Abstract
The environmental conditions and isolation in the Antarctic have driven the evolution of a unique biodiversity at a macro to microorganism scale. Here, we investigated the possible adaptation of the teleost Notothenia coriiceps immune system to the cold environment and unique microbial community of the Southern Ocean. The fish immune system was stimulated through an intraperitoneal injection of lipopolysaccharide (LPS 0111:B4 from E. coli) and the tissue transcriptomic response and plasma biochemistry were analyzed 7 days later and compared to a sham injected control. Gene transcription in the head-kidney, intestine and skin was significantly modified by LPS, although tissues showed different responsiveness, with the duodenum most modified and the skin the least modified. The most modified processes in head-kidney, duodenum and skin were related to cell metabolism (up-regulated) and the immune system (comprising 30% of differentially expressed genes). The immune processes identified were mostly down-regulated, particularly interleukins and pattern recognition receptors (PRRs), nucleotide-binding oligomerization domain-like receptors and mannose receptors, unlike the toll-like receptors response commonly described in other teleost fish. The modified transcriptional response was not mirrored by a modified systemic response, as the circulating levels of enzymes of innate immunity, lysozyme and antiproteases, were not significantly different from the untreated and sham control fish. In conclusion, while the N. coriiceps immune system shares many features with other teleosts there are also some specificities. Further studies should better characterize the PRRs and their role in Antarctic teleosts, as well as the importance of the LPS source and its consequences for immune activation in teleosts.
Keywords:
Antarctic fish; head-kidney; innate immunity; duodenum; lipopolysaccharide; skin; transcriptomics 1. Introduction
The dominant teleost fish fauna in the Southern Ocean belong to the perciform suborder Notothenioidei, which evolved from a benthic ancestor through adaptive radiation modulated by a largely stable cold thermal environment [1,2,3]. The nototheniids have developed unique physiological adaptations, including antifreeze glycoproteins [4,5,6,7] and the loss of hemoglobin and myoglobin in the Channichthyidae [8]. Additional physiological adaptations include buoyancy modifications through reduced bone density and higher whole body lipid levels [9], cold-efficient cellular microtubule assembly [10,11], high mitochondrial densities [12] and the absence of a heat shock response [11,13,14]. The impact of the rapid speciation and isolation of the Antarctic nototheniids on the immune system, however, has been surprisingly little explored. Nonetheless, the frigid Antarctic waters, inhabited by a unique microbial community [8,15], have presumably influenced their immune repertoire and response to pathogens. Furthermore, increased human activity and climate change have been associated with the intrusion of alien organisms into Antarctic waters [16,17,18], raising the possibility of their unwitting exposure to new pathogens not far into the future.
Teleost fish, in common with other vertebrates, possess both innate and acquired components of the immune system [19,20,21]. However, innate immunity in teleost fish is proposed to have a more important role than the acquired immune response presumably because of the constant contact of fish epithelia or mucosal barriers (e.g., gills, skin and intestine) with a micro-organism rich aquatic environment [20,22,23,24].
Studies to assess how water temperature affects the immune response in temperate teleost fish indicate that lower water temperatures generally suppress the immune response [10,20,25,26,27,28,29,30,31]. In contrast, in vitro studies of fish immune cells maintained under reduced temperatures revealed enhanced cytotoxic activity [32] and an enhanced macrophage response [33] and respiratory burst [34]. Low temperatures did not inhibit phagocytosis by macrophages [35] or the inflammatory response in nototheniids [36,37,38]. In the bullhead (Notothenia coriiceps), damage repair of skin occurred at a slower pace than in teleosts from warmer waters with no sign of contamination by pathogens, which was attributed to the protective action of the mucous layer or the low pathogenicity of microorganism in Antarctic seawater [39]. Studies taking a candidate gene approach revealed that the exposure of N. coriiceps and N. rossii to bacterial infection agonist lipopolysaccharide (LPS) modified toll-like receptor gene expression in liver, kidney and spleen [40,41] and modulated iron-related immune genes, including hepcidin 4 (hp4) [42]. A novel hepcidin (hepcidin type II) has been identified in the Antarctic Notothenioids Dissostichus mawsoni and Notothenia angustata and was shown to be under positive selection [43]. Liver of Antarctic bullhead exposed to heat killed bacteria or polyinosinic:polycytidylic acid (poly I:C) was enriched in antigen processing and presentation and bacterial ligand exposure transcripts, which use antigen presentation against bacterial infection, but it may also use other defense mechanisms, such as TNF-mediated apoptosis, against viral infection [44]. Several toll-like receptor genes within the Notothenioid clade were shown to be under positive selection in pathogen recognition domains suggesting adaptation to the specific Antarctic microbiota [41]. A proteome study of the head kidney between white blood fish, Chionodraco hamatus, and two red blood fish, Trematomus bernacchii and N. coriiceps, revealed differences in erythropoiesis, heme biogenesis, leucocyte, and platelet development, with upregulation of lymphoid and megakaryocytic lineage marker proteins in ice fish [45].
The present study was designed to investigate the possible adaptation of the N. coriiceps immune system to the cold environment and unique microbial community of the Southern Ocean. Considering that adaptations might include gene retention and loss and/or response to potential pathogens, the bacterial agonist liposaccharide (LPS), a widely used teleost fish immunogen, was used as an immune challenge. LPS is ubiquitous (including in Antarctica) in the cell-wall of gram-negative bacteria of the Enterobacteriaceae family, although the host response may vary with chemical differences that exist in LPS from different Gram-negative species [46,47,48]. The transcriptional response to LPS was analyzed in the head-kidney, the main hematopoietic organ in fish, and in two mucosal-associated lymphoid tissues, the skin and intestine. The activity of serum enzymes associated with the innate immune response was also analyzed.
2. Materials and Methods
Fish sampling and experimentation were based on a permit issued by the Portuguese Environmental Agency under the regulations of the Madrid Protocol. N. coriiceps specimens (30 ± 2.4 cm total length and 384 ± 93 g weight) were captured using hook-and-line between 5 m and 30 m deep, near the Great Wall Station, King George Island in the Antarctic Peninsula (GPS coordinates: 62°12′57″ S, 58°57′42″ W) in January and February (the Antarctic summer) of 2017. Fish were acclimated for 5 days and maintained indoors in 200 L tanks in a flow-through circuit with seawater pumped from the surrounding bay. They were fed twice daily (morning and evening) with a mixture of limpets, salps, amphipods and fish muscle. Water temperature (2.0 ± 0.8 °C), salinity (28 ± 0.2 psu) and oxygen (11 ± 2 mg/L) were monitored three times a day (7.00 h, 14.00 h and 21.00 h) and were stable throughout the experiments.
LPS from the outer layer of 0111:B4 Gram-negative bacteria (L2630, Sigma-Aldrich, Madrid, Spain), which interacts with toll-like receptors (TLRs) and stimulates cytokine and acute phase protein release in vertebrates (including fish) [49,50] was used in the challenge experiments. Three days before the start of the experiments, fish were randomly allocated between three experimental tanks to create groups corresponding to: (i) the non-injected control (n = 7); (ii) the saline injected sham control (n = 7); and (iii) the LPS-injected treatment (n = 7). The LPS dosage was estimated on the basis of previous studies on teleost fish [29,51,52,53,54]. For injections, fish were lightly anaesthetized in 0.2 mL/L 2-phenoxyethanol (Sigma-Aldrich, Madrid, Spain) and weighed. The fish received an intraperitoneal (i.p.) injection of saline (1.1% NaCl, 2% fish wet weight) in the saline injected control (sham), or an i.p. injection of 1.5 mg/mL LPS in saline (LPS treatment). A second saline and LPS injection was administered to the sham control and to the LPS-treated group, respectively, 48 h after the first injection. Five days after the second injection was administered (day 7 of the experiment), fish were deeply anaesthetized (2-phenoxyethanol, 2.0 mL/L), and the blood was collected by caudal puncture using a heparinized 21-G needle fitted into a 1-mL syringe before sacrificing by cervical section. The blood plasma was separated by centrifugation and frozen at −80 °C until analysis. The head-kidney, skin, and anterior intestine (duodenum region) were dissected out and stored in RNA later (Sigma-Aldrich). Two fish died during the experiment only in the LPS treatment group.
Total plasma protein was quantified using a Quick StartTM Bradford Protein assay kit (Bio-Rad, Portugal) optimized for a 96-well plate [55]. Absorbance was measured at 590 nm at 25 °C in a spectrophotometer (Agilent Technologies, Santa Clara, CA, USA).
The lysozyme and antiprotease activities in blood plasma were measured based on previously described methods [56,57] with modifications [41]. Lyophilized Micrococcus luteus cells, hen egg white lysozyme, trypsin from porcine pancreas, azocasein and buffers were purchased from Sigma-Aldrich. Spectrometric measurements were made at 450 nm in a spectrophotometer (Agilent Technologies).
Plasma cortisol was measured by radioimmunoassay (RIA) using cortisol antiserum 20-CR50 (Fitzgerald Industries International, Concord, USA) as previously described [58]. The tritiated cortisol was purchased from GE Healthcare Europe GmbH (Carnaxide, Portugal). Cross-reactivities of the cortisol RIA were 54% for 11-desoxycortisol, 10% for cortisone, 16% for 17,21-dihydroxy-5β-pregnan-3,11,20-trione, 5% for 11β,17,21-trihydroxy-5β-pregnan-3,20-dione, 0.05% for 11β-hydroxytestosterone and less than 0.001% for testosterone.
Differences in biochemical parameters between the control, sham and LPS-treated groups were assessed using one-way analysis of variance (ANOVA) after verifying that normality and homoscedasticity assumptions were met using SigmaPlot v12.5 (Systat Software Inc, Palo Alto, CA, USA). Graphs were generated using GraphPrism v6.01 (GraphPad Software, San Diego, CA, USA). The threshold for significance was set at p < 0.05 (Figure S1).
