Na+/K+/2Cl– cotransporter (slc12a2), and f—ammonium transporter Rh type C-like, glycoprotein b (rhbg) in the kidney of silver catfish (Rhamdia quelen, n = 12) exposed to different water pHs. Different letters indicate significant differences between treatments. One-way ANOVA and Tukey’s test were used to determine statistical significance (p < 0.05).
Silver catfish exposed to a pH of 9.0 showed a higher expression of slc12a4 in the kidney, compared to those at a pH of 5.5 (Figure 4d), but in the gills, there was no significant differences between fish from different pHs (Figure 3d). The highest expression of branchial slc12a2 was observed at a pH of 5.5 (Figure 3e), and there were no changes in the renal expression of this gene (Figure 4e). The expression of branchial rhbg was higher at acidic pH (5.5) than at neutral pH (Figure 3f); whereas in the kidney, its expression was lower at a pH of 5.5 and 9.0 than at a pH of 7.5 (Figure 4f).
3.3. NKA and H+-ATPase Activities
The activity of NKA in the gills was significantly higher in silver catfish exposed to a pH of 9.0 than in those exposed to 5.5 (Figure 5A). The highest activity of the H+-ATPase in the gills was observed in fish exposed to a pH of 9.0 (Figure 5B). Exposure to acidic and alkaline pHs did not significantly change the activities of these enzymes in the kidney (Figure 5C,D).
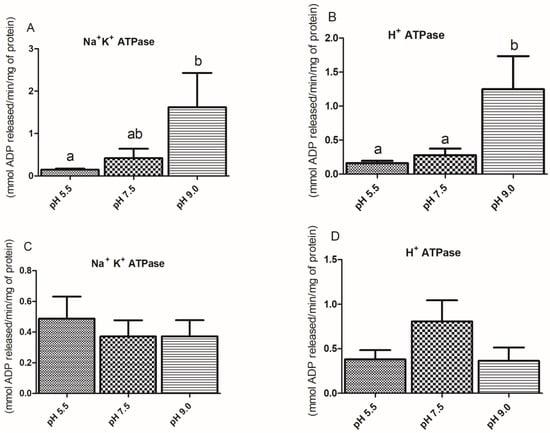
Figure 5.
Activities from Na+/K+-ATPase and H+-ATPase in gill (A,B) and kidney (C,D) of silver catfish (Rhamdia quelen, n = 12) exposed to different water pHs. Different letters indicate significant differences between treatments. One-way ANOVA and Tukey’s test were used to determine statistical significance (p < 0.05).
4. Discussion
In the current study, we identified and analyzed some transporters located in the gills and kidney ionocytes of silver catfish exposed to different pHs. The silver catfish expresses in the gills and kidney several transporters from genes, namely atp1a1, atp6v0a1b, atp6v0a2a, atp6v0a2b, slc9a3, slc12a4, slc12a6, slc12a7a, slc12a7b, slc12a2, and rhbg.
To compensate the ion loss due to diffusion, FW fish use ionocytes [5,6]. Furthermore, ionocytes also act to regulate the release of H+ or HCO3− to water [54,55] to achieve blood pH homeostasis. Freshwater fish went through multiple adaptations and re-adaptations during evolution, and there are probably distinct molecular strategies for obtaining ions and acid-base homeostasis in different species. There are several proposed names for ionocyte subtypes, as well as different patterns in their physiology and morphology, each unique to the species studied [1].
Several transporters, such as atp1a1, slc12a4, slc9a3, and slc12a2, were also expressed in the gills of freshwater-adapted Mozambique tilapia (Oreochromis mossambicus), as well as slc12a10.2 (ncc) [8,39,40,41,42,43,44]. In the gills of zebrafish, the expression of the transporters atp1a1a (nka1a), atp2b2 (pmca2), atp6v1aa rhcg1, slc4a4b (nbc), slc12a10.2 (ncc), slc12a2 (nkcc1) (low expression), slc8a (ncx), and slc9a3 (nhe3) has been identified [45,46]. The genes slc9a2 (nhe2) and slc9a3 (nhe3) were cloned in rainbow trout and are present in ionocytes [47]. Other studies showed the possible involvement of some genes of the same transporters in the gills for the ionoregulation of killifish (Fundulus heteroclitus), such as slc12a10 (ncc2), slc12a1 (nkcc2), and slc12a2 (nkcc1) [11,39,44,48]. Changes in the activity of proteins and in the expression of the genes involved in the osmoregulation of freshwater teleosts illustrate some of the possible responses that animals may exhibit in the face of osmotic alterations.
NKA plays a very important role in the kidney and gills of teleosts, positively regulating their activity in response to environmental changes [13,56,57]. In the present study, there was an increase in Na+/K+-ATPase activity in the gills of silver catfish exposed to alkaline water, compared to those exposed to acidic pH, but there was no difference compared to those at neutral pH. There was no change in renal NKA activity, probably due to the time of exposure. The gills and kidney of fish tend to have normal functioning under exposure to very acidic or alkaline waters [15,18,58].
Alkaline water causes transient decreases in the capacity of the ion transport system by directly acting on Cl− and Na+ transport sites in the gills [17]. Gill NKA activity increased in Amur ide exposed to alkaline water for 5–7 days [26]. Silver catfish exposed to alkaline pH for 24 h showed higher NKA activity in the gills and kidney [59]. Different from enzyme activity, the expression of the NKA transporter (atp1a1) increased at acidic (5.5) and alkaline (9.0) pHs in the gills, suggesting that acute exposure is likely to alter the gene expression differently from the activity of the enzyme. In agreement with this hypothesis, gill NKA activity was higher in pacu (Piaractus mesopotamicus) exposed for 1 day to pH 9.0, but after 15 days it was higher in those exposed to a pH of 5.5 [34]. The expression of atp1a1 was lower in the kidney of silver catfish exposed to a pH of 5.5 and 9.0 than the control (pH 7.5). The opposite was observed in the kidney of common carp (Cyprinus carpio) exposed to a pH of 4.0 for three days [60].
