Microbial and Planktonic Community Characteristics of Eriocheir sinensis Culture Ponds Experiencing Harmful Algal Blooms
Abstract
:1. Introduction
2. Material and Methods
2.1. Sample Collection
2.2. Water Quality Parameters
2.3. DNA Extraction and PCR Amplification
2.4. Illumina Novaseq 6000 Sequencing
2.5. Bioinformatics Analyses
2.5.1. Reads Filtering and Assembly
2.5.2. Raw Tag Filtering
2.5.3. Clustering and Chimera Removal
2.5.4. Taxonomy Annotation, Community Composition and Indicator Species Analysis
2.5.5. Alpha and Beta Diversity Analysis
2.6. Correlation Analysis between Species and Environmental Factors
2.7. Statistical Analysis
3. Results
3.1. Water Quality Parameters
3.2. General Analyses of High-Throughput Sequencing
3.2.1. 16S rDNA Sequencing
3.2.2. 18S rDNA Sequencing
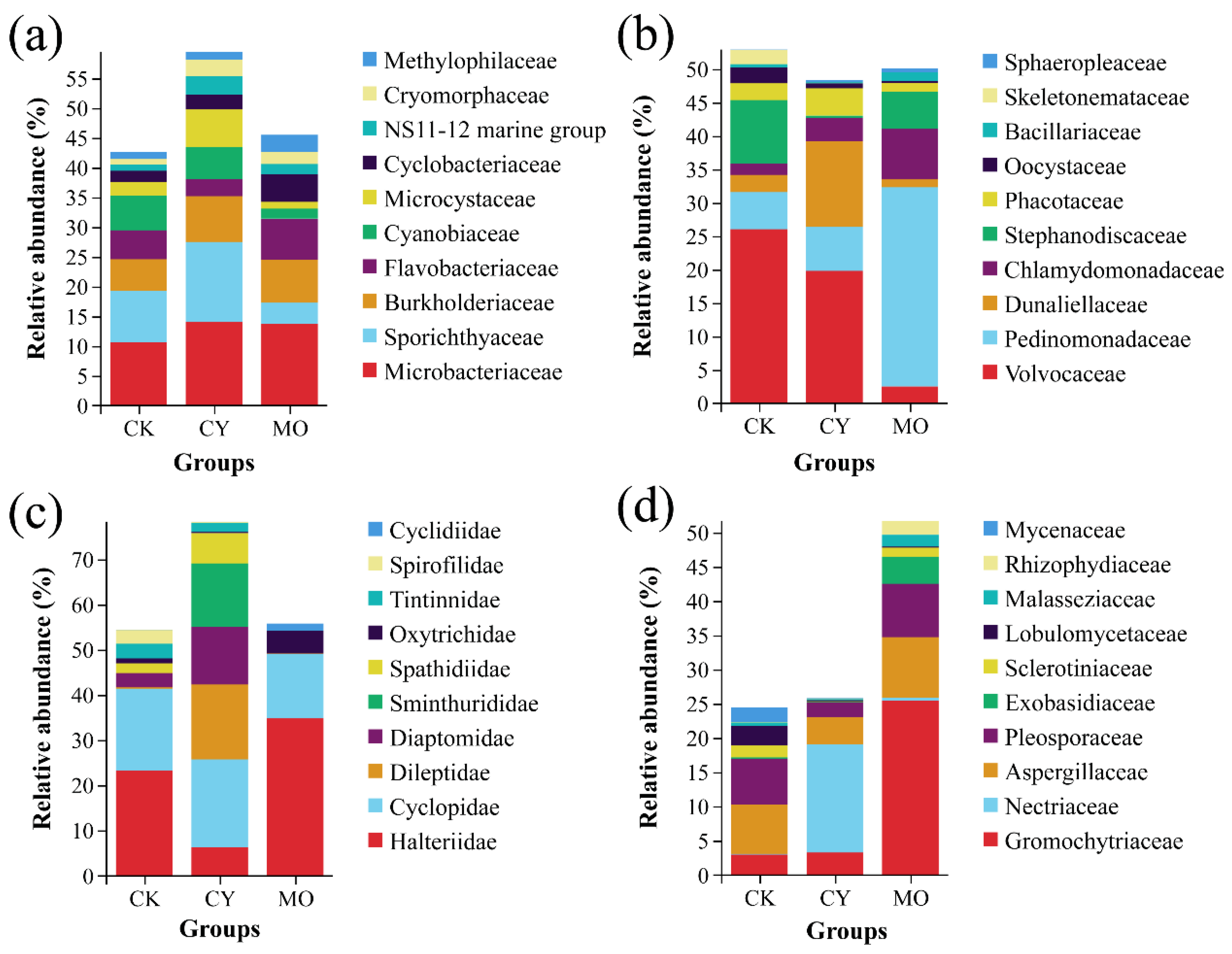
3.3. Community Composition
3.4. Indicator Species
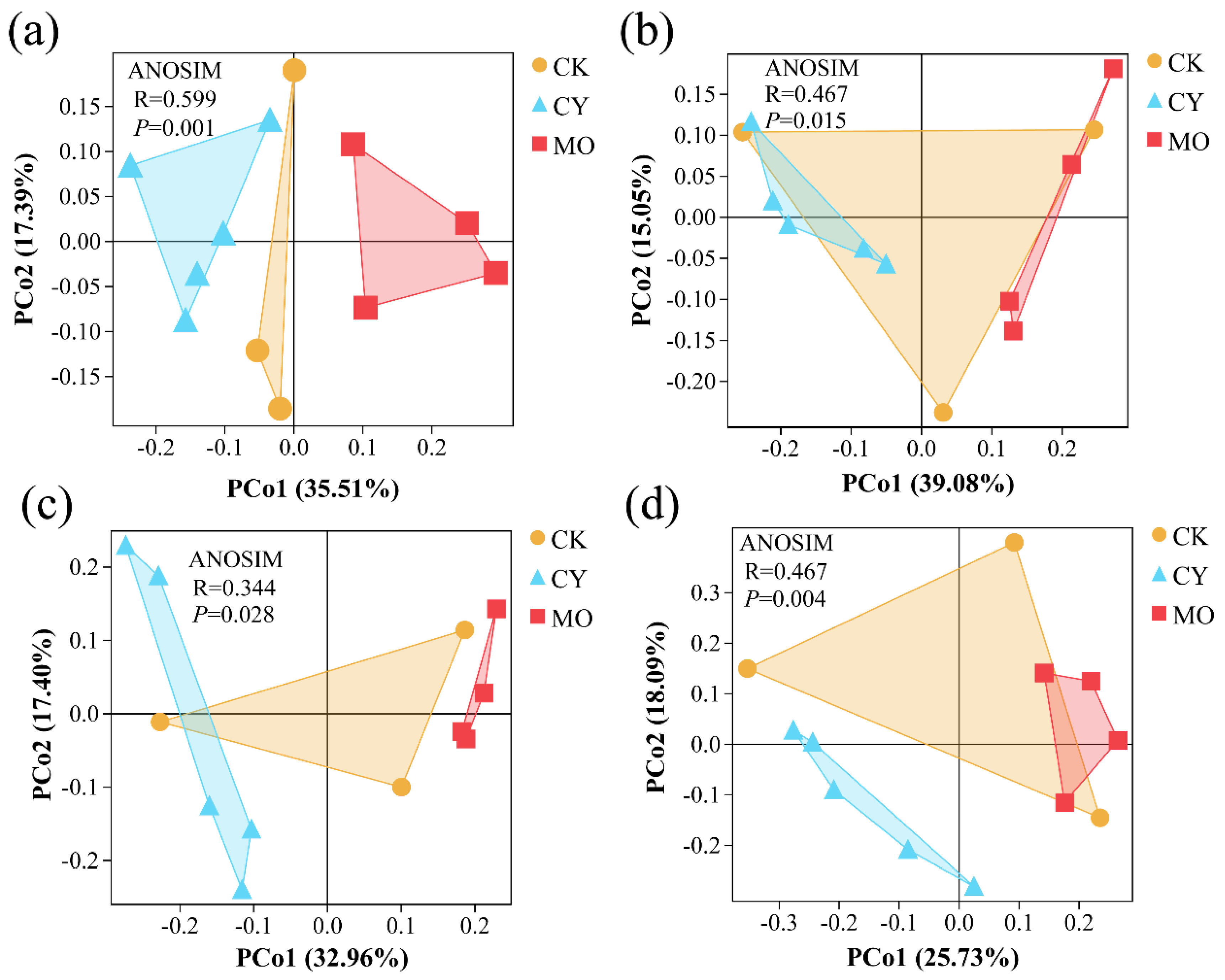
3.5. Alpha and Beta Diversity
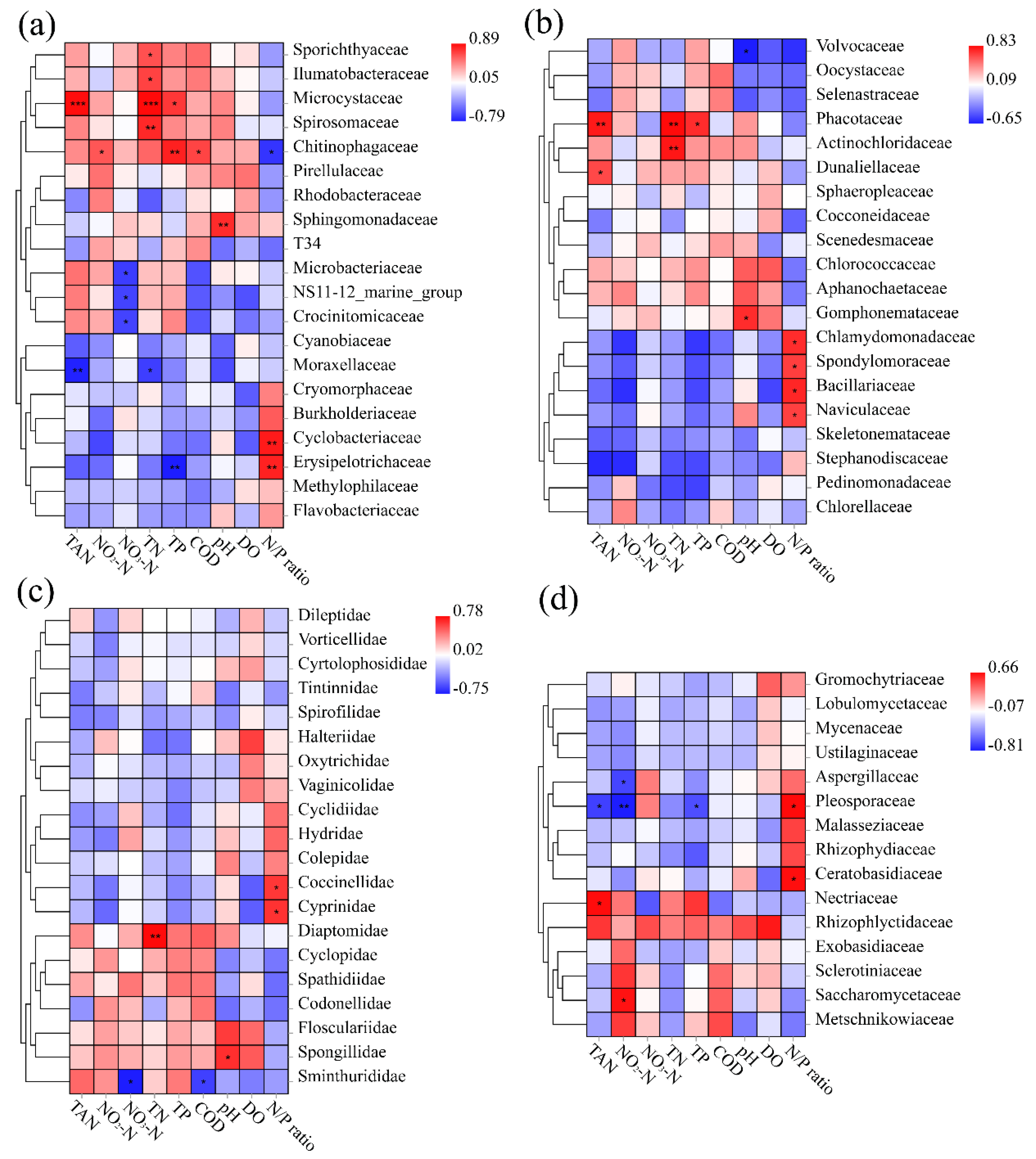
3.6. Correlation Analysis between Species and Environmental Factors
4. Discussion
4.1. Limitations of Water Quality Indicators
4.2. Relationship between NPR and Microalgae
4.3. Biological Indicators for Algal Blooms
4.4. Strategies for Risk Prevention and Control in the Culture of E. sinensis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fishery Administration Bureau of the Ministry of Agriculture and Villages; National Aquatic Products Technology Extension Station; China Society of Fisheries. 2021 China Fishery Statistical Yearbook; Chinese Agricultural Press: Beijing, China, 2021; Volume 34. [Google Scholar]
- Gao, J.; Tai, X.; Shao, N.; Sun, Y.; Nie, Z.; Wang, Y.; Li, Q.; Xu, P.; Xu, G. Effects of effective microorganisms on the growth performance, nutritional composition and flavour quality of the pond-cultured Eriocheir sinensis. Aquac. Res. 2021, 52, 871–880. [Google Scholar] [CrossRef]
- Dai, H.; Sun, Y.; Ren, N.; Lu, X. Investigation of Chinese hairy crab industry and analysis of development strategies. Jiangsu Agric. Sci. 2021, 49, 248–252. [Google Scholar] [CrossRef]
- Ma, X.; Ge, J.; Wang, Z. Study on changes and regulation of ammonia nitrogen and nitrite in Eriocheir sinensis culture. J. Aquac. 2020, 41, 43–45. [Google Scholar]
- Chi, C.; Yu, X.-W.; Zhang, C.-Y.; Liu, J.-D.; Ye, M.-W.; Zhang, D.-D.; Liu, W.-B. Acute exposure to microcystin-LR induces hepatopancreas toxicity in the Chinese mitten crab (Eriocheir sinensis). Arch. Toxicol. 2021, 95, 2551–2570. [Google Scholar] [CrossRef]
- Hasan, M.R.; Chakrabarti, R. Use of Algae and Aquatic Macrophytes as Feed in Small-Scale Aquaculture: A Review. In FAO Fisheries and Aquaculture Technical Paper No. 531; FAO: Rome, Italy, 2009; 123p. [Google Scholar]
- Cannell, R.J.; Farmer, P.; Walker, J.M. Purification and characterization of pentagalloylglucose, and alpha-glucosidase inhibitor/antibiotic from the freshwater green alga Spirogyra varians. Biochem. J. 1988, 255, 937–941. [Google Scholar] [CrossRef]
