Transcriptome Sequencing Analysis Reveals Dynamic Changes in Major Biological Functions during the Early Development of Clearhead Icefish, Protosalanx chinensis
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. RNA Extraction, cDNA Library Construction, and Sequencing
2.3. Genome Mapping and Quantification of Gene Expression Levels
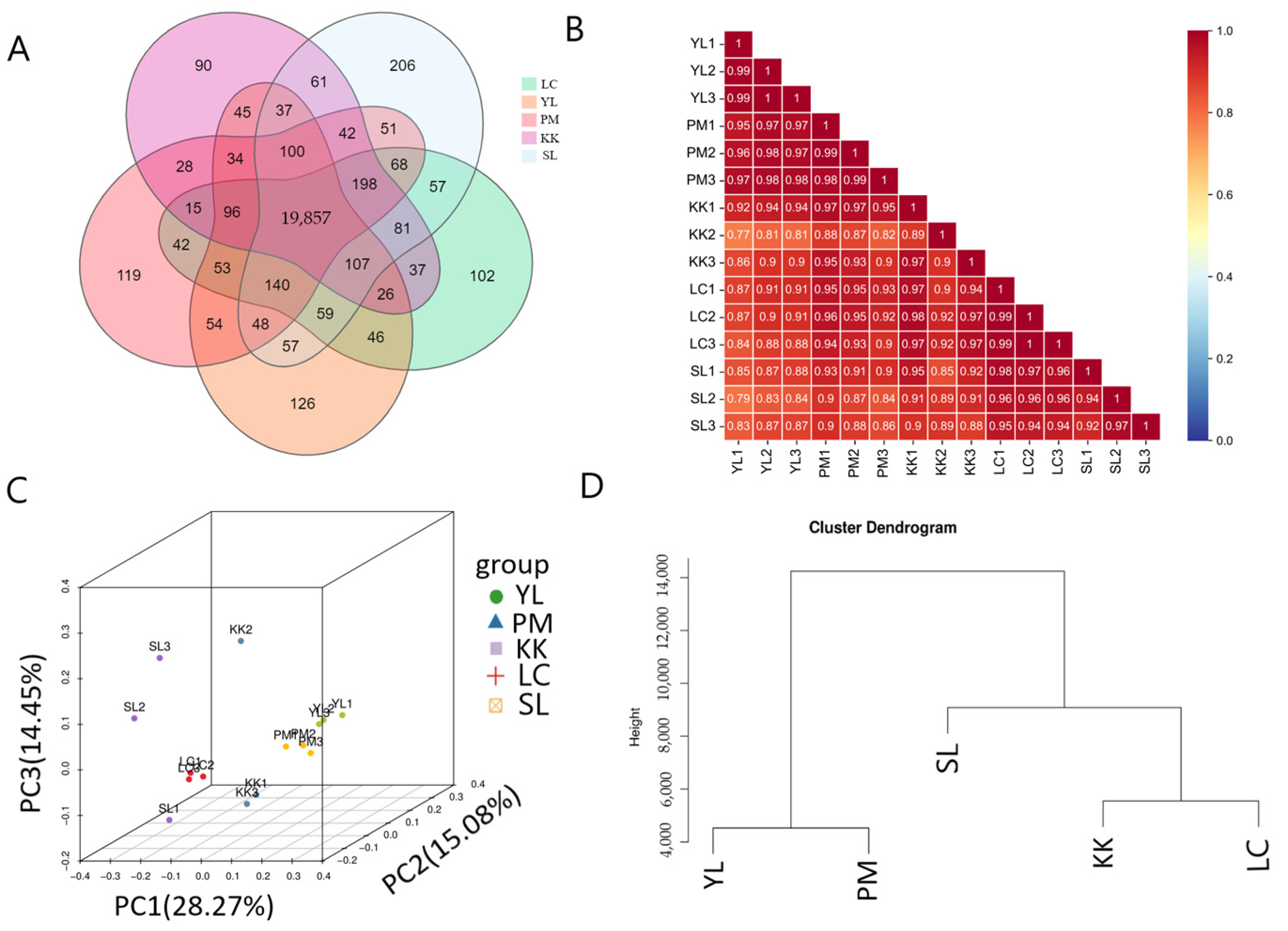
2.4. Sample Correlation and Clustering Analysis
2.5. Trend Analysis of Gene Expression and Weighted Gene Co-Expression Network Analysis (WGCNA)
2.6. Functional Enrichment Analysis
2.7. Identification and Visualization of Hub Genes
2.8. Quantitative Real-Time PCR (qPCR)
3. Results
3.1. Overview of Transcriptome Data of Every Developmental Stage
3.2. Sample Correlation and Cluster Analysis
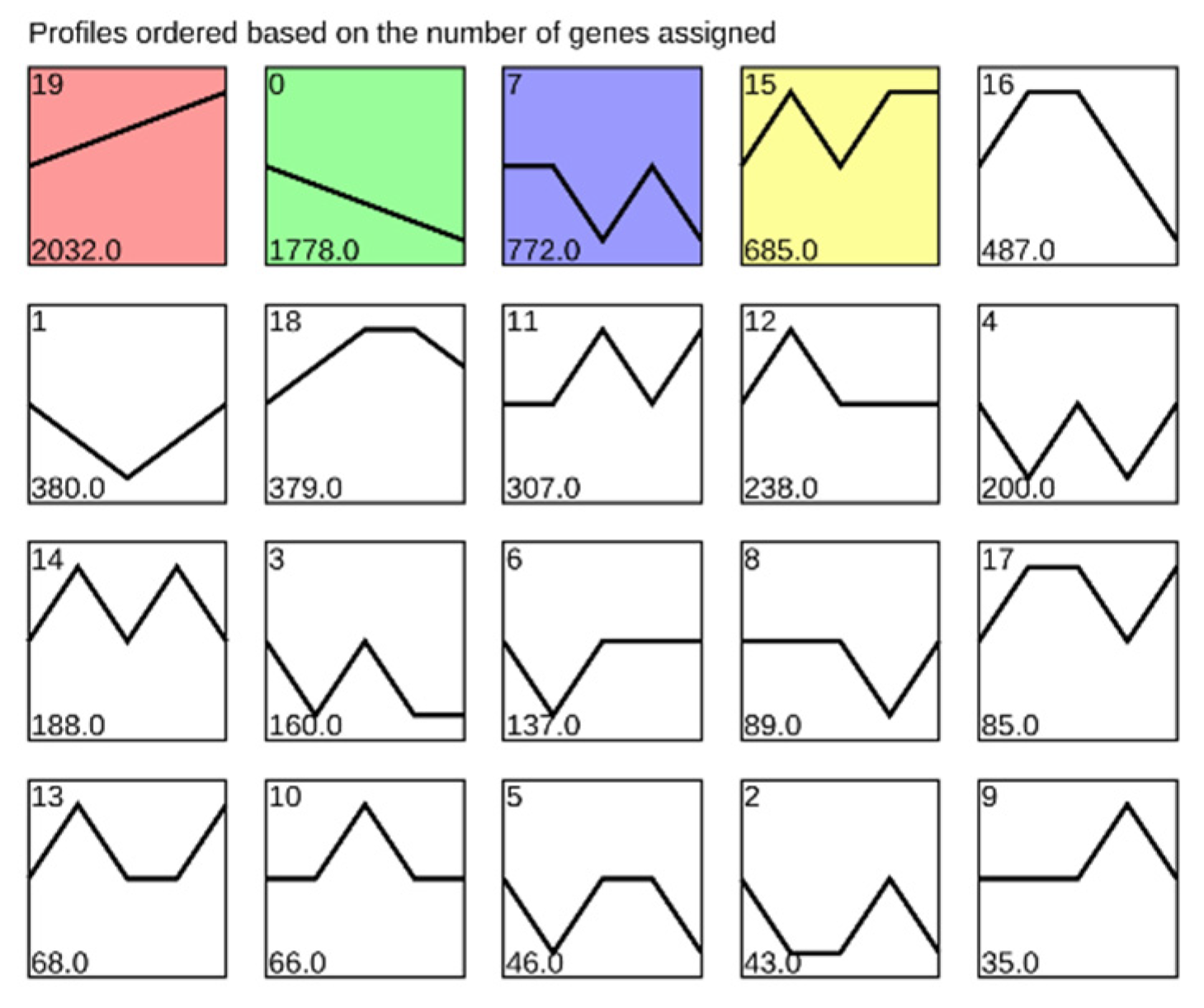
3.3. Gene Expression Trend Analysis and Enrichment Analysis during Early Development of Clearhead Icefish
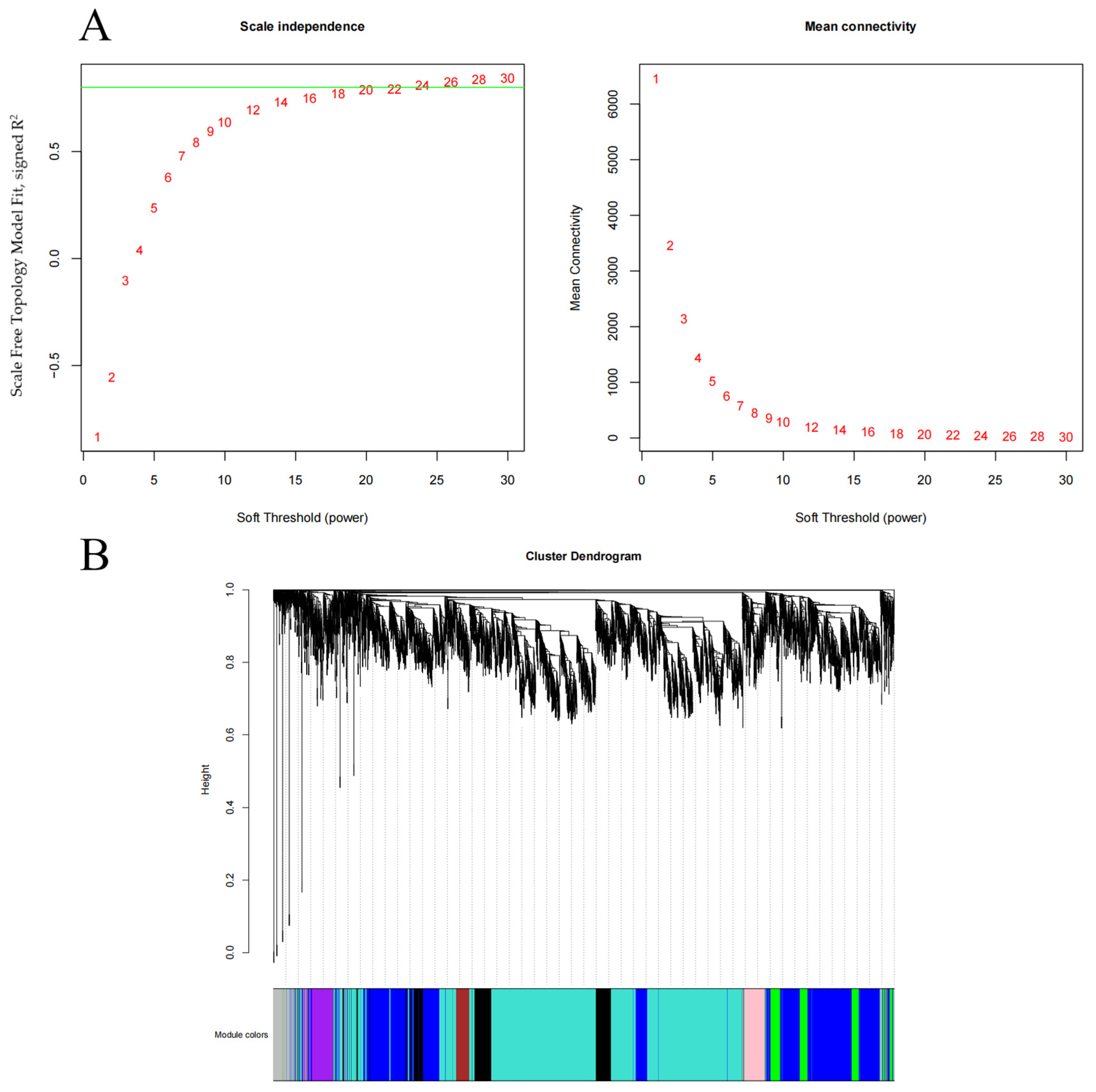
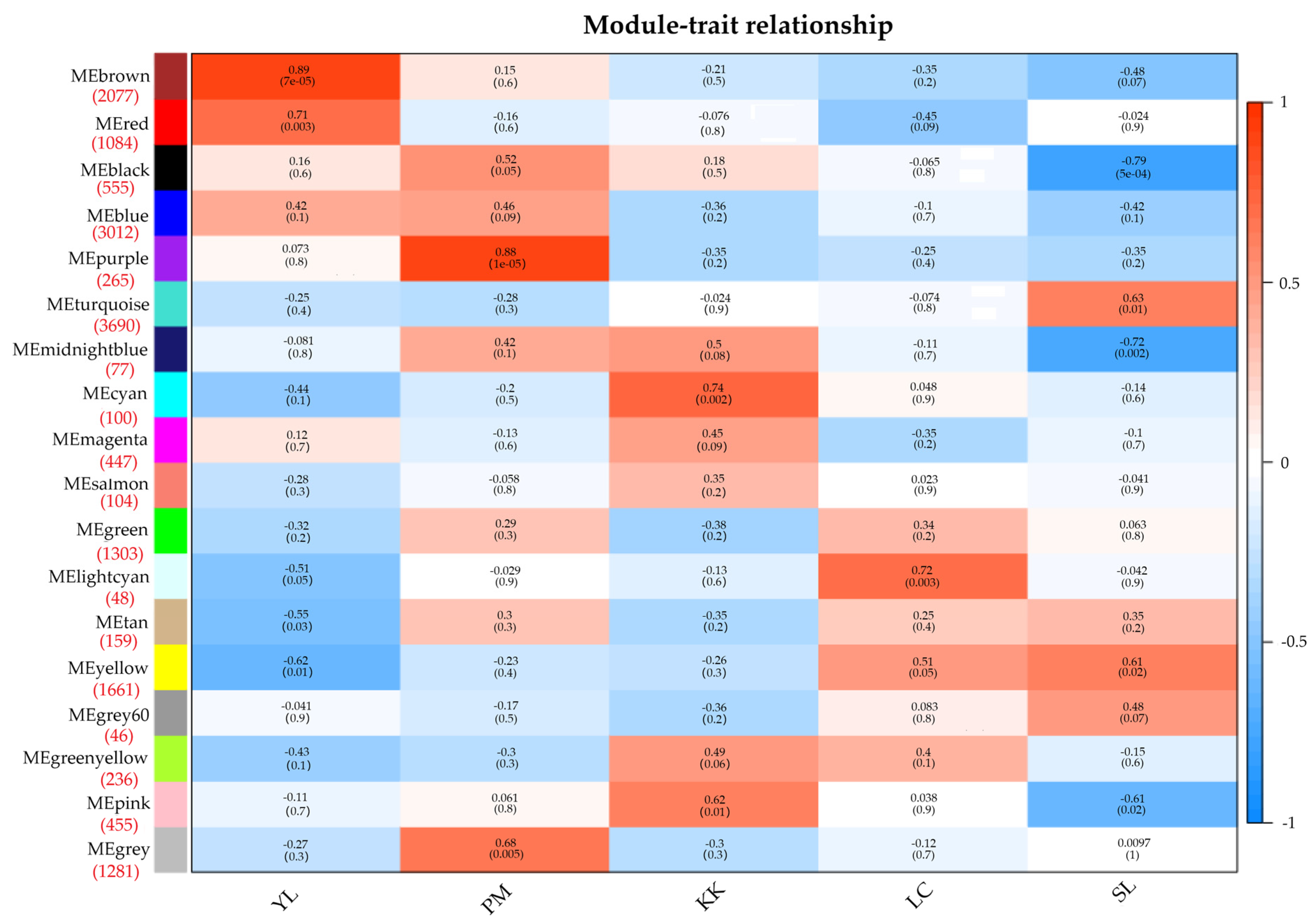
3.4. Weighted Gene Co-Expression Network Analysis and Identification of Modules Related to Early Development
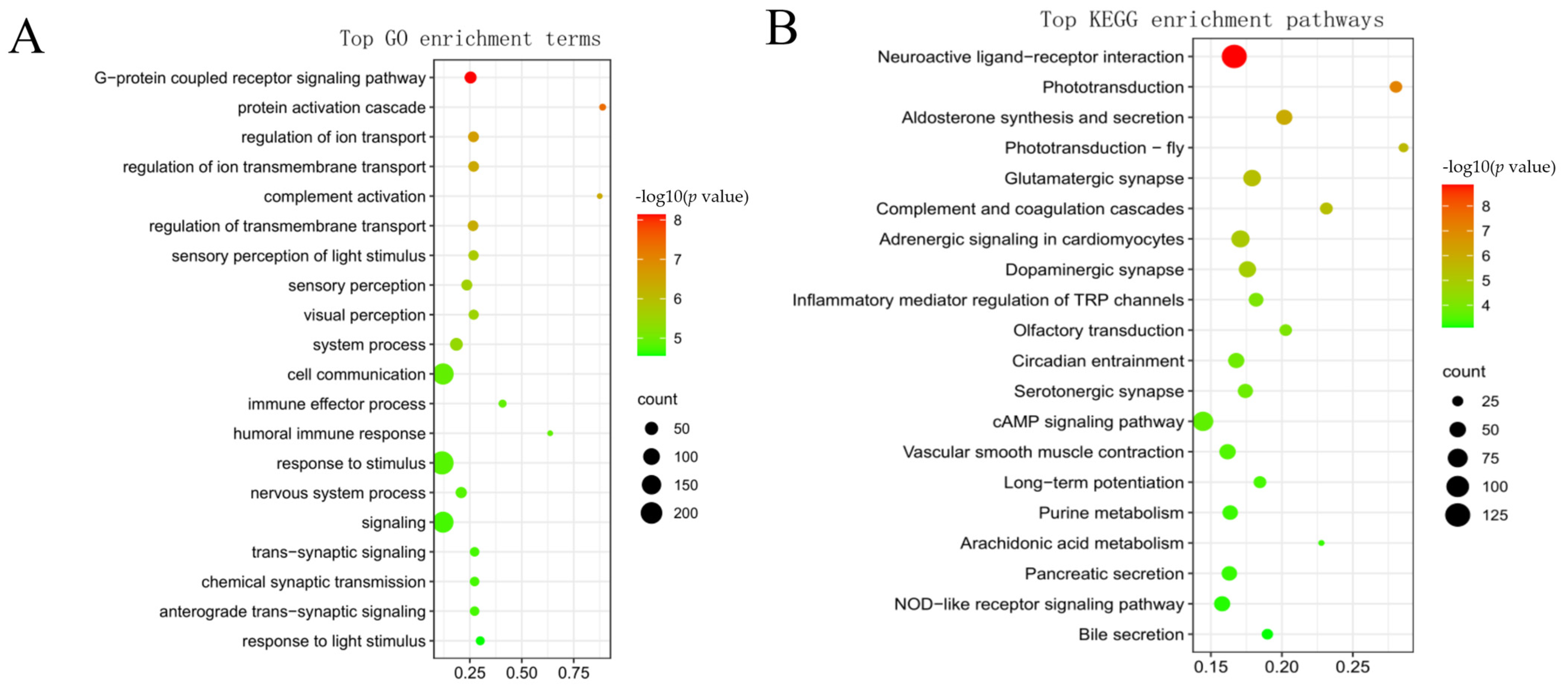
3.5. GO and KEGG Enrichment Analyses of Genes in Each Target Module
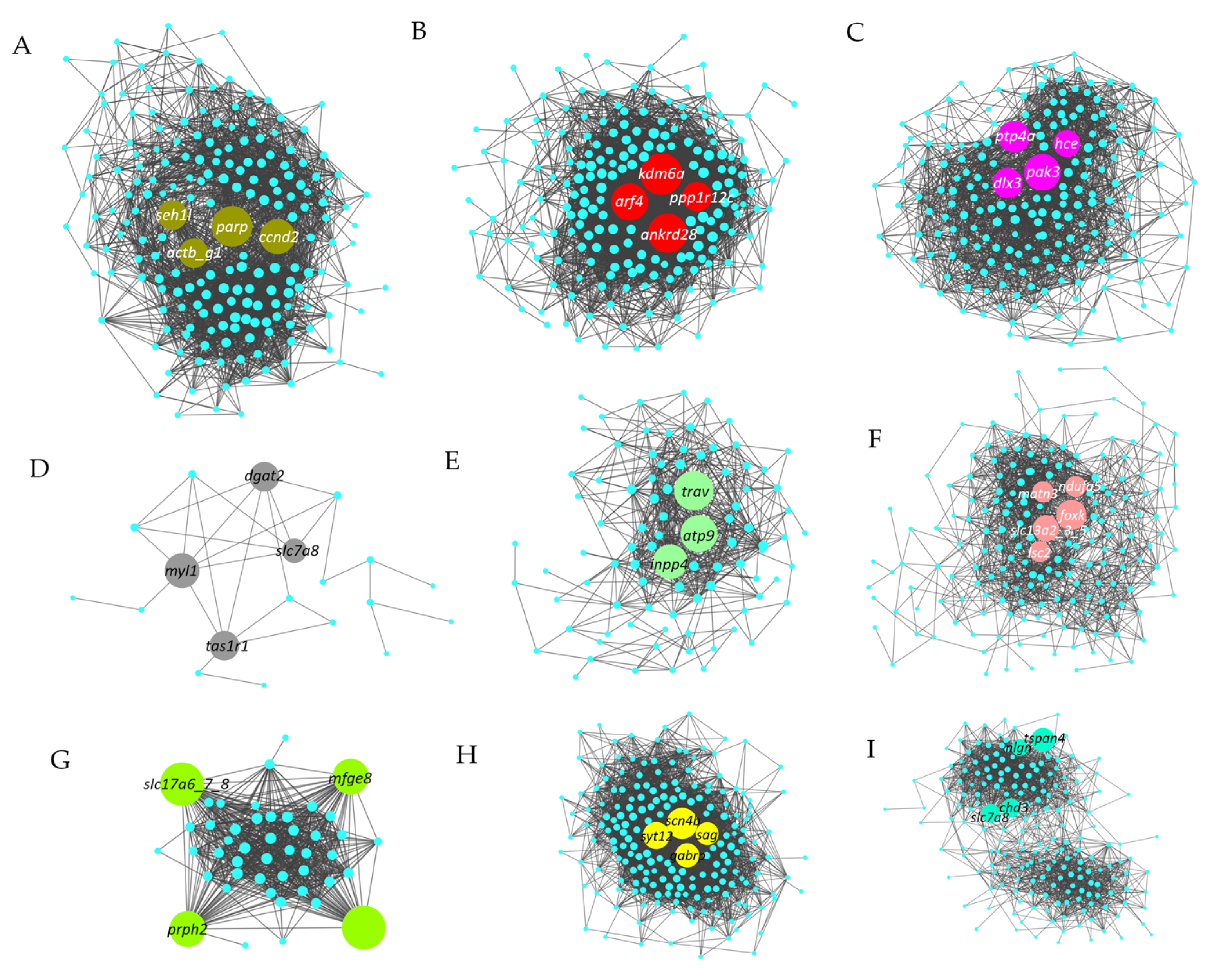
3.6. Identification and Visualization of Hub Genes
4. Discussion
4.1. G Protein-Coupled Receptors Play an Important Role in Early Development of P. chinensis
4.2. Exploration of the Main Biological Functions of the Early Developmental Stages of Clearhead Icefish
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Guo, L. Study on Molecular Phylogeny of Salangidae (Osmeriformes). Master’s Thesis, Fudan University, Shanghai, China, 2014. [Google Scholar]
- Zhang, J.; Qi, J.; Shi, F.; Pan, H.; Liu, M.; Tian, R.; Geng, Y.; Li, H.; Qu, Y.; Chen, J.; et al. Insights into the Evolution of Neoteny from the Genome of the Asian Icefish Protosalanx Chinensis. iScience 2020, 23, 101267. [Google Scholar] [CrossRef] [PubMed]
- Tang, F.; Gao, W.; Li, H.; Liu, W. Biology and fishery ecology of Protosalanx chinensis: A review. J. Fish. China 2020, 44, 2100–2111. [Google Scholar] [CrossRef]
- Wang, Y.; Jiang, Q. Research of growth properties of Protosalanx hyalocranius abbott. J. Lake Sci. 1992, 4, 56–62. [Google Scholar]
- Zhang, K.; Zhuang, D.; Zhang, L.; Gao, L.; Zhang, J.; Xu, A. On the Protosalanx hyalocranius and its Propagation in Hongze HU. J. Fish. China 1981, 5, 29–38. [Google Scholar]
- Tang, F.; Liu, W.; Wang, J.; Li, Z.; Xie, S. Diet composition and transition of clearhead icefish (Protosalanx hyalocranius) in Lake Xingkai. Zool. Res. 2013, 34, 493–498. [Google Scholar] [CrossRef]
- Xie, Y.; Han, X. Classification, Distribution, and Population Ecology of Salangidae Fishes. Chin. J. Fish. 1997, 10, 11–19. [Google Scholar]
- Wang, Z.; Fu, C.; Lei, G. Biodiversity of Chinese Icefishes (Salangidae) and their conserving strategies. Biodiversity 2002, 10, 416–424. [Google Scholar] [CrossRef]
- Shi, W.; Xu, D.; Liu, K.; Duan, J.; Zhang, M. The embryonic development and habit of larvae in large icefish Protosalartx hyalocranius. J. Dalian Ocean Univ. 2011, 26, 391–395. [Google Scholar] [CrossRef]