Vacuolar H+-ATPase (V-ATPase), which also participates in ion regulation and acid-base balance in fish [61,62], secretes H+ by the apical membrane in the H+-ATPase-rich ionocyte of zebrafish gills [46]. In zebrafish larvae exposed to a pH of 4.0 for 4 days, there was an increase in the expression of atp6v1aa in the gills [63]. In killifish, H+-ATPase is expressed in the basolateral membranes of cuboidal cells, the cell type responsible for Na+ uptake, and according to [64], Na+ uptake is most likely not coupled with active proton excretion in this species. In the present study, H+-ATPase has higher activity in the gills of silver catfish exposed to alkaline water compared to the other treatments, but there was no significant difference in the activity of this enzyme in the kidney. The same results were observed in the gills and kidney of pacu exposed to a pH of 9.0 for 1 day [34]. However, the expression of the genes of all three different H+-ATPase isoforms expressed in the gills and kidney of silver catfish were not affected by water pH, which could suggest a clear species-specific regulation of this physiological mechanism.
Na+/H+ exchangers (Slc9a family) are considered to be the main actors in apical Na+ uptake and acid excretion, and can be functionally coupled to ammonia excretion in fish gill ionocytes [3,65]. Slc9a3 (Nhe3) is the major isoform for apical Na+/H+ exchangers in the gill ionocytes of several FW teleosts (killifish, Mozambique tilapia, medaka—Oryzias latipes, and zebrafish, among others) [40,56,66,67,68,69]. However, the presence of the Nhe2 protein (slc9a2) has been shown in cuboidal cells of killifish [70], whereas slc9a2 (nhe2) and slc9a3 (nhe3) have also been identified in the peanut lectin agglutinin (PNA+) ionocytes of rainbow trout [1,71,72]. In [65], slc9a3 (nhe3) was expressed in gills and kidneys of FW- and seawater-adapted (SW) sea bass (Dicentrarchus labrax), with a higher expression in FW-adapted fish.
In zebrafish, the expression of slc9a3.2 (slc9a3 tandem repeat 2) decreased and atp6v0ca increased in the gills after a 7-day acclimation to a pH of 4, suggesting that H+-ATPase most likely plays a more prominent role in the uptake of Na+ in acidic water [67]. However, in silver catfish gills, we observed that there was no significant difference in the expression of the different isoforms and activity of H+-ATPase, but there was an increase in the expression of slc9a3 (nhe3b) after exposure to acidic pH. Consequently, it seems that, in this species, slc9a3 plays a more prominent role in the uptake of Na+ at acidic pHs. Likewise, [66] showed that exposure to acidic pHs led to a significant increase in the expression of gill slc9a3 (nhe3), but only a slight increase in the β-subunit of H+-ATPase in Osorezan dace, Tribolodon hakonensis, suggesting that slc9a3 (nhe3) may play a role in acid secretion, as well as Na+ absorption.
The ammonia transporters (Rhcg1 and/or Rhcg2) present in fish gills [73] form a functional metabolon with Slc9a2 (Nhe2)/Slc9a3 (Nhe3) at acidic pHs [74]. In silver catfish, it was possible to observe a similar response pattern in the slc9a3 and rhbg genes, given there was a significant increase in their expression in the gills of fish exposed to acidic compared to neutral pHs.
The slc12a7a and slc12a4 are the two isoforms tested in silver catfish of this transporter whose expressions are affected by pH because the other two related genes (slc12a6, slc12a7b) did not change. Mozambique tilapia showed no difference in the expression of slc12a7b (kcc4) and slc12a4 (kcc1) between fresh water and fresh water with high potassium concentration [53], which is consistent with the results of SW-acclimated tilapia gills [75]. However, slc12a7a was highly expressed in the kidney compared to the gills, both in the control and in exposure with a high concentration of K+, suggesting a significant renal role of the K+/Cl− symporter [53]. The slc12a4 gene was expressed in the gills and kidneys of silver catfish, but it showed a significant difference only in the kidney, in which fish exposed to a pH of 9.0 showed higher expression compared to a pH of 5.5.
The expression of slc12a2 in the gills of silver catfish was higher at pH 5.5 compared to the other treatments, and its expression in the kidney was not significantly affected by the pH change. The occurrence of the Na+/K+/2Cl− symporter has been demonstrated, not only in the branchial and opercular epithelia [44,48,76,77,78], but also in the renal [79] and intestinal epithelia [80,81]. Bumetamide, an inhibitor of this symporter, did not affect Na+ influx in native cardinal tetra (Paracheirodon axelrodi), hemigrammus tetra (Hemigrammus rhodostomus), and moenkhausia tetra (Moenkhausia diktyota) from the Rio Negro, an acidic, ion-poor, and black water river, but these species have different Na+ uptake mechanisms from the “standard” FW teleosts [82]. This symporter is considered to be mainly related to SW adaptation [8,48,78], and the expression of its gene decreased in the opercular epithelium of killifish one day after transference from seawater to freshwater, but three days later its expression was not significantly different between these two waters [11], demonstrating a high-performance plasticity of this gene/protein to acquire its homeostatic load. The same authors verified that killifish maintained for one year in freshwater presented the same expression of this gene in the opercular epithelium than seawater killifish. The freshwater-adapted Mozambique tilapia presents this symporter in the basolateral membrane of gill ionocytes type III [1]. As of yet, the relationship between the Na+/K+/2Cl− symporter and ion uptake in freshwater has not been explained [3], and consequently, it is not clear if the increased expression of the slc12a2 gene in the gills of silver catfish exposed to acidic pH might be related to a mechanism for compensating the ion loss by diffusion.