- Weber, J.; Schagerl, M. Strategies of Spirogyra against epiphytes. Algol. Stud. 2007, 123, 57–72. [Google Scholar] [CrossRef]
- Mao, G.; Tang, Y. Causes, hazards and prevention of pond moss in crab pond. Sci. Fish Farming 2016, 12, 61–62. [Google Scholar] [CrossRef]
- Huisman, J.; Codd, G.A.; Paerl, H.W.; Ibelings, B.W.; Verspagen, J.M.H.; Visser, P.M. Cyanobacterial blooms. Nat. Rev. Microbiol. 2018, 16, 471–483. [Google Scholar] [CrossRef]
- Zhang, Q.; Zhang, Z.; Lu, T.; Peijnenburg, W.J.G.M.; Gillings, M.; Yang, X.; Chen, J.; Penuelas, J.; Zhu, Y.-G.; Zhou, N.-Y.; et al. Cyanobacterial blooms contribute to the diversity of antibiotic-resistance genes in aquatic ecosystems. Commun. Biol. 2020, 3, 737. [Google Scholar] [CrossRef]
- Havens, K.E. Cyanobacteria blooms: Effects on aquatic ecosystems. Adv. Exp. Med. Biol. 2008, 619, 733–747. [Google Scholar] [CrossRef]
- Wu, J.X.; Huang, H.; Yang, L.; Zhang, X.F.; Zhang, S.S.; Liu, H.H.; Wang, Y.Q.; Yuan, L.; Cheng, X.M.; Zhuang, D.G.; et al. Gastrointestinal toxicity induced by microcystins. World J. Clin. Cases 2018, 6, 344–354. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Pan, L.; Zhang, Y. Formation and countermeasures of harmful filamentous algae in water. South China Fish. Sci. 2011, 7, 77–81. [Google Scholar] [CrossRef]
- Martínez Cruz, P.; Ibáñez, A.L.; Monroy Hermosillo, O.A.; Ramírez Saad, H.C. Use of probiotics in aquaculture. ISRN Microbiol. 2012, 2012, 916845. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, A.; Ran, C.; Wang, Y.; Zhang, Z.; Ding, Q.; Yang, Y.; Olsen, R.E.; Ringø, E.; Bindelle, J.; Zhou, Z. Use of probiotics in aquaculture of China-a review of the past decade. Fish Shellfish. Immunol. 2019, 86, 734–755. [Google Scholar] [CrossRef]
- Kuebutornye, F.K.A.; Abarike, E.D.; Lu, Y. A review on the application of Bacillus as probiotics in aquaculture. Fish Shellfish Immunol. 2019, 87, 820–828. [Google Scholar] [CrossRef]
- Ahmad, M.T.; Shariff, M.; Yusoff, F.M.; Goh, Y.M.; Banerjee, S. Applications of microalga Chlorella vulgaris in aquaculture. Rev. Aquac. 2020, 12, 328–346. [Google Scholar] [CrossRef]
- Sathasivam, R.; Radhakrishnan, R.; Hashem, A.; Abd_Allah, E.F. Microalgae metabolites: A rich source for food and medicine. Saudi J. Biol. Sci. 2019, 26, 709–722. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Subramanian, S.; Faith, J.J.; Gevers, D.; Gordon, J.I.; Knight, R.; Mills, D.A.; Caporaso, J.G. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods 2013, 10, 57–59. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pruesse, E.; Quast, C.; Knittel, K.; Fuchs, B.M.; Ludwig, W.; Peplies, J.; Glöckner, F.O. SILVA: A comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007, 35, 7188–7196. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately maximum-likelihood trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef] [PubMed]
- Lozupone, C.; Knight, R. UniFrac: A new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 2005, 71, 8228–8235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Revelle, W. psych: Procedures for Psychological, Psychometric, and Personality Research; Northwestern University: Evanston, IL, USA, 2021. [Google Scholar]