Total RNA was extracted from approximately 25 mg of head-kidney, skin and anterior intestine (duodenum region) of five specimens of N. coriiceps per experimental group using an E.Z.N.A. Total RNA Kit I (Omega Bio-Tek, Norcross, GA, USA). Total RNA was treated with RNase-free DNase I (Omega Bio-Tek) to remove contaminating genomic DNA before poly(A)+ messenger RNA (mRNA) was purified for sequencing using a DynaBead mRNA Purification Kit (Life Technologies, Carlsbad, CA, USA). Samples with an RNA integrity number of at least eight were used to construct 45 paired-end complementary DNA (cDNA) libraries with an average insert size of 250 base pairs (bp) using a VAHTS stranded mRNA-seq Library Prep Kit from Illumina following the manufacturer’s protocol (Vazyme Biotech Co., Ltd., Nanjing, China). Sequencing library quality was monitored using an Agilent Bioanalyzer DNA 1000 Kit #5067-1504 (Agilent Technologies, Santa Clara, CA, USA). All libraries were sequenced using an Illumina Hiseq X Ten instrument (Illumina, Inc., San Diego, CA, USA) between 30 and149 read length.
A total of 298 million paired-end (PE) raw reads were generated by RNA-seq. Quality control and editing of raw reads to trim adapter sequences and low-quality bases was performed using a Trimgalore wrapper script v0.4.5 and output FastQC quality reports were obtained [59,60]. Mitochondrial and ribosomal reads were removed by aligning reads against the transcripts annotated as ribosomal gene products and the N. coriiceps mitochondrial genome (accession number NC_015653.1) using Bowtie2 v2.3.4 mapping software [61]. One sequencing library (L5) from the head-kidney of LPS challenged N. coriiceps did not produce enough reads to pass the FastQC and was excluded from the analysis (data not shown). Clean reads were quantified using the RSEM package v1.3.1 [62]. The raw sequence reads were deposited to the NCBI Sequence Reads Archive with BioProject accession ID PRJNA822876.
Two different analyses were made: one based on the mapping of clean reads against the N. coriiceps genome (NCBI reference genome NC01) [4], the other based on a reference transcriptome produced from the de novo assembly of all reads from the different treatment groups.
The Galaxy platform v22.01 [63] was used to map reads to the reference genome using Hisat2 v2.2.1 [64], Stringtie v2.1.7 [65] was used to count mapped reads and Deseq2 v1.34.0 [66] to determine differential gene expression.
Transcriptome assembly was performed in Trinity v2.5.1 with the “-normalize reads” parameter defined [67]. TransRate v 1.0.3 was used for quality filtering using default parameters [68]. EdgeR package v3.14.0 was used to determine differentially expressed genes [69]. The contigs were initially automatically annotated against Danio rerio and Homo sapiens genomes available in the Ensembl database using Trinotate v3.1.1 [70] and integrated into a SQLite database v3.34.0 to allow for fast efficient searching for terms with biological meaning. Annotations were based on the best deduced open reading frame (ORF) obtained with Transdecoder v1.03 [68]. Further manual annotation of non-annotated differentially expressed contigs interrogated Notothenioidei (N. coriiceps reference genome NC01, Trematomus bernacchii reference genome fTreBer1.1, Pseudochaenichthys georgianus reference genome fPseGeo1.1, Gymnodraco acuticeps reference genome fGymAcu1.1, Dissostichus mawsoni reference genome KU_Dm_1.0 and Cottoperca gobio reference genome fCotGob3.1), Homo sapiens (reference genome GRCh38.p13) and Danio rerio (reference genome GRCz11) genomes available at the NCBI.
Differentially expressed genes (DEGs) were identified using pairwise comparisons between the three treatment groups (non-injected control, sham and LPS) for head-kidney, duodenum and skin (FDR < 0.05), based on mapped transcripts per million (TPM). A heatmap of DEGs was generated with the “heatmap” function from RStudio v1.0.143. Gene Ontology (GO) enrichment was analysed with GOrilla [71] and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways with KOBAS (FDR < 0.05) [72]. GO terms were summarized using ReViGO [73] and graphical outputs were represented in Rstudio v1.0143.
The benchmarking universal single-copy orthologues (BUSCO) v5.3.2 [74] were used to assess the completeness of the NC01 genome and transcriptome assembly against the eukaryota, vertebrata and actinopterygii databases.
3. Results
3.1. Blood Biochemistry
Plasma protein, plasma total antiprotease activity, plasma lysozyme activity and plasma cortisol levels were not significantly modified between any of the treatment groups (Figure S1).
3.2. Transcriptome Assembly and Differentially Expressed Genes
The analysis using the reference genome yielded few annotated differentially expressed genes (duodenum 33, head kidney eight and skin one) and therefore we resorted to the de novo transcriptome assembly from which we could map more reads and annotate a higher number of differentially expressed genes (see below). Although tissue transcriptomes are unlikely to contain the full set of genes represented in the genome, the BUSCO analysis showed that over 80% of the eukaryote core genes were present in our transcriptomes (Table S1). The average mapping of reads to genome and reference transcriptome were 78% and 84% respectively (Table S2). The assembly of reads from the 45 sequencing libraries (1.1 Gbp) generated 51,506 contigs in the head-kidney, 58,916 in the duodenum and 66,329 in the skin with a N50 of 643 bp (Table S3).
Clustering revealed that individual samples clustered separately according to treatment, but the control and sham samples formed treatment specific sub-clusters separated from the LPS treated group (Figure 1a–c). The percentage annotation of DEGs varied between 47% and 100% (Table S4).
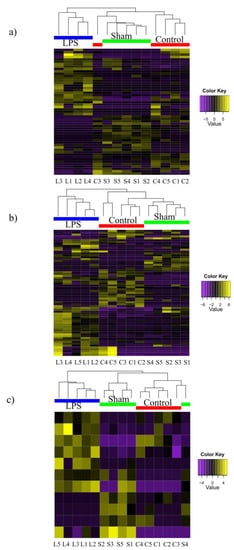
Figure 1.
Heatmap generated from DEGs identified in the (a) head-kidney, (b) duodenum and (c) skin transcriptomes of control and LPS-treated fish. The heatmap of clustered DEGs (log2 expression) between control (C1–C5), sham (S1–S5) and LPS (L1–L4) challenged fish in the three tissue transcriptomes (FDR < 0.05). The yellow color gradient indicates high relative abundance (up-regulation), the purple color gradient indicates low relative abundance (down-regulation) and black indicates equal abundance.
3.3. Control Versus Sham Differentially Expressed Genes
Pairwise comparison of the control versus sham, contigs representing the effect of handling and injection, resulted in 329 DEGs. Of these, 178 DEGs were from the head-kidney (129 up- and 49 down-regulated), 138 from the duodenum (77 up- and 61 down-regulated) and three from the skin (two up and one down-regulated). The skin shared one DEG with head-kidney and duodenum, while duodenum and head-kidney shared six DEGs and one DEG was shared between the three tissues (Figure 2a, Tables S3 and S4).
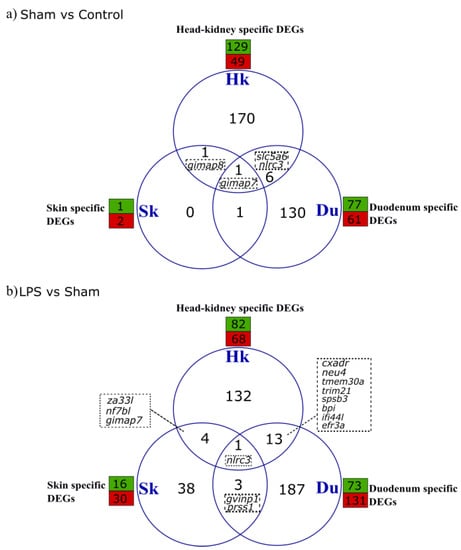
Figure 2.
Venn diagram representing the number of unique and common DEGs in head-kidney (Hk), duodenum (Du) and skin (Sk) in comparisons of (a) Sham versus Control and (b) LPS versus Sham. The green and red boxes represent the total number of genes up- or down-regulated (FDR < 0.05). The dashed boxes contain the name of shared annotated gene transcripts.
In head kidney, the five annotated most upregulated genes were macrophage mannose receptor 1 (mrc1), creatine kinase, muscle a (ttpa), GTPase IMAP family member 7 (gimap7), NLR family, the CARD domain containing 3 (nlrc3) and the kinesin family binding protein (kifbp), and the most downregulated were shootin 3 (shtn3), low affinity immunoglobulin gamma Fc region receptor II (fcgr2b), SLAM family member 7 (slamf7), nuclear factor 7, brain-like (nf7bl), and ciliogenesis associated TTC17 interacting protein (catip). In the duodenum the five annotated most upregulated genes were Acyl-CoA dehydrogenase family, member 11 (acad11), nlrc3, electron transfer flavoprotein dehydrogenase (etfdh), trichohyalin-like (tchhl1) and ladderlectin-like (name n/a), and the five most downregulated genes were gimap7, zinc finger protein 37 (zfp37), interferon-induced very large GTPase 1 (gvinp1), elastase 2-like (ela2l) and C2 calcium-dependent domain containing 2 (c2cd2). In skin only gimap8 was upregulated and gimap7 was downregulated. nlrc3 and sodium-dependent multivitamin transporter slc5a6, were shared between the head-kidney and duodenum, gimap8 was shared between head-kidney and skin and gimap7 was shared by the three tissue transcriptomes (Figure 2a, Table S5).
Functional annotation identified significant enrichment of GO terms in the head kidney and the biological processes (BP) were related to endoplasmic reticulum, translation, and catabolic process (Table S6).
Enriched KEGG pathways of DEGs in the head-kidney were stress and immune response related, including ribosome, adrenergic signaling in cardiomyocytes, cardiac muscle contraction, phagosome, and calcium signaling pathway. In the duodenum, mainly digestive system pathways were represented, including protein digestion and absorption, pancreatic secretion and apoptosis. No KEGG pathway enrichment was obtained in skin (Table S7).
3.4. LPS versus Sham Differentially Expressed Genes
Pairwise comparison of LPS versus sham yielded 401 DEGs, of which 150 were from the head-kidney (82 up- and 68 down-regulated), 204 from the duodenum (73 up- and 131 down-regulated) and 46 from the skin (16 up- and 30 down-regulated) (Figure 2b, Tables S4 and S8). While ca. 30% of DEGs were immune related genes, of these a large number were downregulated: 50% in duodenum, 44% in head kidney and 75% in skin.