- Zhang, K. Observation on embryonic development of Protosalanx hyalocranius. J. Lake Sci. 1992, 4, 25–37. [Google Scholar]
- Liu, K.; Xu, D.; Li, J.; Bian, C.; Duan, J.; Zhou, Y.; Zhang, M.; You, X.; You, Y.; Chen, J.; et al. Whole Genome Sequencing of Chinese Clearhead Icefish, Protosalanx hyalocranius. Gigascience 2017, 6, 1–6. [Google Scholar] [CrossRef]
- Granados-Riveron, J.T.; Aquino-Jarquin, G. CRISPR-Cas13 Precision Transcriptome Engineering in Cancer. Cancer Res. 2018, 78, 4107–4113. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; St John, M.A.R.; Zhou, X.; Kim, Y.; Sinha, U.; Jordan, R.C.K.; Eisele, D.; Abemayor, E.; Elashoff, D.; Park, N.-H.; et al. Salivary Transcriptome Diagnostics for Oral Cancer Detection. Clin. Cancer Res. 2004, 10, 8442–8450. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhao, L.; Yi, S.; Ding, F.; Yang, Y.; Liu, Y.; Wang, Y.; Liu, M.; Xue, C.; Xu, L.; et al. Developmental Temporal Patterns and Molecular Network Features in the Transcriptome of Rat Spinal Cord. Engineering 2021, 7, 1592–1602. [Google Scholar] [CrossRef]
- Graveley, B.R.; Brooks, A.N.; Carlson, J.W.; Duff, M.O.; Landolin, J.M.; Yang, L.; Artieri, C.G.; van Baren, M.J.; Boley, N.; Booth, B.W.; et al. The Developmental Transcriptome of Drosophila Melanogaster. Nature 2011, 471, 473–479. [Google Scholar] [CrossRef] [PubMed]
- Winfield, M.O.; Lu, C.; Wilson, I.D.; Coghill, J.A.; Edwards, K.J. Plant Responses to Cold: Transcriptome Analysis of Wheat: Plant Responses to Cold. Plant Biotechnol. J. 2010, 8, 749–771. [Google Scholar] [CrossRef] [PubMed]
- Dirks, R.P.; Burgerhout, E.; Brittijn, S.A.; de Wijze, D.L.; Ozupek, H.; Tuinhof-Koelma, N.; Minegishi, Y.; Jong-Raadsen, S.A.; Spaink, H.P.; van den Thillart, G.E.E.J.M. Identification of Molecular Markers in Pectoral Fin to Predict Artificial Maturation of Female European Eels (Anguilla anguilla). Gen. Comp. Endocrinol. 2014, 204, 267–276. [Google Scholar] [CrossRef] [PubMed]
- Vesterlund, L.; Jiao, H.; Unneberg, P.; Hovatta, O.; Kere, J. The Zebrafish Transcriptome during Early Development. BMC Dev. Biol. 2011, 11, 30. [Google Scholar] [CrossRef]
- Xu, H.; Liu, E.; Li, Y.; Li, X.; Ding, C. Transcriptome Analysis Reveals Increases in Visceral Lipogenesis and Storage and Activation of the Antigen Processing and Presentation Pathway during the Mouth-Opening Stage in Zebrafish Larvae. Int. J. Mol. Sci. 2017, 18, 1634. [Google Scholar] [CrossRef]
- Fu, J.; Zhu, W.; Wang, L.; Luo, M.; Song, F.; Dong, Z. Dynamic Transcriptome Sequencing and Analysis during Early Development in the Bighead Carp (Hypophthalmichthys nobilis). BMC Genom. 2019, 20, 781. [Google Scholar] [CrossRef]
- Yang, H.; Zhou, Y.; Gu, J.; Xie, S.; Xu, Y.; Zhu, G.; Wang, L.; Huang, J.; Ma, H.; Yao, J. Deep MRNA Sequencing Analysis to Capture the Transcriptome Landscape of Zebrafish Embryos and Larvae. PLoS ONE 2013, 8, e64058. [Google Scholar] [CrossRef]