- Hu, Z.; Li, R.; Xia, X.; Yu, C.; Fan, X.; Zhao, Y. A method overview in smart aquaculture. Environ. Monit. Assess. 2020, 192, 493. [Google Scholar] [CrossRef]
- Cai, C.; Gu, X.; Huang, H.; Dai, X.; Ye, Y.; Shi, C. Water quality, nutrient budget, and pollutant loads in Chinese mitten crab (Eriocheir sinensis) farms around East Taihu Lake. Chin. J. Oceanol. Limnol. 2012, 30, 29–36. [Google Scholar] [CrossRef]
- Güsewell, S.; Gessner, M.O. N: P ratios influence litter decomposition and colonization by fungi and bacteria in microcosms. Funct. Ecol. 2009, 23, 211–219. [Google Scholar] [CrossRef]
- Rhee, G.-Y.; Gotham, I.J. Optimum N:P ratios and coexistence of planktonic algae. J. Phycol. 1980, 16, 486–489. [Google Scholar] [CrossRef]
- Rasdi, N.W.; Qin, J.G. Effect of N:P ratio on growth and chemical composition of Nannochloropsis oculata and Tisochrysis lutea. J. Appl. Phycol. 2015, 27, 2221–2230. [Google Scholar] [CrossRef]
- Palus, J. Effects of N:P Ratio on the Occurrence of Harmful Algal Blooms. BSc Thesis, The Ohio State University, Columbus, OH, USA, 2015. [Google Scholar]
- Kim, H.-S.; Hwang, S.-J.; Shin, J.-K.; An, K.-G.; Yoon, C.G. Effects of limiting nutrients and N:P ratios on the phytoplankton growth in a shallow hypertrophic reservoir. Hydrobiologia 2007, 581, 255–267. [Google Scholar] [CrossRef]
- Falkowski, P.G.; Davis, C.S. Natural proportions. Nature 2004, 431, 131. [Google Scholar] [CrossRef]
- Bergström, A.-K. The use of TN:TP and DIN:TP ratios as indicators for phytoplankton nutrient limitation in oligotrophic lakes affected by N deposition. Aquat. Sci. 2010, 72, 277–281. [Google Scholar] [CrossRef]
- Paerl, H.W.; Xu, H.; Hall, N.S.; Zhu, G.; Qin, B.; Wu, Y.; Rossignol, K.L.; Dong, L.; McCarthy, M.J.; Joyner, A.R. Controlling cyanobacterial blooms in hypertrophic Lake Taihu, China: Will nitrogen reductions cause replacement of non-N2 fixing by N2 fixing taxa? PLoS ONE 2014, 9, e113123. [Google Scholar] [CrossRef] [Green Version]
- Paerl, H.W.; Fulton, R.S., 3rd; Moisander, P.H.; Dyble, J. Harmful freshwater algal blooms, with an emphasis on cyanobacteria. Sci. J. 2001, 1, 76–113. [Google Scholar] [CrossRef]
- Xie, L.; Xie, P.; Li, S.; Tang, H.; Liu, H. The low TN:TP ratio, a cause or a result of Microcystis blooms? Water Res. 2003, 37, 2073–2080. [Google Scholar] [CrossRef]
- Liu, Y.; Li, L.; Jia, R. The Optimum Resource Ratio (N:P) for the Growth of Microcystis Aeruginosa with Abundant Nutrients. Procedia Environ. Sci. 2011, 10, 2134–2140. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.; Hu, G. Effect of nitrogen to phosphorus ratios on cell proliferation in marine micro algae. Chin. J. Oceanol. Limnol. 2011, 29, 739–745. [Google Scholar] [CrossRef]
- Podevin, M.; De Francisci, D.; Holdt, S.L.; Angelidaki, I. Effect of nitrogen source and acclimatization on specific growth rates of microalgae determined by a high-throughput in vivo microplate autofluorescence method. J. Appl. Phycol. 2015, 27, 1415–1423. [Google Scholar] [CrossRef] [Green Version]