In head kidney, the five topmost upregulated genes annotated in response to LPS were shtn3, zinc-binding protein A33-like (za33l), gimap8, disheveled-associated activator of morphogenesis 1 (daam1) and methyltransferase-like protein 12, mitochondrial (cskmt), and the most downregulated were nf7bl, gimap2, EFR3 homolog A (efr3a), gimap7 and aldehyde dehydrogenase 5 family, member A1 (aldh5a1) (Table 1). In duodenum, the 5 most upregulated genes were trypsin-2 (prss2), butyrophilin-like protein 1 (btnl1), trypsin-3 (prss3), chymotrypsin-like elastase family, member 1 (cela1) and high choriolytic enzyme 1-like (hce2l2), and the most downregulated were adaptor related protein complex 5 subunit sigma 1 (ap5s1), bactericidal permeability-increasing protein(bpi), proton-coupled amino acid transporter 1 (slc36a1), protein mono-ADP-ribosyltransferase PARP14 (parp14), cytochrome P450, family 19, subfamily A, polypeptide 1b (cyp19a1). In skin the 5 most upregulated were serine and arginine rich splicing factor 2a (srsf2a), cationic trypsin (prss1), POU domain, class 6, transcription factor 1-like (pou6d1), nf7bl, zinc finger protein 524 (znf524) and the most downregulated were sushi domain containing 2 (susd2), gimap7, nlrc3, cytochrome P450 2K1-like (cy2k1l) and homeobox B9a (hxb9a). The shared DEGs between the head-kidney and skin after LPS challenge group included za33l, nf7bl and gimap7 (Figure 2b, Table 1). The DEGs shared between the head-kidney and duodenum were cell cycle control protein 50A-like (tmem30A), E3 ubiquitin-protein ligase TRIM21-like (trim21), SPRY domain-containing SOCS box protein 3-like (spsb3), bactericidal permeability-increasing protein-like (bpi), interferon-induced protein 44-like (if44l), coxsackievirus and adenovirus receptor (cxadr), sialidase 4 (neu4) and efr3a (Figure 2b, Table 1). Nlrc3 was modified by LPS in the three tissue transcriptomes and gvinp1 and prss1 were differentially expressed in duodenum and skin (Figure 2b, Table 1).

Table 1.
Top 10 most upregulated and top 10 most downregulated differentially expressed genes between LPS and sham-injected fish in each tissue (head-kidney, duodenum and skin).
Functional annotation identified significant enriched GO terms mostly in duodenum and head kidney with little representation in the skin. (Figure 3a–c, Table S9). Enriched GO terms were only identified in duodenum (Figure 3a). The main biological processes were related to the immune system, extracellular matrix disassembly and the regulation of activated T cell differentiation.
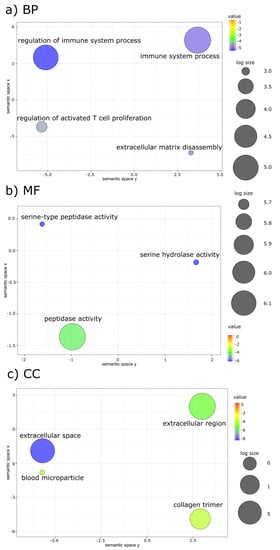
Figure 3.
Enriched GO terms of LPS versus Sham DEGs in Duodenum (a) Biological Process (BP) (b) Molecular Function (MF) and (c) Cellular Component (CC). The sizes of the circles represent the relative abundance, and the color gradient represents values of the log10 p-value.
The main KEGG pathways associated with LPS treatment were identified in the duodenum and included secretory and digestive processes related to protein digestion and absorption, viral responses (viral myocarditis, influenza A) and the immune and hormonal related responses focal adhesion, natural killer cell mediated cytotoxicity, apoptosis, oxytocin signaling pathway (Figure 4a, Table S10). The skin was enriched with viral infection pathway (influenza A) (Figure 4b, Table S10). In the head kidney, no enriched pathways were identified.
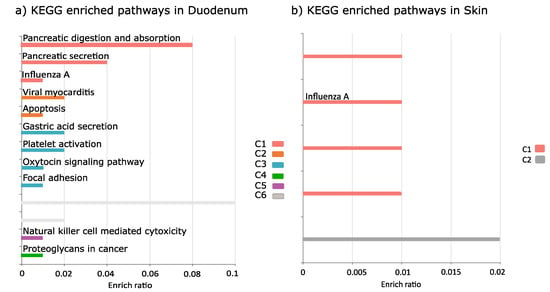
Figure 4.
KEGG enriched pathways of LPS versus Sham DEGs. (a) duodenum, (b) skin. The enriched KEGG pathways that are closely related are grouped in the same cluster (C1–C6) represented by the same color. Different clusters have a different color.
4. Discussion
A challenge with an endotoxin from the cell wall of Gram-negative bacteria (LPS) caused significant modifications of gene transcription 7 days later in the head-kidney, duodenum and skin of N. coriiceps, mostly downregulation, but had no effect on the activity of blood plasma circulating enzymes. However, each tissue showed different responsiveness to LPS, with the duodenum the most modified and the skin the least modified, presumably because of tissue specificities and the route of administration (i.p.) of the LPS that simulated an internal infection, rather than a pathogenic contact with the fishes’ surfaces, as would have been the case if administered through exposure in seawater.
The most modified processes in head-kidney, duodenum and skin in response to LPS were related to cell metabolism (up-regulated) and immune system (comprising 30% of DEGS, many downregulated). The main transcriptional modifications in the immune process category were related to pathogen recognition via receptors such as nucleotide-binding oligomerization domain-like (NOD) receptors, and expression of proteins against bacterial pathogens such as interleukin 6. The modified transcriptional response was not mirrored by a modified systemic response, as the circulating levels of enzymes of innate immunity, lysozyme and antiproteases, were not significantly different between the untreated, sham control and LPS treated fish.
The pairwise comparisons and clustering of the transcriptomes showed a clear effect of the treatments separating saline injected from non-injected controls and these two from LPS. However, although handling and injection modified gene transcription this was not translated in differential cortisol response, possibly because the cortisol peak had subsided after 7 days post injection [75]. The lack of cortisol and plasma protein responses to LPS are also consistent with observations in Atlantic salmon (Salmo salar) exposed to LPS for 1 and 19 days [51]. Based on studies of immunity in fish and shellfish, in which an LPS challenge was administered, we had anticipated a change in plasma lysozyme and antiprotease activity after LPS treatment in N. coriiceps. However, a lack of response of antiprotease activity to LPS has also been reported in gilthead seabream (Sparus aurata) larvae exposed to LPS for 24 h and in blood sera lysozyme activity in rohu (Labeo rohita) injected to LPS for 7–15 days [76,77]. It has been hypothesized that the high resistance and low response of some fish to LPS injections is due to the lack of a TLR4 recognition mechanism that in mammals mediates the down-stream immune activation, including TNF-α- and IL-1β-mediated inflammation, although zebrafish possess TLR4 and not activated by LPS [50,78,79,80]. Taken together, the results suggest that LPS may not always be an effective stimulus of immunity in teleost fish although the specific reasons are not clear.
Although no changes were observed in the activity of plasma circulating enzymes, it was notable the low basal level of blood lysozyme activity in N. coriiceps (50–90 U.mL−1 at 22 °C) compared to other teleosts, including the European sea bass (Dicentrarchus labrax) used as positive control in the present study (270–350 U.mL−1 at 22 °C) [81,82] and the Mozambique tilapia (Oreochromis mossambicus) (770–1000 U.mL−1 at 22 °C) [83]. The reason for the low levels of plasma lysozyme detected in N. coriiceps was not established but it is consistent with the absence of differentially expressed gene transcripts for lysozyme. The observation that the cold-water fishes cusk (Brosme brosme) and Atlantic cod (Gadus morhua) have low levels of lysozyme in the kidney (40 and 150 U.mL−1, respectively) [84] may indicate that lysozyme activity is less important in fishes from colder waters. Furthermore, in the Siberian sturgeon (Acipenser baerii), water temperature and physiological condition (e.g., health status, diet and age) modulate basal lysozyme activity [85]. The relative contribution of water temperature and physiological status to the low basal lysozyme activity in wild captured N. coriiceps remains to be further explored.
The constancy of the plasma biochemical parameters in N. coriiceps exposed to LPS could be related to the timeframe of the experiment (7 days post-i.p. injection), although the source of LPS may also have been a contributing factor. LPS is composed of an O-antigen polysaccharide and lipid A layers. The lipid A moiety varies with the bacterial species [86] and variations in O-antigen affect the virulence and infection process [87]. Temperature has a substantial impact on the structure of LPS and therefore the origin of the bacterial species’ source, from warm, temperate or cold waters, determines its conformation [88], as shown with bacterial LPS from the Siberian permafrost [89,90]. Since innate immune activation occurs through the recognition of microbe associated molecular patterns (MAMPs) [91] changes in LPS structure may interfere with host-bacteria recognition. We hypothesize that since the LPS used in the present study was from E. coli, which colonizes the warm gut of mammals, the relatively low biochemical and transcriptional response of N. coriiceps might be linked to the structure acquired by E. coli LPS 0111:B4 at low temperature making it less recognizable by the immune system of Antarctic fish. This notion seems to be supported by the absence of a serum lysozyme response and should also be reflected in a lower efficacy of humoral mediator recognition [92]. In several temperate fish challenged with LPS at different water temperatures, fish at lower temperatures tended to be more susceptible to pathogen infections, e.g., Paralichthys olivaceus, Oreochromis niloticus, Epinephelus coioides and Danio rerio [93,94,95,96,97]. Furthermore, it has been suggested that cold water fish such as Atlantic cod may have a slower as well as a suppressed immune response [98]. Interestingly, N. coriiceps exposed to fragmented heat killed E. coli 0111:B4, showed a strong transcriptional response of toll-like receptor genes 6 and 12 h post-challenge in the spleen but not in the liver or kidney [40]. Overall, a combination of the duration of the experiment, form of LPS, route of administration, water temperature, and species may have contributed to the absence of a plasma biochemical immune response.
The importance of bacterial infections in aquaculture means that transcriptomics has been used to describe the global immune response to LPS in a range of fish species: e.g., Lates calcarifer, Pelteobagrus fulvidraco, Salmo salar, Ictalarus punctatus [54,99,100,101]. However, there are few immune challenge studies of wild fish such as the present one and those that exist have focused on a single immune tissue [29,99].