Most teleost species presented ammonia as the main nitrogenous excretory product [59,83]. Non-ionized ammonia (NH3) enters the ionocytes with the aid of the Rhbg glycoprotein, while the Rhcg1 and/or Rhcg2 glycoproteins transport ammonia out of ionocytes [74], and the formation of the NH4+ boundary layer from the reaction of NH3 and H+ (from function Nhe) would locally increase pH and decrease NH3, facilitating NHE activity in apparently thermodynamically unfavorable low pH environments [1]. In the current study, the expression of rhbg was higher in the gills of silver catfish exposed to acidic pH (5.5) than in neutral pH (7.5), which could be related to increased ammonia excretion at acidic pHs, but additional studies investigating expression of the rhcg1 and rhcg2 genes must be performed to confirm this hypothesis and to verify if this could be a cause or a consequence of pH and ammonia balance. The increase in ammonia excretion at acidic pHs may be a strategy to facilitate Na+ uptake, as observed in the larvae of D. rerio exposed to acidic pHs [84]. However, in the kidney, there was lower rhbg expression at a pH of 5.5 and 9.0 compared to a neutral pH, suggesting a lower contribution (if any) of this transporter at the renal level during acute pH variations. Lower renal rhbg expression was also observed in common carp exposed for three days to a pH of 4.0 [60].
The genes slc12a7b and slc121a6 did not show significant differences between treatments in the gills and kidney. Overall, the gills of silver catfish exposed to acidic and alkaline pHs showed more significant alterations in the gene expression of ion transporters, probably because this structure is the primary site for ion regulation, acid-base balance, and nitrogen waste excretion [3,7,9]. Longer exposure to acidic and alkaline pHs may yield different results for the enzyme activities and the expression of transporters in the gills and kidney of silver catfish.
In conclusion, silver catfish expressed several transporters in the gills and kidneys that are involved with osmoregulation. Acute pH alterations (24 h) change the atp1a1 gene expression differently from the activity of the enzyme, and the increase in the H+-ATPase activity in the gills in alkaline water seems to be an important strategy to maintain ionic balance. The slc9a3 gene seems to play a more prominent role in the uptake of Na+ at acidic pHs than H+-ATPase in silver catfish. It was possible to observe a response pattern in the slc9a3 and rhbg genes in fish exposed to acidic pH. The slc12a7a and slc12a4 seem to be affected by pH because the other two related genes (slc12a6, slc12a7b) did not change their expression levels. The results of the expression of slc12a4 suggest that it may be involved in the adaptation to alkaline pHs. The slc12a2 may be related to a mechanism to increase ion uptake at acidic pHs to compensate for the loss of ions by diffusion. As expected, rhbg most likely participates in the excretion of ammonia in the gills, and it does not contribute to change renal activity during acute pH variations.
Finally, as the gills are the main target of acidic or alkaline waters and are in close contact with the environment, they showed greater responses than the kidneys to changes in pH. Changes in the activity of proteins and in the expression of the genes involved in the osmoregulation of freshwater teleosts illustrate some of the responses that animals may exhibit in the face of osmotic alteration.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/fishes7050261/s1, Supplementary Figures S1–S8.
Author Contributions
Conceptualization, M.T.S.M., B.B., G.M.-R.; methodology: B.B., G.M.-R., J.A.M.-S., C.d.F.S., A.P.G.A., A.Q.A.; validation: G.M.-R., J.A.M.-S., C.d.F.S., A.P.G.A., A.Q.A.; formal analysis, M.T.S.M., B.B., G.M.-R., J.A.M.-S., C.d.F.S., A.P.G.A., A.Q.A.; investigation, M.T.S.M., C.d.F.S., A.P.G.A., S.N.D., A.E.B.; resources, B.B., G.M.-R., J.A.M.-S., A.Q.A.; data curation, M.T.S.M., C.d.F.S., A.P.G.A., S.N.D., A.E.B.; writing—original draft preparation, M.T.S.M., C.d.F.S., A.P.G.A., S.N.D., A.E.B., B.B., G.M.-R., J.A.M.-S., A.Q.A.; writing—review and editing, M.T.S.M., A.P.G.A., B.B., G.M.-R., J.A.M.-S.; visualization, M.T.S.M., A.P.G.A., B.B., G.M.-R., J.A.M.-S.; supervision, B.B., G.M.-R., J.A.M.-S.; project administration, B.B., G.M.-R., J.A.M.-S.; funding acquisition, B.B., G.M.-R., J.A.M.-S., A.Q.A. All authors have read and agreed to the published version of the manuscript.
Funding
B. Baldisserotto received a research fellowship from the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq, Brazil). This study was also supported by Fundação de Amparo à Pesquisa do Estado do Rio Grande do Sul (FAPERGS, Brazil). The authors also thank Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES, Brazil) for the post-doctoral scholarship for C.F. Souza, CAPES PrInt visiting professor for G. Martínez-Rodríguez and A.P.G. Almeida, and PhD scholarships for S.N. Descovi and A.E. Bianchini, finance code 001.
Institutional Review Board Statement
All procedures were approved by the Ethical Committee on Animal Use from the Federal University of Santa Maria (protocol n. 6472190618/2018).
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest.