- Mohapatra, R.K.; Parhi, P.K.; Thatoi, H.; Panda, C.R. Bioreduction of hexavalent chromium by Exiguobacterium indicum strain MW1 isolated from marine water of Paradip Port, Odisha, India. Chem. Ecol. 2017, 33, 114–130. [Google Scholar] [CrossRef]
- Daims, H.; Wagner, M. Nitrospira. Trends Microbiol. 2018, 26, 462–463. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Ma, L.-J. Chapter Five—Deciphering Pathogenicity of Fusarium oxysporum from a Phylogenomics Perspective. In Advances in Genetics; Townsend, J.P., Wang, Z., Eds.; Academic Press: Cambridge, MA, USA, 2017; Volume 100, pp. 179–209. [Google Scholar]
- Silva, V.; Caniça, M.; Capelo, J.L.; Igrejas, G.; Poeta, P. Diversity and genetic lineages of environmental staphylococci: A surface water overview. FEMS Microbiol. Ecol. 2020, 96, fiaa191. [Google Scholar] [CrossRef]
- Karpov, S.A.; Kobseva, A.A.; Mamkaeva, M.A.; Mamkaeva, K.A.; Mikhailov, K.V.; Mirzaeva, G.S.; Aleoshin, V.V. Gromochytrium mamkaevae gen. & sp. nov. and two new orders: Gromochytriales and Mesochytriales (Chytridiomycetes). Persoonia 2014, 32, 115–126. [Google Scholar] [CrossRef] [Green Version]
- Letcher, P.M.; Vélez, C.G.; Barrantes, M.E.; Powell, M.J.; Churchill, P.F.; Wakefield, W.S. Ultrastructural and molecular analyses of Rhizophydiales (Chytridiomycota) isolates from North America and Argentina. Mycol. Res. 2008, 112, 759–782. [Google Scholar] [CrossRef]
- Chaudhari, V.R.; Vyawahare, A.; Bhattacharjee, S.K.; Rao, B.J. Enhanced excision repair and lack of PSII activity contribute to higher UV survival of Chlamydomonas reinhardtii cells in dark. Plant Physiol. Biochem. 2015, 88, 60–69. [Google Scholar] [CrossRef]
- Dolhi, J.M.; Maxwell, D.P.; Morgan-Kiss, R.M. Review: The Antarctic Chlamydomonas raudensis: An emerging model for cold adaptation of photosynthesis. Extremophiles 2013, 17, 711–722. [Google Scholar] [CrossRef]
- Stahl-Rommel, S.; Kalra, I.; D’Silva, S.; Hahn, M.M.; Popson, D.; Cvetkovska, M.; Morgan-Kiss, R.M. Cyclic electron flow (CEF) and ascorbate pathway activity provide constitutive photoprotection for the photopsychrophile, Chlamydomonas sp. UWO 241 (renamed Chlamydomonas priscuii). Photosynth. Res. 2022, 151, 235–250. [Google Scholar] [CrossRef]
- Allahverdiyeva, Y.; Suorsa, M.; Tikkanen, M.; Aro, E.M. Photoprotection of photosystems in fluctuating light intensities. J. Exp. Bot. 2015, 66, 2427–2436. [Google Scholar] [CrossRef]
- Xu, Y.; Li, A.J.; Qin, J.; Li, Q.; Ho, J.G.; Li, H. Seasonal patterns of water quality and phytoplankton dynamics in surface waters in Guangzhou and Foshan, China. Sci. Total Environ. 2017, 590–591, 361–369. [Google Scholar] [CrossRef] [PubMed]
- Abirhire, O.; North, R.L.; Hunter, K.; Vandergucht, D.M.; Sereda, J.; Hudson, J.J. Environmental factors influencing phytoplankton communities in Lake Diefenbaker, Saskatchewan, Canada. J. Great Lakes Res. 2015, 41, 118–128. [Google Scholar] [CrossRef]