In the present study, the head-kidney and duodenum had a strong transcriptional response and much lower in the skin. In the head kidney biological processes and pathways related energy generation and protein metabolism were mainly modified, consistent with observations in rainbow trout (Oncorhynchus mykiss) after 24 to 72 h after i.p. LPS injection [29] and likely reflect metabolic demand associated with the immune response, since any stress effects of injection and handling have been considered in the comparisons. There was a conserved core response of immune-related DEGs in the head kidney and duodenum, which encompassed PRRs like nlrc3, an intracellular sensor of PAMPs, and cxadr, a viral receptor that also is involved in leukocyte transepithelial migration [102,103], tmem30a, that stimulates phagocytosis and humoral defense-related factors [104,105], bactericidal permeability-increasing protein-like (bpi) and type I interferon stimulated gene (ifi44l). Head kidney-specific DEGs falling within the same general categories were mannose receptors (mrc1) and the humoral response genes pyrin-like (mefv) and peptidoglycan recognition proteins (pgrp-lbl). In Scophthalmus maximus L. [106] and Larimichthys crocea [107] exposed to bacteria, nlrc3 and mrc1 were also modified. Furthermore, in Miichthys miiuy, Paralichthys olivaceus and Cirrhinus mrigala, exposed to pathogen, nod1 expression was also modified [52,108,109,110] and the common response with N. coriiceps suggests it is part of the core antibacterial defence response of teleost fish.
In the duodenum responses were related to blood vessel formation, mitochondrial function, and immune system. Modifications of VEGF signaling were reported in studies with mammals exposed to LPS, an effect linked to the known effects of LPS on angiogenesis [111,112]. Interestingly, in vitro studies reported modifications in IL-1β expression in lumpfish leukocytes [113] and in gilthead seabream macrophages after 24 h bacterial exposure [114] and in IL-6 expression in LPS treated trout IgM+ B cells [115].
The response of immune function related genes to LPS was tissue-specific, compared to the other tissues. Nevertheless, changes in the humoral response due to bacterial pathogen exposure was reported in Ictalurus punctatus [116] and Megalobrama amblycephala [117] and bpi, one of the modified gene transcripts, was also differentially expressed in N. coriiceps. Similarly, peptidoglycan recognition proteins (e.g., pgrp-lbl), that bind multiple components of bacteria, a well characterized part of the innate humoral response in Danio rerio [118] and Cyprinus carpio L. [119], were changed in N. coriiceps. Ifi44l, typically associated with a viral challenge in Danio rerio [120] was differentially expressed in both HK and duodenum of N. coriiceps suggesting it also responds to a bacterial stimulus.
The large percentage of down-regulated immune genes in N. coriiceps were mainly related to the inflammatory response and included the IL-6 receptor and PRRs (NLR and MRC receptors) particularly in the head-kidney and duodenum. IL-6, a pleiotropic cytokine in mammals, is not well-studied in teleosts, but a recent study in Oreochromis niloticus indicated it promoted an inflammatory response to bacterial infection and antibody production [121]. The PRR response to LPS in N. coriiceps diverged from the more commonly described TLR in other teleost fish [122] and instead NLR and MRC receptor genes were more responsive. The lack of TLR response could be linked to the recent observation that TLR5, TLR8, TLR21, TLR22 and TLR23 are under positive selective pressure in Antarctic Notothenioids [41]. Furthermore, a study of deep-sea bacteria revealed pathogen associated molecular patterns (PAMPs) that failed to interact with human TLR, and this was explained by the absence of selective pressure for their recognition during pathogen-recognition receptors (PRR) evolution and so they were “immune silent” [123]. Nonetheless, although the transcriptional response of N. coriiceps to i.p. LPS was not identical to that of other teleosts, the general recruitment of PRRs for pathogen recognition and humoral factors for pathogen neutralization was a well-conserved response.
The DEGs identified in the mucosa-associated lymphoid tissue (MALT), the duodenum and skin, had little in common except for nlrc3. Interestingly this gene was downregulated by LPS in all three tissues suggesting it was important for LPS-mediated bacterial recognition. The duodenum had the highest number of DEGs of all the tissue analyzed, probably due to a more direct and continuous exposure to i.p. LPS. The most notably modified process in N. coriiceps duodenum was mitochondrial depolarization, which has not previously been reported in fish under LPS exposure. It is possible that this reflects the higher cellular mitochondrial density in the Antarctic fish [12], but it may also be related to the upregulation in the duodenum of inflammasome immune related genes, such as nlrp3, which have been associated with mitochondrial depolarization and mitophagy in mammals [124]. In Danio rerio, Nlrp3 is involved in cytokine processing and secretion [125], and a similar function probably exists in N. coriiceps as an elevated number of DEGs for cytokines occurred in the LPS exposed duodenum, as well as a gene for signal transduction (il6st) that respond to a bacterial challenge [126]. Curiously, in the skin gene transcripts linked to mitochondrial function and more specifically the generation of reactive oxygen species (ROS), a process linked with the inflammatory response in fish, were modified [127,128]. In line with this, increased ROS generation was observed in Lobeo rohita, after 7- and 15-days exposure to LPS [77,127,128].
A characteristic transcriptional response to bacterial exposure in a variety of teleost fish is the upregulation of complement [54,122,129] and this was evident in DEGs of the duodenum of N. coriiceps. In Ctenophcuyngodon idellus’ exposure to LPS i.p. increased the expression of adiponectin receptors in the liver [130] and in Trachinotus, ovatus keap1 was upregulated [131]. The hypoferraemic response in response to LPS is also conserved irrespectively [42,51,132].
The results of the present study contrasted with that of Ahn and collaborators who found several TLR genes up-regulated in liver, spleen and kidney after N. coriiceps were immersed in heat killed E. coli (HKEB) for 6–12 h [40]. They identified 567 DEGs in liver and the most enriched immune related process was the antigen processing and presentation pathway and TLR related pathways—TNF pathway, T-cell receptors, B-cells, interleukins and chemokines [44]. Although there was little overlap in the DEGs identified between the previous and the present study, some common processes were identified: antigen processing and presentation in skin, chemokines in the head-kidney and interleukins in both skin and duodenum. Altogether, the results from the present and previous transcriptome studies revealed that, despite its unique evolution and habitat, N. coriiceps respond rapidly to an immune challenge and that the general pattern of immune response is largely similar to teleosts from warmer waters, even taking into consideration some very divergent experimental protocols.
In our study, we selected the results of mapping transcripts to the de novo transcriptome, rather than the available N. coriiceps genome [4] because more genes were mapped and more core genes were annotated. However, we are aware of inherent bias of de novo transcriptome assemblies compared to map-to-reference estimates both in terms of diversity underestimation and positive bias at low expression levels, although these can be minimized using filtering methods, such as we those have used, and other computational methods [133]. It is therefore important that future work should be directed at improving genomes of key species for environmental and evolutionary studies such as N. coriiceps so as to maximize the power of transcriptomics.
5. Conclusions
Among the most striking features of the N. coriiceps response to LPS i.p injection was the unmodified plasma biochemical indicators and the marked difference in the quantitative and qualitative transcriptomic response between tissues. Skin and duodenum are barrier tissues and their difference in response may be related both to immune differences and effective exposure. A unique facet of the response to LPS in N. coriiceps was the up-regulation of genes linked to mitochondria and it is unclear if this is a consequence of adaptation to life in sub-zero conditions or if it is directly linked to the immune response. The immune processes identified were mostly down-regulated, particularly interleukins and pattern recognition receptors NLR and MRC, and the typical TLR response commonly described in other teleost fish was absent. This could have been because of the long period after stimulation (7 days) and/or to specificities derived from very specific pathogens and the cold marine environment of Antarctica. Further studies will be required to better characterize the PRRs and their role in Antarctic teleosts. Moreover, the importance of the LPS source and its consequences for immune activation in teleosts needs to be better understood.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/fishes7040171/s1, Figure S1: Biochemical analysis parameters in blood plasma.; Table S1: BUSCO analysis results from genome and reference transcriptome; Table S2: Alignment rate of the sequenced samples of the control and LPS-treated fish in Head-kidney (Hk), Duodenum (Du) and Skin (Sk) tissues; Table S3: Assemblies input and output data; Table S4: Differential expressed genes annotation statistics; Table S5: Differential expressed genes of Sham versus Control groups in different tissues; Table S6: Gene Ontology enriched terms derived from the analysis of the differentially expressed genes in Sham versus Control groups in different tissues; Table S7: KEGG enriched pathways represented in the sham versus control group in different tissues; Table S8: Differential expressed genes of LPS versus Sham groups in different tissues; Table S9: Gene Ontology enriched terms derived from the analysis of the differentially expressed genes of LPS versus Sham groups in different tissues; Table S10: KEGG enriched pathways represented in the LPS versus Sham group in different tissues.
Author Contributions
Conceptualization, D.M.P., L.C., P.M.G. and A.V.M.C.; formal analysis, C.S.V.S. and B.L.; investigation, C.S.V.S., B.L. and P.M.G.; data curation, C.S.V.S. and B.L.; writing—original draft preparation, C.S.V.S.; writing—D.M.P. and A.V.M.C.; project administration, D.M.P. and A.V.M.C.; funding acquisition, D.M.P., P.M.G. and A.V.M.C. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by Portuguese Foundation for Science and Technology (FCT) through projects FCT-NSFC/0002/2016, PTDC/BIAANM/3484/2014, Portuguese Polar Programme FACC PROPOLAR 2016–2017, UIDB/04326/2020, PhD fellowship SFRH/BD/120040/2016 to CS and Norma transitória- DL 57/2016/CP1361/CT0005 contract with the University of Algarve to BL.
Institutional Review Board Statement
Fish collection and experimental protocols were approved by the Portuguese Environmental Agency, under the regulations set by the Treaty of Madrid for scientific investigation in Antarctica. The experiments performed complied with European Union and Portuguese regulations for animal experimentation.
Data Availability Statement
The data presented in this study are available in Supplementary Figure S1 and Supplementary Tables S1–S10.