Acronyms
| atp1a1 | sodium/potassium-transporting ATPase subunit alpha-1 |
| Atp1a1 | Sodium/potassium-transporting ATPase |
| atp2b2 | Plasma membrane Ca2+ transporting ATPase 2 (previously known as pmca2) |
| Atp2b2 | Plasma membrane Ca2+ transporting ATPase 2 |
| atp6v1aa | ATPase H+ transporting V1 domain |
| Atp6v1aa | V-type proton ATPase V1 |
| atp6v0a1b | ATPase H+ transporting V0 subunit a1b |
| Atp6v0a1b | V-type proton ATPase subunit a1 isoform b |
| atp6v0a2a | ATPase H+ transporting V0 subunit a2a |
| Atp6v0a2a | V-type proton ATPase subunit a2 isoform a |
| atp6v0a2b | ATPase H+ transporting V0 subunit a2b |
| Atp6v0a2b | V-type proton ATPase subunit a2 isoform b |
| atp6v0ca | ATPase H+ transporting V0 subunit c |
| Atp6v0ca | V-type proton ATPase proteolipid subunit |
| rhbg | Rh family B glycoprotein |
| Rhbg | Ammonium transporter Rh type B |
| rhcg1 | Rh family C glycoprotein 1 |
| Rhcg1 | Ammonium transporter Rh type C-like 1 |
| rhcg2 | Rh family type C glycoprotein 2 |
| Rhcg2 | Ammonium transporter Rh type C 2 |
| slc4a4b | solute carrier family 4 member 4b (previously known as nbc) |
| Slc4a4b | solute carrier family 4 member 4b (Na-HCO3 cotransporter) |
| slc8a | solute carrier family 8 (previously known as ncx) |
| Slc8a | solute carrier family 8 (Na/Ca exchanger) |
| slc9a2 | solute carrier family 9 member 2 (previously known as nhe2) |
| Slc9a2 | solute carrier family 9 member 2 (sodium/hydrogen exchanger) |
| slc9a3 | solute carrier family 9 member 3 |
| Slc9a3 | Sodium/hydrogen exchanger (Previously known as Nhe3b) |
| slc12a2 | solute carrier family 12 member 2 (previously known as nkcc1) |
| Slc12a2 | Solute carrier family 12 member 2 (previously known as Na-K-Cl cotransporter 1) |
| slc12a4 | solute carrier family 12 member 4 |
| Slc12a4 | Solute carrier family 12 member 4 (Potassium/chloride transporter) |
| slc12a6 | solute carrier family 12 member 6 |
| Slc12a6 | Solute carrier family 12 member 6 (Potassium/chloride transporter) (previously known as Electroneutral potassium-chloride cotransporter 3, or K-Cl cotransporter 3, Kcc3) |
| slc12a7a | solute carrier family 12 member 7a |
| Slc12a7a | Solute carrier family 12 member 7a (Potassium/chloride transporter) (previously known as Electroneutral potassium-chloride cotransporter 4, K-Cl cotransporter 4, Kcc4) |
| slc12a7b | solute carrier family 12 member 7b |
| Slc12a7b | Solute carrier family 12 member 7b (Potassium/chloride transporter) (previously known as Electroneutral potassium-chloride cotransporter 4, K-Cl cotransporter 4, Kcc4) |
| slc12a10 | solute carrier family 12 member 10 |
| Slc12a10 | Solute carrier family 12 member 10 (previously known as Ncc) |
References
- Dymowska, A.K.; Hwang, P.P.; Goss, G.G. Structure and function of ionocytes in the freshwater fish gill. Respir. Physiol. Neurobiol. 2012, 184, 282–292. [Google Scholar] [CrossRef] [PubMed]
- Hwang, P.P.; Lin, L.Y. Gill ion transport, acid-base regulation and nitrogen excretion. In The Physiology of Fishes, 4th ed.; Evans, D.H., Claiborne, J.B., Currie, S., Eds.; CRC: Boca Raton, FL, USA, 2014; pp. 205–233. [Google Scholar]
- Zimmer, A.M.; Perry, S.F. Physiology and aquaculture: A review of ion and acid-base regulation by the gills of fishes. Fish Fish. 2022, 23, 874–898. [Google Scholar] [CrossRef]
- Robertson, L.M.; Wood, C.M. Measuring gill paracellular permeability with polyethylene glycol-4000 in freely swimming trout: Proof of principle. J. Exp. Biol. 2014, 217, 1425–1429. [Google Scholar] [PubMed]
- Chasiostis, H.; Kolorov, D.; Bui, P.; Kelly, S.P. Tight junctions, tight junction proteins and paracellular permeability across the gill epithelium of fishes: A review. Respir. Physiol. Neurobiol. 2012, 184, 282–292. [Google Scholar]
- Hwang, P.P.; Lee, T.H. New insights into fish ion regulation and mitocondrion-rich cells. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2007, 148, 479–497. [Google Scholar] [CrossRef] [PubMed]
- Evans, D.H.; Piermarini, P.M.; Choe, K.P. The multifuncional fish gill: Dominant site of gas exchange, osmoregulation, acid-base regulation, and excretion of nitrogenous waste. Physiol. Rev. 2005, 85, 97–177. [Google Scholar] [CrossRef] [PubMed]
- Hwang, P.P.; Lee, T.H.; Lin, L.Y. Ion regulation in fish gills: Recent progresses in the cellular and molecular mechanisms. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2011, 301, 28–47. [Google Scholar] [CrossRef] [PubMed]
- Shrivastava, J.; Ndugwa, M.; Caneos, W.; Boeck, G. Physiological trade-offs, acid-base balance and ion-osmoregulatory plasticity in European sea bass (Dicentrarchus labrax) juveniles under complex scenarios of salinity variation, ocean acidification and high ammonia challenge. Aquat. Toxicol. 2019, 212, 54–69. [Google Scholar] [CrossRef] [PubMed]