- Ferraresso, S.; Bonaldo, A.; Parma, L.; Cinotti, S.; Massi, P.; Bargelloni, L.; Gatta, P.P. Exploring the Larval Transcriptome of the Common Sole (Solea Solea L.). BMC Genom. 2013, 14, 315. [Google Scholar] [CrossRef]
- Han, X.; Zhang, L.; Niu, D.; Nan, S.; Miao, X.; Hu, X.; Li, C.; Fu, H. Transcriptome and Co-Expression Network Analysis Reveal Molecular Mechanisms of Mucilage Formation during Seed Development in Artemisia Sphaerocephala. Carbohydr. Polym. 2021, 251, 117044. [Google Scholar] [CrossRef]
- Amiri, A.; Coppola, G.; Scuderi, S.; Wu, F.; Roychowdhury, T.; Liu, F.; Pochareddy, S.; Shin, Y.; Safi, A.; Song, L.; et al. Transcriptome and Epigenome Landscape of Human Cortical Development Modeled in Organoids. Science 2018, 362, eaat6720. [Google Scholar] [CrossRef]
- Ding, H.; Lin, Y.; Zhang, T.; Chen, L.; Zhang, G.; Wang, J.; Xie, K.; Dai, G. Transcriptome Analysis of Differentially Expressed MRNA Related to Pigeon Muscle Development. Animals 2021, 11, 2311. [Google Scholar] [CrossRef]
- Ling, Y.; Zheng, Q.; Jing, J.; Sui, M.; Zhu, L.; Li, Y.; Zhang, Y.; Liu, Y.; Fang, F.; Zhang, X. Switches in Transcriptome Functions during Seven Skeletal Muscle Development Stages from Fetus to Kid in Capra Hircus. J. Integr. Agric. 2021, 20, 212–226. [Google Scholar] [CrossRef]
- Yúfera, M.; Darias, M.J. The Onset of Exogenous Feeding in Marine Fish Larvae. Aquaculture 2007, 268, 53–63. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A Fast Spliced Aligner with Low Memory Requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast Gapped-Read Alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate Transcript Quantification from RNA-Seq Data with or without a Reference Genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R Package for Weighted Correlation Network Analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Han, J.-D.J.; Bertin, N.; Hao, T.; Goldberg, D.S.; Berriz, G.F.; Zhang, L.V.; Dupuy, D.; Walhout, A.J.M.; Cusick, M.E.; Roth, F.P.; et al. Evidence for Dynamically Organized Modularity in the Yeast Protein–Protein Interaction Network. Nature 2004, 430, 88–93. [Google Scholar] [CrossRef]
- Song, H. The Reaserch of Protosalanx Hyalocranius Embryo’s Molecular Adaption Mechanism to Salt Stress Based on Transcriptome Sequencing Technology. Master’s Degree, Shanxi Agricultural University, Shanxi, China, 2014. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Pedersen, B.H.; Nilssen, E.M.; Hjelmeland, K. Variations in the Content of Trypsin and Trysinogen in Larval Herring (Clupea harengus) Digesting Copepod Nauplii. Mar. Biol. 1987, 94, 171–181. [Google Scholar] [CrossRef]
- García-Gasca, A.; Galaviz, M.A.; Gutiérrez, J.N.; García-Ortega, A. Development of the Digestive Tract, Trypsin Activity and Gene Expression in Eggs and Larvae of the Bullseye Puffer Fish Sphoeroides annulatus. Aquaculture 2006, 251, 366–376. [Google Scholar] [CrossRef]
- Srichanun, M.; Tantikitti, C.; Utarabhand, P.; Kortner, T.M. Gene Expression and Activity of Digestive Enzymes during the Larval Development of Asian Seabass (Lates calcarifer). Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2013, 165, 1–9. [Google Scholar] [CrossRef]