- Catherine, A.; Selma, M.; Mouillot, D.; Troussellier, M.; Bernard, C. Patterns and multi-scale drivers of phytoplankton species richness in temperate peri-urban lakes. Sci. Total Environ. 2016, 559, 74–83. [Google Scholar] [CrossRef] [PubMed]
- Romano, N. Chapter 5—Probiotics, prebiotics, biofloc systems, and other biocontrol regimens in fish and shellfish aquaculture. In Aquaculture Pharmacology; Kibenge, F.S.B., Baldisserotto, B., Chong, R.S.-M., Eds.; Academic Press: Cambridge, MA, USA, 2021; pp. 219–242. [Google Scholar]
- Sun, R.; Sun, P.; Zhang, J.; Esquivel-Elizondo, S.; Wu, Y. Microorganisms-based methods for harmful algal blooms control: A review. Bioresour. Technol. 2018, 248, 12–20. [Google Scholar] [CrossRef] [PubMed]
- Gioacchini, G.; Ciani, E.; Pessina, A.; Cecchini, C.; Silvi, S.; Rodiles, A.; Merrifield, D.L.; Olivotto, I.; Carnevali, O. Effects of Lactogen 13, a new probiotic preparation, on gut microbiota and endocrine signals controlling growth and appetite of Oreochromis niloticus Juveniles. Microb. Ecol. 2018, 76, 1063–1074. [Google Scholar] [CrossRef] [Green Version]
- Wang, R.; Guo, Z.; Tang, Y.; Kuang, J.; Duan, Y.; Lin, H.; Jiang, S.; Shu, H.; Huang, J. Effects on development and microbial community of shrimp Litopenaeus vannamei larvae with probiotics treatment. AMB Express 2020, 10, 109. [Google Scholar] [CrossRef]
- Hesni, M.A.; Hedayati, A.; Qadermarzi, A.; Pouladi, M.; Zangiabadi, S.; Naqshbandi, N. Using Chlorella vulgaris and iron oxide nanoparticles in a designed bioreactor for aquaculture effluents purification. Aquac. Eng. 2020, 90, 102069. [Google Scholar] [CrossRef]
- Tejido-Nuñez, Y.; Aymerich, E.; Sancho, L.; Refardt, D. Treatment of aquaculture effluent with Chlorella vulgaris and Tetradesmus obliquus: The effect of pretreatment on microalgae growth and nutrient removal efficiency. Ecol. Eng. 2019, 136, 1–9. [Google Scholar] [CrossRef]
- Daneshvar, E.; Antikainen, L.; Koutra, E.; Kornaros, M.; Bhatnagar, A. Investigation on the feasibility of Chlorella vulgaris cultivation in a mixture of pulp and aquaculture effluents: Treatment of wastewater and lipid extraction. Bioresour. Technol. 2018, 255, 104–110. [Google Scholar] [CrossRef]
- Pakravan, S.; Akbarzadeh, A.; Sajjadi, M.M.; Hajimoradloo, A.; Noori, F. Chlorella vulgaris meal improved growth performance, digestive enzyme activities, fatty acid composition and tolerance of hypoxia and ammonia stress in juvenile Pacific white shrimp Litopenaeus vannamei. Aquac. Nutr. 2018, 24, 594–604. [Google Scholar] [CrossRef]
- Hodač, L.; Hallmann, C.; Spitzer, K.; Elster, J.; Faßhauer, F.; Brinkmann, N.; Lepka, D.; Diwan, V.; Friedl, T. Widespread green algae Chlorella and Stichococcus exhibit polar-temperate and tropical-temperate biogeography. FEMS Microbiol. Ecol. 2016, 92, fiw122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Y.; Shen, Y.; Zhang, S.; Li, Y.; Sun, Z.; Feng, M.; Li, R.; Zhang, J.; Tian, X.; Zhang, W. Characteristics of phytoplankton community structure and indication to water quality in the lake in agricultural areas. Front. Environ. Sci. 2022, 10, 833409. [Google Scholar] [CrossRef]