Acknowledgments
The authors would like to acknowledge the 2017 crew of the Great Wall Chinese Antarctic Station for their hospitability and logistic support and technical support to Elsa Couto.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Nowlin, W.D.; Klinck, J.M. The Physics of the Antarctic Circumpolar Current. Rev. Geophys. 1986, 24, 469–491. [Google Scholar] [CrossRef] [Green Version]
- Clarke, A.; Murphy, E.J.; Meredith, M.P.; King, J.C.; Peck, L.S.; Barnes, D.K.A.; Smith, R.C. Climate Change and the Marine Ecosystem of the Western Antarctic Peninsula. Philos. Trans. R. Soc. B Biol. Sci. 2007, 362, 149–166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rintoul, S.R. Antarctic Circumpolar Current. In Encyclopedia of Ocean Sciences; Elsevier: Amsterdam, The Netherlands, 2009; pp. 178–190. [Google Scholar]
- Shin, S.C.; Ahn, D.H.; Kim, S.J.; Pyo, C.W.; Lee, H.; Kim, M.-K.; Lee, J.; Lee, J.E.; Detrich, H.W.; Postlethwait, J.H.; et al. The Genome Sequence of the Antarctic Bullhead Notothen Reveals Evolutionary Adaptations to a Cold Environment. Genome Biol. 2014, 15, 468. [Google Scholar] [CrossRef] [PubMed]
- DeVries, A.L.; Wohlschlag, D.E. Freezing Resistance in Some Antarctic Fishes. Science 1969, 163, 1073–1075. [Google Scholar] [CrossRef]
- DeVries, A.L.; Cheng, C.-H.C. Antifreeze Proteins and Organismal Freezing Avoidance in Polar Fishes. Fish Physiol. 2005, 22, 155–201. [Google Scholar]
- Cheng, C.-H.C.; Chen, L. Evolution of an Antifreeze Glycoprotein. Nature 1999, 401, 443–444. [Google Scholar] [CrossRef]
- Kim, B.M.; Amores, A.; Kang, S.; Ahn, D.H.; Kim, J.H.; Kim, I.C.; Lee, J.H.; Lee, S.G.; Lee, H.; Lee, J.; et al. Antarctic Blackfin Icefish Genome Reveals Adaptations to Extreme Environments. Nat. Ecol. Evol. 2019, 3, 469–478. [Google Scholar] [CrossRef] [Green Version]
- Albertson, R.C.; Yan, Y.L.; Titus, T.A.; Pisano, E.; Vacchi, M.; Yelick, P.C.; Detrich, H.W.; Postlethwait, J.H. Molecular Pedomorphism Underlies Craniofacial Skeletal Evolution in Antarctic Notothenioid Fishes. BMC Evol. Biol. 2010, 10, 4. [Google Scholar] [CrossRef] [Green Version]
- Detrich, H.W.; Johnson, K.A.; Marchese-Ragona, S.P. Polymerization of Antarctic Fish Tubulins at Low Temperatures: Energetic Aspects. Biochemistry 1989, 28, 10085–10093. [Google Scholar] [CrossRef]
- Detrich, H.W.; Parker, S.K.; Williams, J.; Nogales, E.; Downing, K.H. Cold Adaptation of Microtubule Assembly and Dynamics. Structural Interpretation of Primary Sequence Changes Present in the α- and β-Tubulins of Antarctic Fishes. J. Biol. Chem. 2000, 275, 37038–37047. [Google Scholar] [CrossRef] [Green Version]
- O’Brien, K.M.; Sidell, B.D. The Interplay among Cardiac Ultrastructure, Metabolism and the Expression of Oxygen-Binding Proteins in Antarctic Fishes. J. Exp. Biol. 2000, 203, 1287–1297. [Google Scholar] [CrossRef] [PubMed]
- Place, S.P.; Hofmann, G.E. Comparison of Hsc70 Orthologs from Polar and Temperate Notothenioid Fishes: Differences in Prevention of Aggregation and Refolding of Denatured Proteins. Am. J. Physiol.-Regul. Integr. Comp. Physiol. 2005, 288, R1195–R1202. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, G.E.; Buckley, B.; Airaksinen, S.; Keen, J.E.; Somero, G.N. Heat-Shock Protein Expression Is Absent in the Antarctic Fish Trematomus bernacchii (Family Nototheniidae). J. Exp. Biol. 2000, 203, 2331–2339. [Google Scholar] [CrossRef] [PubMed]
- Furbino, L.E.; Godinho, V.M.; Santiago, I.F.; Pellizari, F.M.; Alves, T.M.A.; Zani, C.L.; Junior, P.A.S.; Romanha, A.J.; Carvalho, A.G.O.; Gil, L.H.V.G.; et al. Diversity Patterns, Ecology and Biological Activities of Fungal Communities Associated with the Endemic Macroalgae Across the Antarctic Peninsula. Microb. Ecol. 2014, 67, 775–787. [Google Scholar] [CrossRef]
- Grimaldi, W.W.; Seddon, P.J.; Lyver, P.O.; Nakagawa, S.; Tompkins, D.M. Infectious Diseases of Antarctic Penguins: Current Status and Future Threats. Polar Biol. 2015, 38, 591–606. [Google Scholar] [CrossRef]
- Duffy, G.A.; Coetzee, B.W.T.; Latombe, G.; Akerman, A.H.; McGeoch, M.A.; Chown, S.L. Barriers to Globally Invasive Species Are Weakening across the Antarctic. Divers. Distrib. 2017, 23, 982–996. [Google Scholar] [CrossRef] [Green Version]
- Cowan, D.A.; Chown, S.L.; Convey, P.; Tuffin, M.; Hughes, K.; Pointing, S.; Vincent, W.F. Non-Indigenous Microorganisms in the Antarctic: Assessing the Risks. Trends Microbiol. 2011, 19, 540–548. [Google Scholar] [CrossRef]
- Magor, B.G.; Magor, K.E. Evolution of Effectors and Receptors of Innate Immunity. Dev. Comp. Immunol. 2001, 25, 651–682. [Google Scholar] [CrossRef]
- Tort, L.; Balasch, J.C.; Mackenzie, S. Fish Immune System. A Crossroads between Innate and Adaptive Responses. Inmunologia 2003, 22, 277–286. [Google Scholar] [CrossRef]
- Magnadottir, B.; Gudmundsdottir, B.K.; Lange, S.; Steinarsson, A.; Oddgeirsson, M.; Bowden, T.; Bricknell, I.; Dalmo, R.A.; Gudmundsdottir, S. Immunostimulation of Larvae and Juveniles of Cod, Gadus Morhua L. J. Fish Dis. 2006, 29, 147–155. [Google Scholar] [CrossRef]
- Gomez, D.; Sunyer, J.O.; Salinas, I. The Mucosal Immune System of Fish: The Evolution of Tolerating Commensals While Fighting Pathogens. Fish Shellfish Immunol. 2013, 35, 1729–1739. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sunyer, J.O. Fishing for Mammalian Paradigms in the Teleost Immune System. Nat. Immunol. 2013, 14, 320–326. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Nie, L.; Zhu, G.; Xiang, L.; Shao, J. Advances in Research of Fish Immune-Relevant Genes: A Comparative Overview of Innate and Adaptive Immunity in Teleosts. Dev. Comp. Immunol. 2013, 39, 39–62. [Google Scholar] [CrossRef]
- Bonneaud, C.; Wilson, R.S.; Seebacher, F. Immune-Challenged Fish up-Regulate Their Metabolic Scope to Support Locomotion. PLoS ONE 2016, 11, e0166028. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abram, Q.H.; Dixon, B.; Katzenback, B.A. Impacts of Low Temperature on the Teleost Immune System. Biology 2017, 6, 39. [Google Scholar] [CrossRef] [Green Version]
- Bly, J.E.; Clem, L.W. Temperature and Teleost Immune Functions. Fish Shellfish Immunol. 1992, 2, 159–171. [Google Scholar] [CrossRef]
- Le Morvan, C.; Troutaud, D.; Deschaux, P. Differential Effects of Temperature on Specific and Nonspecific Immune Defences in Fish. J. Exp. Biol. 1998, 201, 165–168. [Google Scholar] [CrossRef]
- MacKenzie, S.; Balasch, J.C.; Novoa, B.; Ribas, L.; Roher, N.; Krasnov, A.; Figueras, A. Comparative Analysis of the Acute Response of the Trout, O. mykiss, Head Kidney to in Vivo Challenge with Virulent and Attenuated Infectious Hematopoietic Necrosis Virus and LPS-Induced Inflammation. BMC Genom. 2008, 9, 141. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Zhou, S.; Jiang, Z.; Wang, X.; Liu, Y. Chemokine Receptor CXCR3 in Turbot (Scophthalmus maximus): Cloning, Characterization and Its Responses to Lipopolysaccharide. Fish Physiol. Biochem. 2016, 42, 659–671. [Google Scholar] [CrossRef]
- Magnoni, L.; Roher, N.; Crespo, D.; Krasnov, A.; Planas, J. In Vivo Molecular Responses of Fast and Slow Muscle Fibers to Lipopolysaccharide in a Teleost Fish, the Rainbow Trout (Oncorhynchus mykiss). Biology 2015, 4, 67–87. [Google Scholar] [CrossRef] [Green Version]
- Le Morvan-Rocher, C.; Troutaud, D.; Deschaux, P. Effects of Temperature on Carp Leukocyte Mitogen-Induced Proliferation and Nonspecific Cytotoxic Activity. Dev. Comp. Immunol. 1995, 19, 87–95. [Google Scholar] [CrossRef]
- Le Morvan, C.; Clerton, P.; Deschaux, P.; Troutaud, D. Effects of Environmental Temperature on Macrophage Activities in Carp. Fish Shellfish Immunol. 1997, 7, 209–212. [Google Scholar] [CrossRef]
- Dexiang, C.; Ainsworth, A.J. Effect of Temperature on the Immune System of Channel Catfish (Ictalurus punctatus)—II. Adaptation of Anterior Kidney Phagocytes to 10 °C. Comp. Biochem. Physiol. Part A Physiol. 1991, 100, 913–918. [Google Scholar] [CrossRef]
- Silva, J. Phagocytosis and Giant Cell Formation at 0 °C by Macrophage (MØ) of Notothenia coriiceps. J. Fish Biol. 2002, 60, 466–478. [Google Scholar] [CrossRef]