- Kortner, T.M.; Overrein, I.; Øie, G.; Kjørsvik, E.; Bardal, T.; Wold, P.-A.; Arukwe, A. Molecular Ontogenesis of Digestive Capability and Associated Endocrine Control in Atlantic Cod (Gadus morhua) Larvae. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2011, 160, 190–199. [Google Scholar] [CrossRef]
- Takahashi, A.; Kobayashi, Y.; Amano, M.; Yamanome, T. Structural and Functional Diversity of Proopiomelanocortin in Fish with Special Reference to Barfin Flounder. Peptides 2009, 30, 1374–1382. [Google Scholar] [CrossRef]
- Butler, A.A. The Melanocortin System and Energy Balance. Peptides 2006, 27, 281–290. [Google Scholar] [CrossRef]
- López-Patiño, M.A.; Guijarro, A.I.; Isorna, E.; Delgado, M.J.; Alonso-Bedate, M.; de Pedro, N. Neuropeptide Y Has a Stimulatory Action on Feeding Behavior in Goldfish (Carassius auratus). Eur. J. Pharmacol. 1999, 377, 147–153. [Google Scholar] [CrossRef]
- Cerdá-Reverter, J.M.; Sorbera, L.A.; Carrillo, M.; Zanuy, S. Energetic Dependence of NPY-Induced LH Secretion in a Teleost Fish (Dicentrarchus Labrax). Am. J. Physiol. 1999, 277, R1627–R1634. [Google Scholar] [CrossRef]
- Cerdá-Reverter, J.M.; Anglade, I.; Martínez-Rodríguez, G.; Mazurais, D.; Muñoz-Cueto, J.A.; Carrillo, M.; Kah, O.; Zanuy, S. Characterization of Neuropeptide Y Expression in the Brain of a Perciform Fish, the Sea Bass (Dicentrarchus labrax). J. Chem. Neuroanat. 2000, 19, 197–210. [Google Scholar] [CrossRef]
- Yang, L.; Sun, C.; Li, W. Neuropeptide B in Nile Tilapia Oreochromis Niloticus: Molecular Cloning and Its Effects on the Regulation of Food Intake and MRNA Expression of Growth Hormone and Prolactin. Gen. Comp. Endocrinol. 2014, 200, 27–34. [Google Scholar] [CrossRef]
- Matsuda, K. Regulation of Feeding Behavior and Psychomotor Activity by Corticotropin-Releasing Hormone (CRH) in Fish. Front. Neurosci. 2013, 7, 91. [Google Scholar] [CrossRef]
- McGowan, B.M.C.; Stanley, S.A.; Smith, K.L.; White, N.E.; Connolly, M.M.; Thompson, E.L.; Gardiner, J.V.; Murphy, K.G.; Ghatei, M.A.; Bloom, S.R. Central Relaxin-3 Administration Causes Hyperphagia in Male Wistar Rats. Endocrinology 2005, 146, 3295–3300. [Google Scholar] [CrossRef]
- Assou, S.; Boumela, I.; Haouzi, D.; Anahory, T.; Dechaud, H.; De Vos, J.; Hamamah, S. Dynamic Changes in Gene Expression during Human Early Embryo Development: From Fundamental Aspects to Clinical Applications. Hum. Reprod. Update 2010, 17, 272–290. [Google Scholar] [CrossRef]
- Ko, M.S.H. Expression Profiling of the Mouse Early Embryo: Reflections and Perspectives. Dev. Dyn. 2006, 235, 2437–2448. [Google Scholar] [CrossRef][Green Version]
- D’Amato, G.; Luxán, G.; del Monte-Nieto, G.; Martínez-Poveda, B.; Torroja, C.; Walter, W.; Bochter, M.S.; Benedito, R.; Cole, S.; Martinez, F.; et al. Sequential Notch Activation Regulates Ventricular Chamber Development. Nat. Cell Biol. 2016, 18, 7–20. [Google Scholar] [CrossRef]
- Xiong, H.; Huang, Y.; Mao, Y.; Liu, C.; Wang, J. Inhibition in Growth and Cardiotoxicity of Tris (2-Butoxyethyl) Phosphate through down-Regulating Wnt Signaling Pathway in Early Developmental Stage of Zebrafish (Danio rerio). Ecotoxicol. Environ. Saf. 2021, 208, 111431. [Google Scholar] [CrossRef]