- Zhang, Y.; Gao, W.; Li, Y.; Jiang, Y.; Chen, X.; Yao, Y.; Messyasz, B.; Yin, K.; He, W.; Chen, Y. Characteristics of the phytoplankton community structure and water quality evaluation in autumn in the Huaihe river (China). Int. J. Environ. Res. Public Health 2021, 18, 12092. [Google Scholar] [CrossRef]
- Fuentes, J.L.; Garbayo, I.; Cuaresma, M.; Montero, Z.; González-Del-Valle, M.; Vílchez, C. Impact of microalgae-bacteria interactions on the production of algal biomass and associated compounds. Mar. Drugs 2016, 14, 100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hasan, K.N.; Banerjee, G. Recent studies on probiotics as beneficial mediator in aquaculture: A review. J. Basic Appl. Zool. 2020, 81, 53. [Google Scholar] [CrossRef]



| Group | CK | CY | MO |
|---|---|---|---|
| Total ammonia nitrogen (TAN) | 0.10 ± 0.13 ab | 0.37 ± 0.04 a | 0.09 ± 0.04 b |
| Nitrite nitrogen (NO2-N) | 0.002 ± 0.001 | 0.002 ± 0.001 | 0.002 ± 0.001 |
| Nitrate nitrogen (NO3-N) | 0.22 ± 0.02 | 0.21 ± 0.06 | 0.21 ± 0.02 |
| Total nitrogen (TN) | 0.88 ± 0.41 b | 1.82 ± 0.58 a | 0.77 ± 0.20 b |
| Total phosphorus (TP) | 0.16 ± 0.08 ab | 0.22 ± 0.01 a | 0.06 ± 0.01 b |
| Chemical oxygen demand (COD) | 9.66 ± 0.75 | 9.32 ± 1.26 | 9.04 ± 0.29 |
| pH | 9.37 ± 1.08 | 9.50 ± 0.66 | 9.40 ± 0.29 |
| Dissolved oxygen (DO) | 12.29 ± 3.70 | 10.19 ± 3.47 | 10.70 ± 4.10 |
| N/P ratio (NPR) | 6.41 ± 2.86 b | 8.31 ± 2.54 ab | 14.32 ± 4.62 a |
| Sequencing Type | Sample ID | Domain | Phylum | Class | Order | Family | Genus | Species |
|---|---|---|---|---|---|---|---|---|
| (a) 16S rDNA | CK1 | 67,241 | 66,492 (98.89%) | 66,271 (98.56%) | 61,369 (91.27%) | 54,342 (80.82%) | 39,314 (58.47%) | 7641 (11.36%) |
| CK2 | 115,727 | 115,348 (99.67%) | 115,062 (99.43%) | 104,470 (90.27%) | 89,308 (77.17%) | 71,261 (61.58%) | 7902 (6.83%) | |
| CK3 | 94,083 | 93,649 (99.54%) | 93,369 (99.24%) | 88,040 (93.58%) | 71,858 (76.38%) | 44,265 (47.05%) | 5299 (5.63%) | |
| CKmean | 92,350.33 | 91,829.67 (99.44%) | 91,567.33 (99.15%) | 84,626.33 (91.64%) | 71,836.00 (77.79%) | 51,613.33 (55.89%) | 6947.33 (7.52%) | |
| CY1 | 96,216 | 95,995 (99.77%) | 94,320 (98.03%) | 88,079 (91.54%) | 82,684 (85.94%) | 53,475 (55.58%) | 9955 (10.35%) | |
| CY2 | 64,237 | 64,025 (99.67%) | 63,920 (99.51%) | 58,677 (91.34%) | 48,519 (75.53%) | 37,198 (57.91%) | 7498 (11.67%) | |
| CY3 | 70,039 | 69,689 (99.50%) | 69,554 (99.31%) | 66,061 (94.32%) | 63,490 (90.65%) | 47,345 (67.60%) | 10,991 (15.69%) | |
| CY4 | 69,558 | 69,290 (99.61%) | 69,258 (99.57%) | 66,936 (96.23%) | 60,453 (86.91%) | 43,103 (61.97%) | 1585 (2.28%) | |
| CY5 | 58,450 | 58,015 (99.26%) | 57,924 (99.10%) | 55,798 (95.46%) | 50,875 (87.04%) | 36,710 (62.81%) | 2370 (4.05%) | |
| CYmean | 71,700.00 | 71,402.80 (99.59%) | 70,995.20 (99.02%) | 67,110.20 (93.60%) | 61,204.20 (85.36%) | 43,566.20 (60.76%) | 6479.80 (9.04%) | |