- Atli, G.; Canli, M. Essential metal (Cu, Zn) exposures the activity of ATPases in gill, kidney and muscle of tilapia Oreochromis niloticus. Ecotoxicology 2011, 20, 1861–1869. [Google Scholar] [CrossRef] [PubMed]
- Breves, J.P.; Starling, J.A.; Popovski, C.M.; Doud, J.M.; Tipsmark, C.K. Salinity-dependent expression of ncc2 in opercular epithelium and gill of mummichog (Fundulus heteroclitus). J. Comp. Physiol. B 2020, 190, 219–230. [Google Scholar] [CrossRef]
- Takvam, M.; Wood, C.M.; Kryvi, H.; Nilsen, T.O. Ion transporters and osmorregulation in the kidney of teleost fishes as a function of salinity. Front. Physiol. 2021, 12, 664588. [Google Scholar] [CrossRef] [PubMed]
- Evans, D.H. Freshwater fish gill ion transport: August Krogh to morpholinos and microprobes. Acta Physiol. 2011, 202, 349–359. [Google Scholar] [CrossRef]
- Nebel, C.L.; Boulo, V.; Bodinier, C.; Charmantier, G. The Na+/K+/2Cl− cotransporter in the sea bass Dicentrarchus labrax during ontogeny: Involvement in osmoregulation. J. Exp. Biol. 2006, 209, 4908–4922. [Google Scholar] [CrossRef] [PubMed]
- Parra, J.E.G.; Baldisserotto, B. Effect of water pH and hardness on survival and growth of freshwater teleosts. In Fish Osmoregulation; Baldisserotto, B., Mancera, J.M., Kapoor, B.G., Eds.; Science Publishers: Boca Raton, FL, USA, 2007; pp. 135–150. [Google Scholar]
- Baldisserotto, B. Water pH and hardness affect growth of freshwater teleosts. Braz. J. Anim. Sci. 2011, 40, 138–144. [Google Scholar]
- Wilkie, M.P.; Wood, C.M. The adaptations of fish to extremely alkaline environments. Comp. Biochem. Physiol. Part B 1996, 113, 665–673. [Google Scholar] [CrossRef]
- Bolner, K.C.S.; Baldisserotto, B. Water pH and urinary excretion in silver catfish Rhamdia quelen. J. Fish Biol. 2007, 70, 50–64. [Google Scholar] [CrossRef]
- Baldisserotto, B.; Copatti, C.E.; Gomes, L.C.; Chagas, E.C.; Brinn, R.P.; Roubach, R. Net ion fluxes in the facultative air-breather Hoplosternum littorale (tamoata) and the obligate air-breather Arapaima gigas (pirarucu) exposed to different Amazonian waters. Fish. Physiol. Biochem. 2008, 34, 405–412. [Google Scholar] [CrossRef] [PubMed]
- Baldisserotto, B.; Copatti, C.E.; Gomes, L.C.; Chagas, E.; Brinn, R.P.; Roubach, R. Calcium fluxes in Hoplosternum littorale (tamoata) exposed to different types of Amazonian waters. Neotrop. Ichthyol. 2009, 7, 465–470. [Google Scholar] [CrossRef]
- Freda, J.; McDonald, D.G. Physiological correlates of interspecific variation in acid tolerance in fish. J. Exp. Biol. 1998, 136, 243–258. [Google Scholar] [CrossRef]
- Gonzalez, R.J. Ion regulation in ion poor waters of low pH. In Physiology and Biochemistry of the Fishes of the Amazon; Val, A.L., Almeida-Val, V.M.F., Randall, D.J., Eds.; INPA: Manaus, Brazil, 1996; pp. 111–121. [Google Scholar]
- Wood, C.M. Toxic responses of the gill. In Target Organ Toxicity in Marine and Freshwater Teleosts: Organs; Schlenk, D., Benson, W.H., Eds.; Taylor & Francis: New York, NY, USA, 2001; Volume 1, pp. 33–37. [Google Scholar]
- Heath, A.G. Water Pollution and Fish Physiology, 2nd ed.; Lewis Publishers: Boca Raton, FL, USA, 1995. [Google Scholar]
- Xu, J.; Li, J.T.; Jiang, Y.L.; Peng, W.Z.; Yao, Z.L.; Chen, B.H. Genomic basis of adaptive evolution: The survival of Amur Ide (Leuciscus waleckii) in an extremely alkaline environment. Mol. Biol. Evol. 2017, 34, 145–159. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.M.; Zhao, X.F.; Liew, H.J.; Sun, B.; Wang, S.Y.; Luo, L.; Zhang, L.M.; Liang, L.Q. Effects of bicarbonate stress on serum ions and gill transporters in alkali and freshwater forms of Amur Ide (Leuciscus waleckii). Front. Physiol. 2021, 12, 676096. [Google Scholar] [CrossRef] [PubMed]
- Wright, P.A.; Wood, C.M. Seven things fish know about ammonia and we don’t. Respir. Physiol. Neurobiol. 2012, 184, 231–240. [Google Scholar] [CrossRef] [PubMed]
- Thompson, W.A.; Rodela, T.M.; Richards, J.G. The effects of strain and ploidy on the physiological response of rainbow trout (Oncorhynchus mykiss) to pH 9.5 exposure. Comp. Biochem. Physiol. Part B 2015, 183, 22–29. [Google Scholar] [CrossRef]
- Yesaki, T.; Iwama, G.K. Survival, acid-base regulation, ion regulation and ammonia excretion in rainbow trout in highly alkaline hard water. Physiol. Biochem. Zool. 1992, 65, 763–787. [Google Scholar] [CrossRef]
- Gomes, L.C.; Golombieski, J.I.; Gomes, A.R.C.; Baldisserotto, B. Biologia do jundiá Rhamdia quelen (TELEOSTEI, PIMELODIDAE). Ciência Rural. 2000, 10, 179–185. [Google Scholar] [CrossRef]