- Duan, P.; Bonewald, L.F. The Role of the Wnt/β-Catenin Signaling Pathway in Formation and Maintenance of Bone and Teeth. Int. J. Biochem. Cell Biol. 2016, 77, 23–29. [Google Scholar] [CrossRef]
- Fairbairn, E.A.; Bonthius, J.; Cherr, G.N. Polycyclic Aromatic Hydrocarbons and Dibutyl Phthalate Disrupt Dorsal–Ventral Axis Determination via the Wnt/β-Catenin Signaling Pathway in Zebrafish Embryos. Aquat. Toxicol. 2012, 124–125, 188–196. [Google Scholar] [CrossRef]
- Goessling, W.; North, T.E.; Lord, A.M.; Ceol, C.; Lee, S.; Weidinger, G.; Bourque, C.; Strijbosch, R.; Haramis, A.-P.; Puder, M.; et al. APC Mutant Zebrafish Uncover a Changing Temporal Requirement for Wnt Signaling in Liver Development. Dev. Biol. 2008, 320, 161–174. [Google Scholar] [CrossRef]
- Liu, Z.; Guo, S.; Sun, H.; Bai, Y.; Song, Z.; Liu, X. Circular RNA CircHIPK3 Elevates CCND2 Expression and Promotes Cell Proliferation and Invasion Through MiR-124 in Glioma. Front. Genet. 2020, 11, 1013. [Google Scholar] [CrossRef]
- Platani, M.; Samejima, I.; Samejima, K.; Kanemaki, M.T.; Earnshaw, W.C. Seh1 Targets GATOR2 and Nup153 to Mitotic Chromosomes. J. Cell Sci. 2018, 131, jcs213140. [Google Scholar] [CrossRef]
- Tachibana, M.; Kiyokawa, E.; Hara, S.; Iemura, S.-I.; Natsume, T.; Manabe, T.; Matsuda, M. Ankyrin Repeat Domain 28 (ANKRD28), a Novel Binding Partner of DOCK180, Promotes Cell Migration by Regulating Focal Adhesion Formation. Exp. Cell Res. 2009, 315, 863–876. [Google Scholar] [CrossRef]
- Li, X.; Duan, Y.; Hao, Y. Identification of Super Enhancer-Associated Key Genes for Prognosis of Germinal Center B-Cell Type Diffuse Large B-Cell Lymphoma by Integrated Analysis. BMC Med. Genom. 2021, 14, 69. [Google Scholar] [CrossRef]
- Follit, J.A.; San Agustin, J.T.; Jonassen, J.A.; Huang, T.; Rivera-Perez, J.A.; Tremblay, K.D.; Pazour, G.J. Arf4 Is Required for Mammalian Development but Dispensable for Ciliary Assembly. PLoS Genet. 2014, 10, e1004170. [Google Scholar] [CrossRef]
- Lan, F.; Bayliss, P.E.; Rinn, J.L.; Whetstine, J.R.; Wang, J.K.; Chen, S.; Iwase, S.; Alpatov, R.; Issaeva, I.; Canaani, E.; et al. A Histone H3 Lysine 27 Demethylase Regulates Animal Posterior Development. Nature 2007, 449, 689–694. [Google Scholar] [CrossRef]
- Pascolini, G.; Gaudioso, F.; Passarelli, C.; Novelli, A.; Di Giosaffatte, N.; Majore, S.; Grammatico, P. Clinical and Molecular Aspects of the Neurodevelopmental Disorder Associated with PAK3 Perturbation. J. Mol. Neurosci. 2021, 71, 2474–2481. [Google Scholar] [CrossRef]
- Zhao, N.; Han, D.; Liu, Y.; Li, Y.; Zeng, L.; Wang, Y.; Feng, H. DLX3 Negatively Regulates Osteoclastic Differentiation through MicroRNA-124. Exp. Cell Res. 2016, 341, 166–176. [Google Scholar] [CrossRef]
- Pang, L.; Zhang, Z.; Shen, Y.; Cheng, Z.; Gao, X.; Zhang, B.; Wang, X.; Tian, H. Mutant Dlx3b Disturbs Normal Tooth Mineralization and Bone Formation in Zebrafish. PeerJ 2020, 8, e8515. [Google Scholar] [CrossRef]