| MO1 | 104,543 | 104,253 (99.72%) | 103,999 (99.48%) | 95,491 (91.34%) | 76,683 (73.35%) | 50,391 (48.20%) | 3000 (2.87%) | |
| MO2 | 79,867 | 79,101 (99.04%) | 78,843 (98.72%) | 72,527 (90.81%) | 58,087 (72.73%) | 39,720 (49.73%) | 7101 (8.89%) | |
| MO3 | 101,091 | 100,759 (99.67%) | 100,609 (99.52%) | 88,947 (87.99%) | 72,576 (71.79%) | 53,235 (52.66%) | 12,908 (12.77%) | |
| MO4 | 104,645 | 104,255 (99.63%) | 103,888 (99.28%) | 94,707 (90.50%) | 75,193 (71.86%) | 51,555 (49.27%) | 4865 (4.65%) | |
| MOmean | 97,536.50 | 97,092.00 (99.54%) | 96,834.75 (99.28%) | 87,918.00 (90.14%) | 70,634.75 (72.42%) | 48,725.25 (49.96%) | 6968.50 (7.14%) | |
| (b) 18S rDNA | CK1 | 81,121 | 75,557 (93.14%) | 67,328 (83.00%) | 48,553 (59.85%) | 39,688 (48.92%) | 34,071 (42.00%) | 22,643 (27.91%) |
| CK2 | 98,335 | 97,345 (98.99%) | 93,351 (94.93%) | 79,612 (80.96%) | 62,979 (64.05%) | 58,996 (59.99%) | 50,642 (51.50%) | |
| CK3 | 105,828 | 104,404 (98.65%) | 100,742 (95.19%) | 87,007 (82.22%) | 77,651 (73.37%) | 66,507 (62.84%) | 57,388 (54.23%) | |
| CKmean | 95,094.67 | 92,435.33 (97.20%) | 87,140.33 (91.64%) | 71,724.00 (75.42%) | 60,106.00 (63.21%) | 53,191.33 (55.94%) | 43,557.67 (45.80%) | |
| CY1 | 108,732 | 107,926 (99.26%) | 105,512 (97.04%) | 89,218 (82.05%) | 84,700 (77.90%) | 82,683 (76.04%) | 77,331 (71.12%) | |
| CY2 | 60,595 | 59,973 (98.97%) | 58,800 (97.04%) | 54,832 (90.49%) | 35,878 (59.21%) | 32,964 (54.40%) | 6093 (10.06%) | |
| CY3 | 108,869 | 105,713 (97.10%) | 97,828 (89.86%) | 89,936 (82.61%) | 87,373 (80.26%) | 83,170 (76.39%) | 78,928 (72.50%) | |
| CY4 | 112,691 | 111,388 (98.84%) | 99,026 (87.87%) | 83,779 (74.34%) | 55,383 (49.15%) | 50,548 (44.86%) | 25,269 (22.42%) | |
| CY5 | 97,828 | 96,991 (99.14%) | 61,628 (63.00%) | 58,405 (59.70%) | 51,231 (52.37%) | 47,761 (48.82%) | 42,149 (43.08%) | |
| CYmean | 97,743.00 | 96,398.20 (98.62%) | 84,558.80 (86.51%) | 75,234.00 (76.97%) | 62,913.00 (64.37%) | 59,425.20 (60.80%) | 45,954.00 (47.02%) | |
| MO1 | 96,828 | 95,199 (98.32%) | 78,525 (81.10%) | 69,429 (71.70%) | 62,588 (64.64%) | 43,185 (44.60%) | 20,838 (21.52%) | |
| MO2 | 99,288 | 97,175 (97.87%) | 91,076 (91.73%) | 71,515 (72.03%) | 69,638 (70.14%) | 64,130 (64.59%) | 14,049 (14.15%) | |
| MO3 | 108,951 | 107,673 (98.83%) | 105,916 (97.21%) | 84,000 (77.10%) | 72,832 (66.85%) | 66,802 (61.31%) | 56,694 (52.04%) | |
| MO4 | 87,864 | 84,904 (96.63%) | 81,274 (92.50%) | 71,157 (80.99%) | 34,887 (39.71%) | 24,659 (28.06%) | 16,340 (18.60%) | |
| MOmean | 98,232.75 | 96,237.75 (97.97%) | 89,197.75 (90.80%) | 74,025.25 (75.36%) | 59,986.25 (61.07%) | 49,694.00 (50.59%) | 26,980.25 (27.47%) |
| Index | CK | CY | MO | |
|---|---|---|---|---|
| (a) bacteria | Species richness | 1514.00 ± 663.52 | 1269.00 ± 107.04 | 1439.00 ± 493.03 |
| Shannon | 7.35 ± 0.16 | 6.68 ± 0.40 | 7.16 ± 0.53 | |
| Simpson (×10−2) | 97.79 ± 0.11 | 97.07 ± 1.08 | 97.56 ± 0.84 | |
| Chao1 | 1609.38 ± 626.19 | 1375.82 ± 112.65 | 1535.81 ± 459.37 | |