- Zaions, M.I.; Baldisserotto, B. Na+ and K+ body levels and survival of juveniles of Rhamdia quelen (Siluriformes, Pimelodidae) exposed to acute changes of water pH. Ciência Rural. 2000, 30, 1041–1045. [Google Scholar] [CrossRef]
- Lemos, C.H.P.; Ribeiro, C.V.M.; Oliveira, C.P.B.; Couto, R.D.; Copatti, C.E. Effects of interaction between pH and stocking density on the growth, haematological and biochemical response of Nile tilapia juveniles. Aquaculture 2018, 495, 62–67. [Google Scholar] [CrossRef]
- Lemos, C.H.P.; Chung, S.; Ribeiro, C.V.M.; Copatti, C.E. Growth and biochemical variables in Amazon catfish (Pseudoplatystoma reticulatum ♀ x Leiarius marmoratus ♂) under different water pH. An. Braz. Acad. Sci. 2018, 90, 3573–3581. [Google Scholar] [CrossRef]
- Copatti, C.E.; Baldisserotto, B.; Souza, C.F.; Monserrat, J.M.; Garcia, L.O. Water pH and hardness alter ATPase and oxidative stress in the gills and kidney of Pacu (Piaractus mesopotamicus). Neotrop. Ichthyol. 2019, 17, e190032. [Google Scholar] [CrossRef]
- Copatti, C.E.; Garcia, L.O.; Cunha, M.A.; Baldisserotto, B.; Kochhann, D. Interaction of water hardness and pH on growth of silver catfish, Rhamdia quelen, juveniles. J. World. Aquac. Soc. 2011, 42, 580–585. [Google Scholar] [CrossRef]
- Cunha, M.A.; Zeppenfeld, C.C.; Garcia, L.O.; Loro, V.L.; Fonseca, M.B.; Emanuelli, T.; Veeck, A.P.L.; Copatti, C.E.; Baldisserotto, B. Anesthesia of silver catfish with eugenol: Time of induction, cortisol response and sensory analysis of fillet. Ciência Rural. 2010, 40, 2107–2114. [Google Scholar] [CrossRef]
- Souza, C.F.; Descovi, S.; Baldissera, M.D.; Bertolin, K.; Bianchini, A.; Mourão, R.H.V.; Schmidt, D.; Heinzmann, B.M.; Antoniazzi, A.; Baldisserotto, B.; et al. Involvement of HPI-axis in anesthesia with Lippia alba essential oil citral and linalool chemotypes: Gene expression in the secondary responses in silver catfish. Fish Physiol. Biochem. 2019, 45, 155–166. [Google Scholar] [CrossRef]
- Baldisserotto, B.; Martos-Sitcha, J.A.; Menezes, C.C.; Toni, C.; Prati, R.L.; Garcia, L.O.; Salbego, J.; Mancera, J.M.; Martínez-Rodríguez, G. The effects of ammonia and water hardness on the hormonal, osmoregulatory and metabolic responses of the freshwater silver catfish Rhamdia quelen. Aquat. Toxicol. 2014, 152, 341–352. [Google Scholar] [CrossRef]
- Hiroi, J.; McCormick, S.D.; Ohtani-Kaneko, R.; Kaneko, T. Functional classification of mitochondrion-rich cells in euryhaline Mozambique tilapia (Oreochromis mossambicus) embryos, by means of triple immunofluorescence staining for Na+/K+-ATPase Na+/K+/2Cl− cotransporter and CFTR anion channel. J. Exp. Biol. 2005, 208, 2023–2036. [Google Scholar] [CrossRef]
- Inokuchi, M.; Hiroi, J.; Watanabe, S.; Hwang, P.P.; Kaneko, T. Morphological and functional classification of ion-absorbing mitochondria-rich cells in the gills of Mozambique tilapia. J. Exp. Biol. 2009, 212, 1003–1010. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.H.; Lee, K.M.; Inokuchi, M.; Kaneko, T. Acute responses of gill mitochondria-rich cells in Mozambique tilapia Oreochromis mossambicus following transfer from normal freshwater to deionized freshwater. Fish. Sci. 2010, 76, 101–109. [Google Scholar] [CrossRef]
- Zikos, A.; Seale, A.P.; Lerner, D.T.; Grau, E.G.; Korsmeyer, K.E. Effects of salinity on metabolic rate and branchial expression of genes involved in ion transport and metabolism in Mozambique tilapia (Oreochromis mossambicus). Comp. Biochem. Physiol. A 2014, 178, 121–131. [Google Scholar] [CrossRef]
- Pavlosky, K.K.; Yamaguchi, Y.; Lerner, D.T.; Seale, A.P. The effects of transfer from steady-state to tidally-changing salinities on plasma and branchial osmoregulatory variables in adult Mozambique tilapia. Comp. Biochem. Physiol. A 2019, 227, 134–145. [Google Scholar] [CrossRef]
- Breves, J.P.; Nelson, N.N.; Koltenyuk, V.; Petro-Sakuma, C.K.; Celino-Brady, F.T.; Sele, A.P. Enhanced expression of ncc1 and clc2c in the kidney and urinary bladder accompanies freshwater acclimation in Mozambique tilapia. Comp. Biochem. Physiol. A 2021, 260, 111021. [Google Scholar] [CrossRef]
- Shih, T.H.; Horng, J.L.; Liu, S.T.; Hwanh, P.P.; Lin, L.Y. Rhcg1 and NHE3b are involved in ammonium-dependent sodium uptake by zebrafish larvae acclimated to low-sodium water. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2012, 302, 84–93. [Google Scholar] [CrossRef]
- Guh, Y.J.; Lin, C.H.; Hwang, P.P. Osmoregulation in zebrafish: Ion transport mechanisms and functional regulation. EXCLI J. 2015, 4, 627–659. [Google Scholar]