- Gunter, H.M.; Koppermann, C.; Meyer, A. Revisiting de Beer’s Textbook Example of Heterochrony and Jaw Elongation in Fish: Calmodulin Expression Reflects Heterochronic Growth, and Underlies Morphological Innovation in the Jaws of Belonoid Fishes. EvoDevo 2014, 5, 8. [Google Scholar] [CrossRef]
- Caprio, J.; Brand, J.G.; Teeter, J.H.; Valentincic, T.; Kalinoski, D.L.; Kohbara, J.; Kumazawa, T.; Wegert, S. The Taste System of the Channel Catfish: From Biophysics to Behavior. Trends Neurosci. 1993, 16, 192–197. [Google Scholar] [CrossRef]
- Barreiro-Iglesias, A.; Anadón, R.; Rodicio, M.C. The Gustatory System of Lampreys. Brain Behav. Evol. 2010, 75, 241–250. [Google Scholar] [CrossRef]
- Nakamura, T.; Matsuyama, N.; Kirino, M.; Kasai, M.; Kiyohara, S.; Ikenaga, T. Distribution, Innervation, and Cellular Organization of Taste Buds in the Sea Catfish, Plotosus japonicus. Brain Behav. Evol. 2017, 89, 209–218. [Google Scholar] [CrossRef]
- Sato, J.J.; Wolsan, M. Loss or Major Reduction of Umami Taste Sensation in Pinnipeds. Naturwissenschaften 2012, 99, 655–659. [Google Scholar] [CrossRef]
- Calo, J.; Blanco, A.M.; Comesaña, S.; Conde-Sieira, M.; Morais, S.; Soengas, J.L. First Evidence for the Presence of Amino Acid Sensing Mechanisms in the Fish Gastrointestinal Tract. Sci. Rep. 2021, 11, 4933. [Google Scholar] [CrossRef]
- Eriksson, L.; Esberg, A.; Haworth, S.; Holgerson, P.L.; Johansson, I. Allelic Variation in Taste Genes Is Associated with Taste and Diet Preferences and Dental Caries. Nutrients 2019, 11, 1491. [Google Scholar] [CrossRef]
- Chitraju, C.; Walther, T.C.; Farese, R.V. The Triglyceride Synthesis Enzymes DGAT1 and DGAT2 Have Distinct and Overlapping Functions in Adipocytes. J. Lipid Res. 2019, 60, 1112–1120. [Google Scholar] [CrossRef]
- Bhatt-Wessel, B.; Jordan, T.W.; Miller, J.H.; Peng, L. Role of DGAT Enzymes in Triacylglycerol Metabolism. Arch. Biochem. Biophys. 2018, 655, 1–11. [Google Scholar] [CrossRef]
- Sukonina, V.; Ma, H.; Zhang, W.; Bartesaghi, S.; Subhash, S.; Heglind, M.; Foyn, H.; Betz, M.J.; Nilsson, D.; Lidell, M.E.; et al. FOXK1 and FOXK2 Regulate Aerobic Glycolysis. Nature 2019, 566, 279–283. [Google Scholar] [CrossRef] [PubMed]
- Bowman, C.J.; Ayer, D.E.; Dynlacht, B.D. Foxk Proteins Repress the Initiation of Starvation-Induced Atrophy and Autophagy Programs. Nat. Cell Biol. 2014, 16, 1202–1214. [Google Scholar] [CrossRef] [PubMed]
- Pajor, A.M. Sodium-Coupled Dicarboxylate and Citrate Transporters from the SLC13 Family. Pflug. Arch.-Eur. J. Physiol. 2014, 466, 119–130. [Google Scholar] [CrossRef]
- Rak, M.; Rustin, P. Supernumerary Subunits NDUFA3, NDUFA5 and NDUFA12 Are Required for the Formation of the Extramembrane Arm of Human Mitochondrial Complex I. FEBS Lett. 2014, 588, 1832–1838. [Google Scholar] [CrossRef] [PubMed]
- Phillips, D.; Aponte, A.M.; French, S.A.; Chess, D.J.; Balaban, R.S. Succinyl-CoA Synthetase Is a Phosphate Target for the Activation of Mitochondrial Metabolism. Biochemistry 2009, 48, 7140–7149. [Google Scholar] [CrossRef]
- Ottaway, J.H.; McClellan, J.A.; Saunderson, C.L. Succinic Thiokinase and Metabolic Control. Int. J. Biochem. 1981, 13, 401–410. [Google Scholar] [CrossRef]