- Silva, J.R.M.C.; Hernadez-Blazquez, F.J.; Barbieri, R.L. Induced Inflammatory Process in the Antarctic Fish Notothenia neglecta. Polar Biol. 1998, 20, 206–212. [Google Scholar] [CrossRef]
- Silva, J.R.M.C.; Staines, N.A.; Parra, O.M.; Hernandez-Blazquez, F.J. Experimental Studies on the Response of the Fish (Notothenia coriiceps Richardson, 1844) to Parasite (Pseudoterranova decipiens Krabbe, 1878) and Other Irritant Stimuli at Antarctic Temperatures. Polar Biol. 1999, 22, 417–424. [Google Scholar] [CrossRef]
- O’Neill, J.G.; White, M.G.; Sims, T.A.; Barber, D.L. The Inflammatory Response of the Antarctic Silverfish, Pleuragramma antarcticum Boulenger, 1902, to a Plerocercoid (Species Unknown) Infestation. J. Fish Biol. 1987, 31, 231–232. [Google Scholar] [CrossRef]
- Cunha da Silva, J.R.M.; Cooper, E.L.; Sinhorini, I.L.; Borges, J.C.S.; Jensch-Junior, B.E.; Porto-Neto, L.R.; Hernandez-Blazquez, F.J.; Vellutini, B.C.; Pressinotti, L.N.; Costa-Pinto, F.A. Microscopical Study of Experimental Wound Healing in Notothenia coriiceps (Cabeçuda) at 0 Degrees C. Cell Tissue Res. 2005, 321, 401–410. [Google Scholar] [CrossRef]
- Ahn, D.H.; Shin, S.C.; Park, H. Characterization of Toll-like Receptor Gene Expression and the Pathogen Agonist Response in the Antarctic Bullhead Notothen Notothenia coriiceps. Immunogenetics 2014, 66, 563–573. [Google Scholar] [CrossRef]
- Sousa, C.; Fernandes, S.A.; Cardoso, J.C.R.; Wang, Y.; Zhai, W.; Guerreiro, P.M.; Chen, L.; Canário, A.V.M.; Power, D.M. Toll-Like Receptor Evolution: Does Temperature Matter? Front. Immunol. 2022, 13, 27. [Google Scholar] [CrossRef]
- Martínez, D.P.; Sousa, C.; Oyarzún, R.; Pontigo, J.P.; Canario, A.V.M.; Power, D.M.; Vargas-Chacoff, L.; Guerreiro, P.M. LPS Modulates the Expression of Iron-Related Immune Genes in Two Antarctic Notothenoids. Front. Physiol. 2020, 11, 102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Q.; Cheng, C.-H.C.; Hu, P.; Ye, H.; Chen, Z.; Cao, L.; Chen, L.; Shen, Y.; Chen, L. Adaptive Evolution of Hepcidin Genes in Antarctic Notothenioid Fishes. Mol. Biol. Evol. 2008, 25, 1099–1112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahn, D.-H.; Kang, S.; Park, H. Transcriptome Analysis of Immune Response Genes Induced by Pathogen Agonists in the Antarctic Bullhead Notothen Notothenia coriiceps. Fish Shellfish Immunol. 2016, 55, 315–322. [Google Scholar] [CrossRef] [PubMed]
- Jia, R.; Huang, S.; Zhai, W.; Jiang, S.; Li, W.; Wang, F.; Xu, Q. Comparative Proteomic Analysis of Head Kidney among Three Antarctic Fishes. Mar. Biol. 2022, 169, 58. [Google Scholar] [CrossRef]
- Nilsson, C.; Skoglund, A.; Moran, A.P.; Annuk, H.; Engstrand, L.; Normark, S. Lipopolysaccharide Diversity Evolving in Helicobacter Pylori Communities through Genetic Modifications in Fucosyltransferases. PLoS ONE 2008, 3, e3811. [Google Scholar] [CrossRef]
- Araya, M.A.; Valenzuela, T.; Inostroza, N.G.; Maruyama, F.; Jorquera, M.A.; Acuña, J.J. Isolation and Characterization of Cold-Tolerant Hyper-ACC-Degrading Bacteria from the Rhizosphere, Endosphere, and Phyllosphere of Antarctic Vascular Plants. Microorganisms 2020, 8, 1788. [Google Scholar] [CrossRef]
- Meredith, T.C.; Aggarwal, P.; Mamat, U.; Lindner, B.; Woodard, R.W. Redefining the Requisite Lipopolysaccharide Structure in Escherichia coli. ACS Chem. Biol. 2006, 1, 33–42. [Google Scholar] [CrossRef]
- Bishop, R.E. Fundamentals of Endotoxin Structure and Function. In Concepts in Bacterial Virulence; KARGER: Basel, Switzerland, 2004; Volume 12, pp. 1–27. [Google Scholar]
- Swain, P.; Nayak, S.K.; Nanda, P.K.; Dash, S. Biological Effects of Bacterial Lipopolysaccharide (Endotoxin) in Fish: A Review. Fish Shellfish Immunol. 2008, 25, 191–201. [Google Scholar] [CrossRef]
- Langston, A.L.; Johnstone, R.; Ellis, A.E. The Kinetics of the Hypoferraemic Response and Changes in Levels of Alternative Complement Activity in Diploid and Triploid Atlantic Salmon, Following Injection of Lipopolysaccharide. Fish Shellfish Immunol. 2001, 11, 333–345. [Google Scholar] [CrossRef]
- Chu, Q.; Bi, D.; Zheng, W.; Xu, T. MicroRNA Negatively Regulates NF-ΚB-Mediated Immune Responses by Targeting NOD1 in the Teleost Fish Miichthys miiuy. Sci. China Life Sci. 2021, 64, 803–815. [Google Scholar] [CrossRef]
- Haukenes, A.H.; Barton, B.A. Characterization of the Cortisol Response Following an Acute Challenge with Lipopolysaccharide in Yellow Perch and the Influence of Rearing Density. J. Fish Biol. 2004, 64, 851–862. [Google Scholar] [CrossRef]
- Jiang, H.; Wang, M.; Fu, L.; Zhong, L.; Liu, G.; Zheng, Y.; Chen, X.; Bian, W. Liver Transcriptome Analysis and Cortisol Immune-Response Modulation in Lipopolysaccharide-Stimulated in Channel Catfish (Ictalurus punctatus). Fish Shellfish Immunol. 2020, 101, 19–50. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Ellis, A.E. Lysozyme Assays. In Techniques in Fish Immunology; Stolen, J.S., Fletcher, T.C., Anderson, D.P., Roberson, B.S., Van Muiswinkel, W.B., Eds.; SOS Publications: Cambridge, UK, 1990; pp. 101–103. [Google Scholar]
- Ellis, A.E. Innate Host Defense Mechanisms of Fish against Viruses and Bacteria. Dev. Comp. Immunol. 2001, 25, 827–839. [Google Scholar] [CrossRef]
- Rotllant, J.; Guerreiro, P.M.; Anjos, L.; Redruello, B.; Canario, A.V.M.; Power, D.M. Stimulation of Cortisol Release by the N Terminus of Teleost Parathyroid Hormone-Related Protein in Interrenal Cells in Vitro. Endocrinology 2005, 146, 71–76. [Google Scholar] [CrossRef]
- Andrews, S. FASTQC A Quality Control Tool for High Throughput Sequence Data. Babraham Inst. 2015. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 15 January 2022).
- Krueger, F. Trim Galore: A Wrapper Tool around Cutadapt and FastQC. Babraham Inst. 2015. Available online: https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/ (accessed on 15 January 2022).
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and Memory-Efficient Alignment of Short DNA Sequences to the Human Genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef] [Green Version]
- Li, B.; Dewey, C.N. RSEM: Accurate Transcript Quantification from RNA-Seq Data with or without a Reference Genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [Green Version]
- Afgan, E.; Baker, D.; Batut, B.; van den Beek, M.; Bouvier, D.; Čech, M.; Chilton, J.; Clements, D.; Coraor, N.; Grüning, B.A.; et al. The Galaxy Platform for Accessible, Reproducible and Collaborative Biomedical Analyses: 2018 Update. Nucleic Acids Res. 2018, 46, W537–W544. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A Fast Spliced Aligner with Low Memory Requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie Enables Improved Reconstruction of a Transcriptome from RNA-Seq Reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De Novo Transcript Sequence Reconstruction from RNA-Seq Using the Trinity Platform for Reference Generation and Analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef]
- Smith-Unna, R.; Boursnell, C.; Patro, R.; Hibberd, J.M.; Kelly, S. TransRate: Reference-Free Quality Assessment of de Novo Transcriptome Assemblies. Genome Res. 2016, 26, 1134–1144. [Google Scholar] [CrossRef] [Green Version]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics 2009, 26, 139–140. [Google Scholar] [CrossRef] [Green Version]
- Bryant, D.M.; Johnson, K.; DiTommaso, T.; Tickle, T.; Couger, M.B.; Payzin-Dogru, D.; Lee, T.J.; Leigh, N.D.; Kuo, T.H.; Davis, F.G.; et al. A Tissue-Mapped Axolotl De Novo Transcriptome Enables Identification of Limb Regeneration Factors. Cell Rep. 2017, 18, 762–776. [Google Scholar] [CrossRef] [Green Version]
- Eden, E.; Navon, R.; Steinfeld, I.; Lipson, D.; Yakhini, Z. GOrilla: A Tool for Discovery and Visualization of Enriched GO Terms in Ranked Gene Lists. BMC Bioinform. 2009, 10, 48. [Google Scholar] [CrossRef] [Green Version]
- Xie, C.; Mao, X.; Huang, J.; Ding, Y.; Wu, J.; Dong, S.; Kong, L.; Gao, G.; Li, C.Y.; Wei, L. KOBAS 2.0: A Web Server for Annotation and Identification of Enriched Pathways and Diseases. Nucleic Acids Res. 2011, 39, W316–W322. [Google Scholar] [CrossRef] [Green Version]