- Ivanis, G.; Esbaugh, A.J.; Perry, S.F. Branchial expression and localization of SLC9A2 and SLC9A3 sodium/hydrogen exchangers and their possible role in acid–base regulation in freshwater rainbow trout (Oncorhynchus mykiss). J. Exp. Biol. 2008, 211, 2467–2477. [Google Scholar] [CrossRef]
- Scott, G.R.; Richards, J.G.; Forbush, B.; Isenring, P.; Schulte, P.M. Changes in gene expression in gills of the euryhaline killifish Fundulus heteroclitus after abrupt salinity transfer. Am. J. Physiol. 2004, 287, 300–309. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T. Analysis relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, 2002–2007. [Google Scholar] [CrossRef]
- Vandesompele, J.; Preter, K.; Pattyn, F.; Poppe, B.; Roy, N.V.; Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, 34.1–34.11. [Google Scholar] [CrossRef]
- Ruiz-Jarabo, I.; González-Wevar, C.A.; Oyarzún, R.; Fuentes, J.; Poulin, E.; Bertrán, C. Isolation driven divergence in osmoregulation in Galaxias maculatus (Jenyns, 1848) (Actinopterygii: Osmeriformes). PLoS ONE 2016, 11, e0154766. [Google Scholar] [CrossRef]
- Furukawa, F.; Watanabe, S.; Kakumura, K.; Hiroi, J.; Kaneko, T. Gene expression and cellular localization of ROMKs in the gills and kidney of Mozambique tilapia acclimated to fresh water with high potassium concentration. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2014, 307, 1303–1312. [Google Scholar] [CrossRef]
- Goss, G.G.; Wood, C.M. Na+ and C1 uptake kinetics, diffusive effluxes, and acidic equivalent fluxes across the gills of rainbow trout: I. Responses to environmental hyperoxia. J. Exp. Biol. 1990, 152, 521–547. [Google Scholar] [CrossRef]
- Goss, G.G.; Wood, C.M. Na+ and Cl− uptake kinetics, diffusive effluxes, and acidic equivalent fluxes across the gills of rainbow trout: II. Responses to bicarbonate infusion. J. Exp. Biol. 1990, 152, 549–571. [Google Scholar] [CrossRef]
- Morgan, J.D.; Iwama, G.K. Effects of salinity on growth metabolism and ion regulation in juvenille rainbow and steelhead trout (Oncorhychus mykiss) and fall Chinook salmon (Oncorhynchus tshawytscha). Can. J. Fish. Aquat. Sci. 2011, 48, 2083–2094. [Google Scholar] [CrossRef]
- Imsland, A.; Gunnarsson, S.; Foss, A.; Stefansson, S.O. Gill Na+/K+-ATPase activity, plasma chloride and osmolality in juvenile turbot (Scophthalmus maximus) reared at different temperatures and salinities. Aquaculture 2003, 218, 671–683. [Google Scholar] [CrossRef]
- Wood, C.M.; Milligan, C.L.; Walsh, P.J. Renal responses of trout to chronic respiratory and metabolic acidosis and metabolic alkalosis. Am. J. Physiol.-Reg. Integr. Comp. Physiol. 1999, 277, 482–492. [Google Scholar] [CrossRef] [PubMed]
- Bolner, K.C.S.; Copatti, C.E.; Rosso, F.L.; Loro, V.L.; Baldisserotto, B. Water pH and metabolic parameters in silver catfish (Rhamdia quelen). Biochem. Syst. Ecol. 2014, 56, 202–208. [Google Scholar] [CrossRef]
- Wright, P.A.; Wood, C.M.; Wilson, J.M. Rh versus pH: The role of Rhesus glycoproteins in renal ammonia excretion during metabolic acidosis in a freshwater teleost fish. J. Exp. Biol. 2014, 16, 2855–2865. [Google Scholar]
- Perry, S.F.; Furimsky, M.; Bayaa, M.; Georgalis, T.; Shahsavarani, A.; Nickerson, J.G.; Moon, T.W. Integrated responses of Na+/HCO3) cotransporters and V-type H+-ATPases in the fish gill and kidney during respiratory acidosis. Biochim. Biophys. Acta 2003, 1618, 175–184. [Google Scholar] [CrossRef]
- Perry, S.F.; Shahsavarani, A.; Georgalis, T.; Bayaa, M.; Furimsky, M.; Thomas, S.L. Channels, pumps, and exchangers in the gill and kidney of freshwater fishes: Their role in ionic and acid-base regulation. J. Exp. Zool. A 2003, 300, 53–62. [Google Scholar] [CrossRef]
- Shir-Mohammadi, K.; Perry, S.F. Expression of ion transport genes in ionocytes isolated from larval zebrafish (Danio rerio) exposed to acidic or Na+-deficient water. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2020, 319, 412–4277. [Google Scholar] [CrossRef]
- Katoh, F.; Hyodo, S.; Kaneko, T. Vacuolar-type proton pump in the basolateral plasma membrane energizes ion uptake in branchial mitochondria-rich cells of killifish Fundulus heteroclitus, adapted to a low ion environment. J. Exp. Biol. 2003, 206, 793–803. [Google Scholar] [CrossRef]
- Blondeau-Bidet, E.; Hiroi, J.; Lorin-Nebel, C. Ion uptake pathways in European sea bass Dicentrarchus labrax. Gene 2019, 692, 126–137. [Google Scholar] [CrossRef]