- Akram, M. Citric Acid Cycle and Role of Its Intermediates in Metabolism. Cell Biochem. Biophys. 2014, 68, 475–478. [Google Scholar] [CrossRef]
- Ruzicka, J.J.; Gallager, S.M. The Importance of the Cost of Swimming to the Foraging Behavior and Ecology of Larval Cod (Gadus morhua) on Georges Bank. Deep Sea Res. Part II Top. Stud. Oceanogr. 2006, 53, 2708–2734. [Google Scholar] [CrossRef]
- Birkenfeld, A.L.; Lee, H.Y.; Guebre-Egziabher, F.; Alves, T.C.; Jurczak, M.J.; Jornayvaz, F.R.; Zhang, D.; Hsiao, J.J.; Martin-Montalvo, A.; Fischer-Rosinsky, A.; et al. Deletion of the Mammalian INDY Homolog Mimics Aspects of Dietary Restriction and Protects against Adiposity and Insulin Resistance in Mice. Cell Metabol. 2011, 14, 184–195. [Google Scholar] [CrossRef]
- Gao, J.; Xu, G.; Xu, P. Comparative Transcriptome Analysis Reveals Metabolism Transformation in Coilia Nasus Larvae during the Mouth-Open Period. Comp. Biochem. Physiol. Part D Genom. Proteom. 2020, 36, 100712. [Google Scholar] [CrossRef]
- Lu, Z.; Hu, X.; Reilly, J.; Jia, D.; Liu, F.; Yu, S.; Liu, X.; Xie, S.; Qu, Z.; Qin, Y.; et al. Deletion of the Transmembrane Protein Prom1b in Zebrafish Disrupts Outer-Segment Morphogenesis and Causes Photoreceptor Degeneration. J. Biol. Chem. 2019, 294, 13953–13963. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Hu, X.; Liu, F.; Soares, D.C.; Liu, X.; Yu, S.; Gao, M.; Han, S.; Qin, Y.; Li, C.; et al. Ablation of EYS in Zebrafish Causes Mislocalisation of Outer Segment Proteins, F-Actin Disruption and Cone-Rod Dystrophy. Sci. Rep. 2017, 7, 46098. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, O.N.P.; Böhm, S.; Gießl, A.; Butz, E.S.; Wolfrum, U.; Brandstätter, J.H.; Wahl-Schott, C.; Biel, M.; Becirovic, E. Peripherin-2 Differentially Interacts with Cone Opsins in Outer Segments of Cone Photoreceptors. Hum. Mol. Genet. 2016, 25, 2367–2377. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Stuck, M.W.; Conley, S.M.; Naash, M.I. PRPH2/RDS and ROM-1: Historical Context, Current Views and Future Considerations. Progr. Retin. Eye Res. 2016, 52, 47–63. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.-S.; Lee, C.-H.; Lee, Y.-D. Retinal Development and Opsin Gene Expression during the Juvenile Development in Red Spotted Grouper (Epinephelus akaara). Dev. Reprod. 2019, 23, 171–181. [Google Scholar] [CrossRef] [PubMed]
- Matsuura, R.; Sawada, Y.; Ishibashi, Y. Development of Visual Cells in the Pacific Bluefin Tuna Thunnus orientalis. Fish. Physiol. Biochem. 2010, 36, 391–402. [Google Scholar] [CrossRef]
- Miranda, V.; Cohen, S.; Díaz, A.O.; Diaz, M.V. Development of the Visual System of Anchovy Larvae, Engraulis anchoita: A Microanatomical Description. J. Morphol. 2020, 281, 465–475. [Google Scholar] [CrossRef]
- Butler, J.M.; Field, K.E.; Maruska, K.P. Cobalt Chloride Treatment Used to Ablate the Lateral Line System Also Impairs the Olfactory System in Three Freshwater Fishes. PLoS ONE 2016, 11, e0159521. [Google Scholar] [CrossRef]