| Ace | 1635.49 ± 625.40 | 1414.71 ± 119.70 | 1539.02 ± 469.03 | |
| Good’s coverage (×10−2) | 99.79 ± 0.11 | 99.67 ± 0.10 | 99.84 ± 0.05 | |
| Pielou’s evenness (×10−2) | 70.33 ± 2.82 | 64.85 ± 3.73 | 68.60 ± 3.08 | |
| PD-whole tree | 212.72 ± 98.17 | 184.27 ± 15.49 | 196.83 ± 71.28 | |
| (b) phytoplankton | Species richness | 117.67 ± 10.97b | 146.80 ± 17.54a | 117.75 ± 19.75b |
| Shannon | 4.38 ± 0.70 | 4.10 ± 1.15 | 3.96 ± 0.97 | |
| Simpson (×10−2) | 89.18 ± 7.56 | 84.63 ± 12.51 | 80.68 ± 15.07 | |
| Chao1 | 147.31 ± 6.60 | 169.45 ± 15.95 | 137.03 ± 22.86 | |
| Ace | 152.99 ± 8.15 | 168.17 ± 17.39 | 138.09 ± 21.90 | |
| Good’s coverage (×10−2) | 99.67 ± 0.08 | 99.66 ± 0.08 | 99.75 ± 0.06 | |
| Pielou’s evenness (×10−2) | 63.91 ± 11.33 | 56.74 ± 15.04 | 57.49 ± 12.62 | |
| PD-whole tree | 9.85 ± 1.19 | 10.66 ± 0.89 | 10.24 ± 1.10 | |
| (c) zooplankton | Species richness | 56.33 ± 5.13 | 62.20 ± 12.58 | 55.00 ± 6.00 |
| Shannon | 3.52 ± 0.56 | 2.87 ± 0.77 | 2.81 ± 0.27 | |
| Simpson (×10−2) | 82.95 ± 7.51 | 72.85 ± 13.72 | 76.23 ± 5.55 | |
| Chao1 | 61.72 ± 6.40 | 64.75 ± 13.03 | 63.60 ± 7.22 | |
| Ace | 65.55 ± 8.09 | 66.85 ± 11.53 | 61.05 ± 5.88 | |
| Good’s coverage (×10−2) | 99.92 ± 0.02 | 99.93 ± 0.01 | 99.92 ± 0.01 | |
| Pielou’s evenness (×10−2) | 60.42 ± 8.15 | 48.04 ± 11.54 | 48.87 ± 6.04 | |
| PD-whole tree | 6.92 ± 0.67 | 7.68 ± 1.50 | 7.22 ± 0.62 | |
| (d) fungi | Species richness | 43.67 ± 13.80 | 50.60 ± 7.96 | 46.00 ± 9.59 |
| Shannon | 4.52 ± 0.70 | 3.71 ± 0.97 | 3.91 ± 0.88 | |
| Simpson (×10−2) | 93.89 ± 3.03 | 82.97 ± 11.99 | 84.57 ± 14.46 | |
| Chao1 | 51.53 ± 12.61 | 61.51 ± 10.69 | 53.83 ± 12.81 | |
| Ace | 54.04 ± 7.06 | 59.89 ± 7.52 | 55.33 ± 14.75 | |
| Good’s coverage (×10−2) | 99.02 ± 0.07 | 98.61 ± 0.25 | 98.84 ± 0.38 | |
| Pielou’s evenness (×10−2) | 83.55 ± 5.50 | 65.34 ± 14.96 | 70.66 ± 13.22 | |
| PD-whole tree | 5.42 ± 1.26 | 5.95 ± 0.66 | 5.61 ± 1.11 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, J.; Shen, L.; Nie, Z.; Zhu, H.; Cao, L.; Du, J.; Dai, F.; Xu, G. Microbial and Planktonic Community Characteristics of Eriocheir sinensis Culture Ponds Experiencing Harmful Algal Blooms. Fishes 2022, 7, 180. https://doi.org/10.3390/fishes7040180
Gao J, Shen L, Nie Z, Zhu H, Cao L, Du J, Dai F, Xu G. Microbial and Planktonic Community Characteristics of Eriocheir sinensis Culture Ponds Experiencing Harmful Algal Blooms. Fishes. 2022; 7(4):180. https://doi.org/10.3390/fishes7040180
Chicago/Turabian StyleGao, Jiancao, Lei Shen, Zhijuan Nie, Haojun Zhu, Liping Cao, Jinliang Du, Fei Dai, and Gangchun Xu. 2022. "Microbial and Planktonic Community Characteristics of Eriocheir sinensis Culture Ponds Experiencing Harmful Algal Blooms" Fishes 7, no. 4: 180. https://doi.org/10.3390/fishes7040180
APA StyleGao, J., Shen, L., Nie, Z., Zhu, H., Cao, L., Du, J., Dai, F., & Xu, G. (2022). Microbial and Planktonic Community Characteristics of Eriocheir sinensis Culture Ponds Experiencing Harmful Algal Blooms. Fishes, 7(4), 180. https://doi.org/10.3390/fishes7040180