- Supek, F.; Bošnjak, M.; Škunca, N.; Šmuc, T. REVIGO Summarizes and Visualizes Long Lists of Gene Ontology Terms. PLoS ONE 2011, 6, e21800. [Google Scholar] [CrossRef] [Green Version]
- Manni, M.; Berkeley, M.R.; Seppey, M.; Simão, F.A.; Zdobnov, E.M. BUSCO Update: Novel and Streamlined Workflows along with Broader and Deeper Phylogenetic Coverage for Scoring of Eukaryotic, Prokaryotic, and Viral Genomes. Mol. Biol. Evol. 2021, 38, 4647–4654. [Google Scholar] [CrossRef] [PubMed]
- Kandalski, P.K.; de Souza, M.R.D.P.; Herrerias, T.; Machado, C.; Zaleski, T.; Forgati, M.; Guillen, A.C.; Viana, D.; Moura, M.O.; Donatti, L. Effects of Short-Term Thermal Stress on the Plasma Biochemical Profiles of Two Antarctic Nototheniid Species. Rev. Fish Biol. Fish. 2018, 28, 925–940. [Google Scholar] [CrossRef]
- Hanif, A.; Bakopoulos, V.; Leonardos, I.; Dimitriadis, G.J. The Effect of Sea Bream (Sparus aurata) Broodstock and Larval Vaccination on the Susceptibility by Photobacterium damsela Subsp. Piscicida and on the Humoral Immune Parameters. Fish Shellfish Immunol. 2005, 19, 345–361. [Google Scholar] [CrossRef] [PubMed]
- Nayak, S.K.; Swain, P.; Nanda, P.K.; Dash, S.; Shukla, S.; Meher, P.K.; Maiti, N.K. Effect of Endotoxin on the Immunity of Indian Major Carp, Labeo rohita. Fish Shellfish Immunol. 2008, 24, 394–399. [Google Scholar] [CrossRef] [PubMed]
- Sepulcre, M.P.; Alcaraz-Pérez, F.; López-Muñoz, A.; Roca, F.J.; Meseguer, J.; Cayuela, M.L.; Mulero, V. Evolution of Lipopolysaccharide (LPS) Recognition and Signaling: Fish TLR4 Does Not Recognize LPS and Negatively Regulates NF-ΚB Activation. J. Immunol. 2009, 182, 1836–1845. [Google Scholar] [CrossRef] [Green Version]
- Berczi, I.; Bertók, L.; Bereznai, T. Comparative Studies on the Toxicity of Escherichia Coli Lipopolysaccharide Endotoxin in Various Animal Species. Can. J. Microbiol. 1966, 12, 1070–1071. [Google Scholar] [CrossRef] [PubMed]
- Park, B.S.; Lee, J.-O. Recognition of Lipopolysaccharide Pattern by TLR4 Complexes. Exp. Mol. Med. 2013, 45, e66. [Google Scholar] [CrossRef] [Green Version]
- Obach, A.; Quentel, C.; Baudin Laurencin, F. Effects of Alpha-Tocopherol and Dietary Oxidized Fish Oil on the Immune Response of Sea Bass Dicentrarchus labrax. Dis. Aquat. Org. 1993, 15, 175–185. [Google Scholar] [CrossRef]
- Yildiz, H.Y.; Altunay, S. Physiological Stress and Innate Immune Response in Gilthead Sea Bream (Sparus aurata) and Sea Bass (Dicentrarchus labrax) Exposed to Combination of Trimethoprim and Sulfamethoxazole (TMP-SMX). Fish Physiol. Biochem. 2011, 37, 401–409. [Google Scholar] [CrossRef]
- Christybapita, D.; Divyagnaneswari, M.; Dinakaran Michael, R. Oral Administration of Eclipta Alba Leaf Aqueous Extract Enhances the Non-Specific Immune Responses and Disease Resistance of Oreochromis mossambicus. Fish Shellfish Immunol. 2007, 23, 840–852. [Google Scholar] [CrossRef]
- Lie, Ø.; Evensen, Ø.; Sørensen, A.; Frøysadal, E. Study on Lysozyme Activity in Some Fish Species. Dis. Aquat. Org. 1989, 6, 55–61. [Google Scholar] [CrossRef]
- Simide, R.; Richard, S.; Prévot-D’Alvise, N.; Miard, T.; Gaillard, S. Assessment of the Accuracy of Physiological Blood Indicators for the Evaluation of Stress, Health Status and Welfare in Siberian Sturgeon (Acipenser baerii) Subject to Chronic Heat Stress and Dietary Supplementation. Int. Aquat. Res. 2016, 8, 121–135. [Google Scholar] [CrossRef] [Green Version]
- Garate, J.A.; Oostenbrink, C. Lipid a from Lipopolysaccharide Recognition: Structure, Dynamics and Cooperativity by Molecular Dynamics Simulations. Proteins Struct. Funct. Bioinform. 2013, 81, 658–674. [Google Scholar] [CrossRef] [PubMed]
- Lerouge, I. O-Antigen Structural Variation: Mechanisms and Possible Roles in Animal/Plant–Microbe Interactions. FEMS Microbiol. Rev. 2002, 26, 17–47. [Google Scholar] [CrossRef] [Green Version]
- Kumar, G.S.; Jagannadham, M.V.; Ray, M.K. Low-Temperature-Induced Changes in Composition and Fluidity of Lipopolysaccharides in the Antarctic Psychrotrophic Bacterium Pseudomonas syringae. J. Bacteriol. 2002, 184, 6746–6749. [Google Scholar] [CrossRef] [Green Version]
- Wu, E.L.; Engström, O.; Jo, S.; Stuhlsatz, D.; Yeom, M.S.; Klauda, J.B.; Widmalm, G.; Im, W. Molecular Dynamics and NMR Spectroscopy Studies of E. Coli Lipopolysaccharide Structure and Dynamics. Biophys. J. 2013, 105, 1444–1455. [Google Scholar] [CrossRef] [Green Version]
- Chattopadhyay, M.K. Mechanism of Bacterial Adaptation to Low Temperature. J. Biosci. 2006, 31, 157–165. [Google Scholar] [CrossRef]
- Akira, S.; Uematsu, S.; Takeuchi, O. Pathogen Recognition and Innate Immunity. Cell 2006, 124, 783–801. [Google Scholar] [CrossRef] [Green Version]
- Ohno, N.; Morrison, D.C. Lipopolysaccharide Interactions with Lysozyme Differentially Affect Lipopolysaccharide Immunostimulatory Activity. Eur. J. Biochem. 1989, 186, 629–636. [Google Scholar] [CrossRef]
- Sanders, G.E.; Batts, W.N.; Winton, J.R. Susceptibility of Zebrafish (Danio rerio) to a Model Pathogen, Spring Viremia of Carp Virus. Comp. Med. 2003, 53, 514–521. [Google Scholar]
- Cheng, A.C.; Cheng, S.A.; Chen, Y.Y.; Chen, J.C. Effects of Temperature Change on the Innate Cellular and Humoral Immune Responses of Orange-Spotted Grouper Epinephelus coioides and Its Susceptibility to Vibrio alginolyticus. Fish Shellfish Immunol. 2009, 26, 768–772. [Google Scholar] [CrossRef] [PubMed]
- Ndong, D.; Chen, Y.-Y.; Lin, Y.-H.; Vaseeharan, B.; Chen, J.-C. The Immune Response of Tilapia Oreochromis mossambicus and Its Susceptibility to Streptococcus iniae under Stress in Low and High Temperatures. Fish Shellfish Immunol. 2007, 22, 686–694. [Google Scholar] [CrossRef] [PubMed]
- Avunje, S.; Oh, M.J.; Jung, S.J. Impaired TLR2 and TLR7 Response in Olive Flounder Infected with Viral Haemorrhagic Septicaemia Virus at Host Susceptible 15 °C but High at Non-Susceptible 20 °C. Fish Shellfish Immunol. 2013, 34, 1236–1243. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Xu, D.; Tian, L.; Chen, R.; Wang, L.; Tan, P.; You, Q. Overwinter Mortality in Yellow Drum (Nibea albiflora): Insights from Growth and Immune Responses to Cold and Starvation Stress. Fish Shellfish Immunol. 2019, 92, 341–347. [Google Scholar] [CrossRef]
- Magnadottir, B.; Audunsdottir, S.S.; Bragason, B.T.; Gisladottir, B.; Jonsson, Z.O.; Gudmundsdottir, S. The Acute Phase Response of Atlantic Cod (Gadus morhua): Humoral and Cellular Response. Fish Shellfish Immunol. 2011, 30, 1124–1130. [Google Scholar] [CrossRef]
- Palstra, A.P.; Kals, J.; Garcia, A.B.; Dirks, R.P.; Poelman, M. Immunomodulatory Effects of Dietary Seaweeds in LPS Challenged Atlantic Salmon Salmo salar as Determined by Deep RNA Sequencing of the Head Kidney Transcriptome. Front. Physiol. 2018, 9, 625. [Google Scholar] [CrossRef]
- Lu, X.-J.; Ning, Y.-J.; Liu, H.; Nie, L.; Chen, J. A Novel Lipopolysaccharide Recognition Mechanism Mediated by Internalization in Teleost Macrophages. Front. Immunol. 2018, 9, 2758. [Google Scholar] [CrossRef] [Green Version]
- Zoccola, E.; Kellie, S.; Barnes, A.C. Immune Transcriptome Reveals the Mincle C-Type Lectin Receptor Acts as a Partial Replacement for TLR4 in Lipopolysaccharide-Mediated Inflammatory Response in Barramundi (Lates calcarifer). Mol. Immunol. 2017, 83, 33–45. [Google Scholar] [CrossRef]
- Chiaranunt, P.; Burrows, K.; Ngai, L.; Mortha, A. Isolation of mononuclear phagocytes from the mouse gut. In Methods in Enzymology; Academic Press Inc.: Cambridge, MA, USA, 2020; Volume 632, pp. 67–90. [Google Scholar] [CrossRef]
- Fritz, J.H.; Girardin, S.E. How Toll-like Receptors and Nod-like Receptors Contribute to Innate Immunity in Mammals. J. Endotoxin Res. 2005, 11, 390–394. [Google Scholar] [CrossRef]
- Hong, J.; Yoon, D.; Nam, Y.; Seo, D.; Kim, J.H.; Kim, M.S.; Lee, T.Y.; Kim, K.S.; Ko, P.W.; Lee, H.W.; et al. Lipopolysaccharide Administration for a Mouse Model of Cerebellar Ataxia with Neuroinflammation. Sci. Rep. 2020, 10, 13337. [Google Scholar] [CrossRef]