- Howe, H.B.; McIntyre, P.B.; Wolman, M.A. Adult Zebrafish Primarily Use Vision to Guide Piscivorous Foraging Behavior. Behav. Processes 2018, 157, 230–237. [Google Scholar] [CrossRef]
- Chiu, L.; Chang, E. Role of Sensory Mechanisms in Predatory Feeding Behaviorof Juvenile Red Drum Sciaenops ocellatus. Fish. Sci. 2003, 69, 317–322. [Google Scholar] [CrossRef]
- Zhou, H.; Hylemon, P.B. Bile Acids Are Nutrient Signaling Hormones. Steroids 2014, 86, 62–68. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.-J.; Oh, D.-H.; Khosravi, S.; Cha, J.-H.; Park, S.-H.; Kim, K.-W.; Lee, K.-J. Taurine Is an Essential Nutrient for Juvenile Parrot Fish Oplegnathus fasciatus. Aquaculture 2013, 414–415, 274–279. [Google Scholar] [CrossRef]
- Kim, S.-K.; Matsunari, H.; Nomura, K.; Tanaka, H.; Yokoyama, M.; Murata, Y.; Ishihara, K.; Takeuchi, T. Effect of Dietary Taurine and Lipid Contents on Conjugated Bile Acid Composition and Growth Performance of Juvenile Japanese Flounder Paralichthys olivaceus. Fish. Sci. 2008, 74, 875–881. [Google Scholar] [CrossRef]
- Takagi, S.; Murata, H.; Goto, T.; Hatate, H.; Endo, M.; Yamashita, H.; Miyatake, H.; Ukawa, M. Necessity of Dietary Taurine Supplementation for Preventing Green Liver Symptom and Improving Growth Performance in Yearling Red Sea Bream Pagrus Major Fed Nonfishmeal Diets Based on Soy Protein Concentrate. Fish. Sci. 2009, 76, 119. [Google Scholar] [CrossRef]
- Gibson Gaylord, T.; Barrows, F.T.; Teague, A.M.; Johansen, K.A.; Overturf, K.E.; Shepherd, B. Supplementation of Taurine and Methionine to All-Plant Protein Diets for Rainbow Trout (Oncorhynchus mykiss). Aquaculture 2007, 269, 514–524. [Google Scholar] [CrossRef]
- Kidd, D.; Liu, Y.; Cravatt, B.F. Profiling Serine Hydrolase Activities in Complex Proteomes. Biochemistry 2001, 40, 4005–4015. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tang, X.; Jiang, S.; Wang, H.; Zhou, Y.; Peng, F.; Zhang, X.; Zhou, Y.; Guo, S.; You, Y. Transcriptome Sequencing Analysis Reveals Dynamic Changes in Major Biological Functions during the Early Development of Clearhead Icefish, Protosalanx chinensis. Fishes 2022, 7, 115. https://doi.org/10.3390/fishes7030115
Tang X, Jiang S, Wang H, Zhou Y, Peng F, Zhang X, Zhou Y, Guo S, You Y. Transcriptome Sequencing Analysis Reveals Dynamic Changes in Major Biological Functions during the Early Development of Clearhead Icefish, Protosalanx chinensis. Fishes. 2022; 7(3):115. https://doi.org/10.3390/fishes7030115
Chicago/Turabian StyleTang, Xuemei, Shulun Jiang, Henglin Wang, Yanfeng Zhou, Fei Peng, Xizhao Zhang, Yifan Zhou, Shiyue Guo, and Yang You. 2022. "Transcriptome Sequencing Analysis Reveals Dynamic Changes in Major Biological Functions during the Early Development of Clearhead Icefish, Protosalanx chinensis" Fishes 7, no. 3: 115. https://doi.org/10.3390/fishes7030115
APA StyleTang, X., Jiang, S., Wang, H., Zhou, Y., Peng, F., Zhang, X., Zhou, Y., Guo, S., & You, Y. (2022). Transcriptome Sequencing Analysis Reveals Dynamic Changes in Major Biological Functions during the Early Development of Clearhead Icefish, Protosalanx chinensis. Fishes, 7(3), 115. https://doi.org/10.3390/fishes7030115