- Hirata, T.; Kaneko, T.; Ono, T.; Nakazato, T.; Furukawa, N.; Hasegawa, S.; Wakabayashi, S.; Shigekawa, M.; Chang, M.H.; Romero, M.F. Mechanism of acid adaptation of a fish living in a pH 3.5 lake. Am. J. Physiol. 2003, 284, 1199–1212. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.J.; Chou, M.Y.; Kaneko, T.; Hwang, P.P. Gene expression of Na+/H+ exchanger in zebrafish H+-ATPase-rich cells during acclimation to low-Na+ and acidic environments. Am. J. Physiol. Cell. Physiol. 2007, 293, C1814–C1823. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.C.; Horng, J.L.; Liu, S.T.; Hwang, P.P.; Wen, Z.H.; Lin, C.S.; Lin, L.Y. Ammonium-dependent sodium uptake in mitochondrion-rich cells of medaka (Oryzias latipes) larvae. Am. J. Physiol. Cell. Physiol. 2010, 298, 237–250. [Google Scholar] [CrossRef] [PubMed]
- Seo, M.Y.; Mekuchi, M.; Teranishi, K.; Kaneko, T. Expression of ion transporters in gill mitochondrion-rich cells in Japanese eel acclimated to a wide range of environmental salinity. Comp. Biochem. Physiol. Part A 2013, 166, 323–332. [Google Scholar]
- Edwards, S.L.; Wall, B.P.; Shetlar-Morrison, A.; Sligh, S.; Weakley, J.C.; Claiborne, J.B. The effect of environmental hypercapnia and salinity on the expression of NHE-like isoforms in the gills of a euryhaline fish (Fundulus heteroclitus). J. Exp. Zool. A 2010, 303, 464–475. [Google Scholar] [CrossRef]
- Zimmer, A.M.; Nawata, C.M.; Wood, C.M. Physiological and molecular analysis of the interactive effects of feeding and high environmental ammonia on branchial ammonia excretion and Na+ uptake in freshwater rainbow trout. J. Comp. Physiol. B 2010, 180, 1191–1204. [Google Scholar] [CrossRef]
- Zimmer, A.M.; Wilson, J.M.; Wright, P.A.; Hiroi, J.; Wood, C.M. Different mechanisms of Na+ uptake and ammonia excretion by the gill and yolk sac epithelium of early life stage rainbow trout. J. Exp. Biol. 2017, 220, 775–786. [Google Scholar] [CrossRef]
- Nawata, C.M.; Hung, C.C.Y.; Tsui, T.K.N.; Wilson, J.M.; Wright, P.A.; Wood, C.M. Ammonia excretion in rainbow trout (Oncorhynchus mykiss): Evidence for Rh glycoprotein and H+-ATPase involvement. Physiol. Genom. 2007, 31, 463–474. [Google Scholar] [CrossRef]
- Wright, P.A.; Wood, C.M. A new paradigm for ammonia excretion in aquatic animals: Role of Rhesus (Rh) glycoproteins. J. Exp. Biol. 2009, 212, 2303–2312. [Google Scholar] [CrossRef]
- Furukawa, F.; Watanabe, S.; Kimura, S.; Kaneko, T. Potassium excretion through ROMK potassium channel expressed in gill mitochondrion-rich cells of Mozambique tilapia. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2012, 302, 568–576. [Google Scholar] [CrossRef]
- Marshall, W.S.; Ossum, C.G.; Hoffmann, E.K. Hypotonic shock mediation by p38 MAPK, JNK, PKC, FAK, OSR1 and SPAK in osmosensing chloride secreting cells of killifish opercular epithelium. J. Exp. Biol. 2005, 208, 1063–1077. [Google Scholar] [CrossRef]
- Martos-Sitcha, J.A.; Campinho, M.A.; Mancera, J.M.; Martínez-Rodriguez, G.; Fuentes, J. Vasotocin and isotocin regulate aquaporin 1 function in the sea bream. J. Exp. Biol. 2015, 218, 684–693. [Google Scholar] [CrossRef]
- Marshall, W.S. Na+, Cl−, Ca2+, and Zn2+ transport by fish gills: Retrospective review and prospective synthesis. J. Exp. Zool. 2002, 293, 264–283. [Google Scholar] [CrossRef]
- Cutler, C.P.; Cramb, G. Two isoforms of the Na+/K+/2Cl− cotransporter are expressed in the European eel (Anguilla anguilla). Biochim. Biophys. Acta 2002, 1566, 92–103. [Google Scholar] [CrossRef]
- Baldisserotto, B.; Mimura, O.M. Ion transport across the isolated intestinal mucosa of Anguilla anguilla (Pisces). Comp. Biochem. Phys. A 1994, 108, 297–302. [Google Scholar] [CrossRef]
- Martos-Sitcha, J.A.; Wunderink, Y.S.; Gozdowska, M.; Kulczykowska, E.; Mancera, J.M.; Martínez-Rodríguez, G. Vasotocinergic and isotocinergic systems in the gilthead sea bream (Sparus aurata): An osmoregulatory story. Comp. Biochem. Physiol. Part A 2013, 166, 571–581. [Google Scholar] [CrossRef] [PubMed]
- Wood, C.M.; Robertson, L.M.; Johannsson, O.E.; Val, A.L. Mechanisms of Na+ uptake, ammonia excretion, and their potential linkage in native Rio Negro tetras (Paracheirodon axelrodi, Hemigrammus rhodostomus, and Moenkhausia diktyota). J. Comp. Physiol. B 2014, 184, 877–890. [Google Scholar] [CrossRef]
- Golombieski, J.I.; Koakoski, G.; Becker, A.J.; Almeida, A.P.G.; Toni, C.; Finamor, I.A.; Pavanato, M.A.; Almeida, T.M.; Baldisserotto, B. Nitrogenous and phosphorus excretions in juvenile silver catfish (Rhamdia quelen) exposed to different water hardness, humic acid, and pH levels. Fish Physiol. Biochem. 2013, 39, 837–849. [Google Scholar] [CrossRef]
- Kumai, Y.; Perry, S.F. Ammonia excretion via Rhcg1 facilitates Na+ uptake in larval zebrafish, Danio rerio, in acidic water. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2011, 301, 1517–1528. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).