- Yang, I.V.; Wade, C.M.; Kang, H.M.; Alper, S.; Rutledge, H.; Lackford, B.; Eskin, E.; Daly, M.J.; Schwartz, D.A. Identification of Novel Genes That Mediate Innate Immunity Using Inbred Mice. Genetics 2009, 183, 1535–1544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hou, Z.; Ye, Z.; Zhang, D.; Gao, C.; Su, B.; Song, L.; Tan, F.; Song, H.; Wang, Y.; Li, C. Characterization and Expression Profiling of NOD-like Receptor C3 (NLRC3) in Mucosal Tissues of Turbot (Scophthalmus maximus L.) Following Bacterial Challenge. Fish Shellfish Immunol. 2017, 66, 231–239. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Li, J.; He, J.; Liu, W.; Jiang, L.; Ye, Y.; Wu, C. Anti-Infective Mannose Receptor Immune Mechanism in Large Yellow Croaker (Larimichthys crocea). Fish Shellfish Immunol. 2016, 54, 257–265. [Google Scholar] [CrossRef] [PubMed]
- Bin Park, S.; Hikima, J.-I.; Suzuki, Y.; Ohtani, M.; Nho, S.W.; Cha, I.S.; Bin Jang, H.; Kondo, H.; Hirono, I.; Aoki, T.; et al. Molecular cloning and functional analysis of nucleotide-binding oligomerization domain 1 (NOD1) in olive flounder, Paralichthys olivaceus. Dev. Comp. Immunol. 2011, 36, 680–687. [Google Scholar] [CrossRef]
- Swain, B.; Basu, M.; Samanta, M. NOD1 and NOD2 Receptors in Mrigal (Cirrhinus mrigala): Inductive Expression and Downstream Signalling in Ligand Stimulation and Bacterial Infections. J. Biosci. 2013, 38, 533–548. [Google Scholar] [CrossRef]
- Li, J.; Kong, L.; Gao, Y.; Wu, C.; Xu, T. Characterization of NLR-A Subfamily Members in Miiuy Croaker and Comparative Genomics Revealed NLRX1 Underwent Duplication and Lose in Actinopterygii. Fish Shellfish Immunol. 2015, 47, 397–406. [Google Scholar] [CrossRef]
- Aplin, A.C.; Ligresti, G.; Fogel, E.; Zorzi, P.; Smith, K.; Nicosia, R.F. Regulation of Angiogenesis, Mural Cell Recruitment and Adventitial Macrophage Behavior by Toll-like Receptors. Angiogenesis 2014, 17, 147–161. [Google Scholar] [CrossRef]
- Ni, H.; Zhao, W.; Kong, X.; Li, H.; Ouyang, J. Celastrol Inhibits Lipopolysaccharide-Induced Angiogenesis by Suppressing TLR4-Triggered Nuclear Factor-Kappa B Activation. Acta Haematol. 2014, 131, 102–111. [Google Scholar] [CrossRef]
- Eggestøl, H.; Lunde, H.S.; Knutsen, T.M.; Haugland, G.T. Interleukin-1 Ligands and Receptors in Lumpfish (Cyclopterus lumpus L.): Molecular Characterization, Phylogeny, Gene Expression, and Transcriptome Analyses. Front. Immunol. 2020, 11, 502. [Google Scholar] [CrossRef]
- Pelegrín, P.; García-Castillo, J.; Mulero, V.; Meseguer, J. Interleukin-1β Isolated from a Marine Fish Reveals up-Regulated Expression in Macrophages Following Activation with Lipopolysaccharide and Lymphokines. Cytokine 2001, 16, 67–72. [Google Scholar] [CrossRef] [Green Version]
- Abos, B.; Wang, T.; Castro, R.; Granja, A.G.; Leal, E.; Havixbeck, J.; Luque, A.; Barreda, D.R.; Secombes, C.J.; Tafalla, C. Distinct Differentiation Programs Triggered by IL-6 and LPS in Teleost IgM+ B Cells in the Absence of Germinal Centers. Sci. Rep. 2016, 6, 30004. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, P.; Bao, B.; He, Q.; Peatman, E.; He, C.; Liu, Z. Characterization and Expression Analysis of Bactericidal Permeability-Increasing Protein (BPI) Antimicrobial Peptide Gene from Channel Catfish Ictalurus punctatus. Dev. Comp. Immunol. 2005, 29, 865–878. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.; Liang, Y.; Jiang, Y.; Liu, S.; Zhang, F.; He, X.; Wang, T.; Zhou, Y.; Zhong, H.; Yan, J. Identification and Expression Analysis on Bactericidal Permeability-Increasing Protein/Lipopolysaccharide-Binding Protein of Blunt Snout Bream, Megalobrama amblycephala. Fish Shellfish Immunol. 2015, 45, 630–640. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wang, S.; Qi, J.; Echtenkamp, S.F.; Chatterjee, R.; Wang, M.; Boons, G.-J.; Dziarski, R.; Gupta, D. Zebrafish Peptidoglycan Recognition Proteins Are Bactericidal Amidases Essential for Defense against Bacterial Infections. Immunity 2007, 27, 518–529. [Google Scholar] [CrossRef] [Green Version]
- Zhang, F.; Shan, S.; Xu, X.; Wang, Y.; Zhang, Y.; Yin, M.; Yang, G. Molecular Characterization and Expression Analysis of Two Peptidoglycan Recognition Proteins (CcPGRP5, CcPGRP6) in Larvae Ontogeny of Common Carp Cyprinus carpio L. and upon Immune Stimulation by Bacteria. BMC Vet. Res. 2019, 15, 10. [Google Scholar] [CrossRef] [Green Version]
- Briolat, V.; Jouneau, L.; Carvalho, R.; Palha, N.; Langevin, C.; Herbomel, P.; Schwartz, O.; Spaink, H.P.; Levraud, J.-P.; Boudinot, P. Contrasted Innate Responses to Two Viruses in Zebrafish: Insights into the Ancestral Repertoire of Vertebrate IFN-Stimulated Genes. J. Immunol. 2014, 192, 4328–4341. [Google Scholar] [CrossRef] [Green Version]
- Wei, X.; Li, B.; Wu, L.; Yin, X.; Zhong, X.; Li, Y.; Wang, Y.; Guo, Z.; Ye, J. Interleukin-6 Gets Involved in Response to Bacterial Infection and Promotes Antibody Production in Nile Tilapia (Oreochromis niloticus). Dev. Comp. Immunol. 2018, 89, 141–151. [Google Scholar] [CrossRef]
- Liu, Q.-N.; Xin, Z.-Z.; Chai, X.-Y.; Jiang, S.-H.; Li, C.-F.; Zhang, H.-B.; Ge, B.-M.; Zhang, D.-Z.; Zhou, C.-L.; Tang, B.-P. Characterization of Immune-Related Genes in the Yellow Catfish Pelteobagrus fulvidraco in Response to LPS Challenge. Fish Shellfish Immunol. 2016, 56, 248–254. [Google Scholar] [CrossRef]
- Gauthier, A.E.; Chandler, C.E.; Poli, V.; Gardner, F.M.; Tekiau, A.; Smith, R.; Bonham, K.S.; Cordes, E.E.; Shank, T.M.; Zanoni, I.; et al. Deep-Sea Microbes as Tools to Refine the Rules of Innate Immune Pattern Recognition. Sci. Immunol. 2021, 6, eabe0531. [Google Scholar] [CrossRef]
- Gurung, P.; Lukens, J.R.; Kanneganti, T.-D. Mitochondria: Diversity in the Regulation of the NLRP3 Inflammasome. Trends Mol. Med. 2015, 21, 193–201. [Google Scholar] [CrossRef] [Green Version]
- Li, J.-Y.; Wang, Y.-Y.; Shao, T.; Fan, D.-D.; Lin, A.-F.; Xiang, L.-X.; Shao, J.-Z. The Zebrafish NLRP3 Inflammasome Has Functional Roles in ASC-Dependent Interleukin-1β Maturation and Gasdermin E–Mediated Pyroptosis. J. Biol. Chem. 2020, 295, 1120–1141. [Google Scholar] [CrossRef]
- Reyes-Cerpa, S.; Maisey, K.; Reyes-Lpez, F.; Toro-Ascuy, D.; Mara, A.; Imarai, M. Fish Cytokines and Immune Response. In New Advances and Contributions to Fish Biology; IntechOpen: London, UK, 2012. [Google Scholar] [CrossRef] [Green Version]
- Lulijwa, R.; Alfaro, A.C.; Merien, F.; Burdass, M.; Venter, L.; Young, T. In Vitro Immune Response of Chinook Salmon (Oncorhynchus tshawytscha) Peripheral Blood Mononuclear Cells Stimulated by Bacterial Lipopolysaccharide. Fish Shellfish Immunol. 2019, 94, 190–198. [Google Scholar] [CrossRef] [PubMed]
- Ko, E.Y.; Cho, S.H.; Kwon, S.H.; Eom, C.Y.; Jeong, M.S.; Lee, W.W.; Kim, S.Y.; Heo, S.J.; Ahn, G.; Lee, K.P.; et al. The Roles of NF-ΚB and ROS in Regulation of pro-Inflammatory Mediators of Inflammation Induction in LPS-Stimulated Zebrafish Embryos. Fish Shellfish Immunol. 2017, 68, 525–529. [Google Scholar] [CrossRef] [PubMed]
- Maekawa, S.; Pulpipat, T.; Wang, P.C.; Chen, S.C. Transcriptome Analysis of Immune- and Iron-Related Genes after Francisella noatunensis Subsp. Orientalis Infection in Nile Tilapia (Oreochromis niloticus). Fish Shellfish Immunol. 2021, 111, 36–48. [Google Scholar] [CrossRef]
- Qin, C.; Zhao, W.; Deng, D.; Lu, R.; Yang, G.; Yan, X.; Meng, X.; Nie, G. Effects of Glucose, Insulin, Glucagon, LPS and Poly (I:C) on Adiponectin Receptors Expression in Grass Carp (Ctenophcuyngodon idellus). Aquaculture 2020, 528, 735501. [Google Scholar] [CrossRef]
- Xie, J.; Chen, X.; He, X.; Niu, J. Cloning and Expression Analysis of Keap1 of Golden Pompano (Trachinotus ovatus) and Response to Oxidized Fish Oil and LPS Administration. Aquac. Rep. 2020, 18, 100527. [Google Scholar] [CrossRef]
- Congleton, J.L.; Wagner, E.J. Acute-Phase Hypoferremic Response to Lipopolysaccharide in Rainbow Trout (Oncorhynchus mykiss). Comp. Biochem. Physiol.-Part A Physiol. 1991, 98, 195–200. [Google Scholar] [CrossRef]
- Freedman, A.H.; Clamp, M.; Sackton, T.B. Error, Noise and Bias in de Novo Transcriptome Assemblies. Mol. Ecol. Resour. 2021, 21, 18–29. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).