Cloning and Expression Profiling of the Gene vasa during First Annual Gonadal Development of Cobia (Rachycentron canadum)
Abstract
1. Introduction
2. Materials and Methods
2.1. Biological Samples
2.2. Total RNA Extraction, Cloning, and Phylogenetic Analysis of the vasa Gene
2.3. Semiquantitative RT-PCR
2.4. Quantitative Real-Time PCR (qRT-PCR)
2.5. Chromogenic In Situ Hybridization (CISH)
3. Results
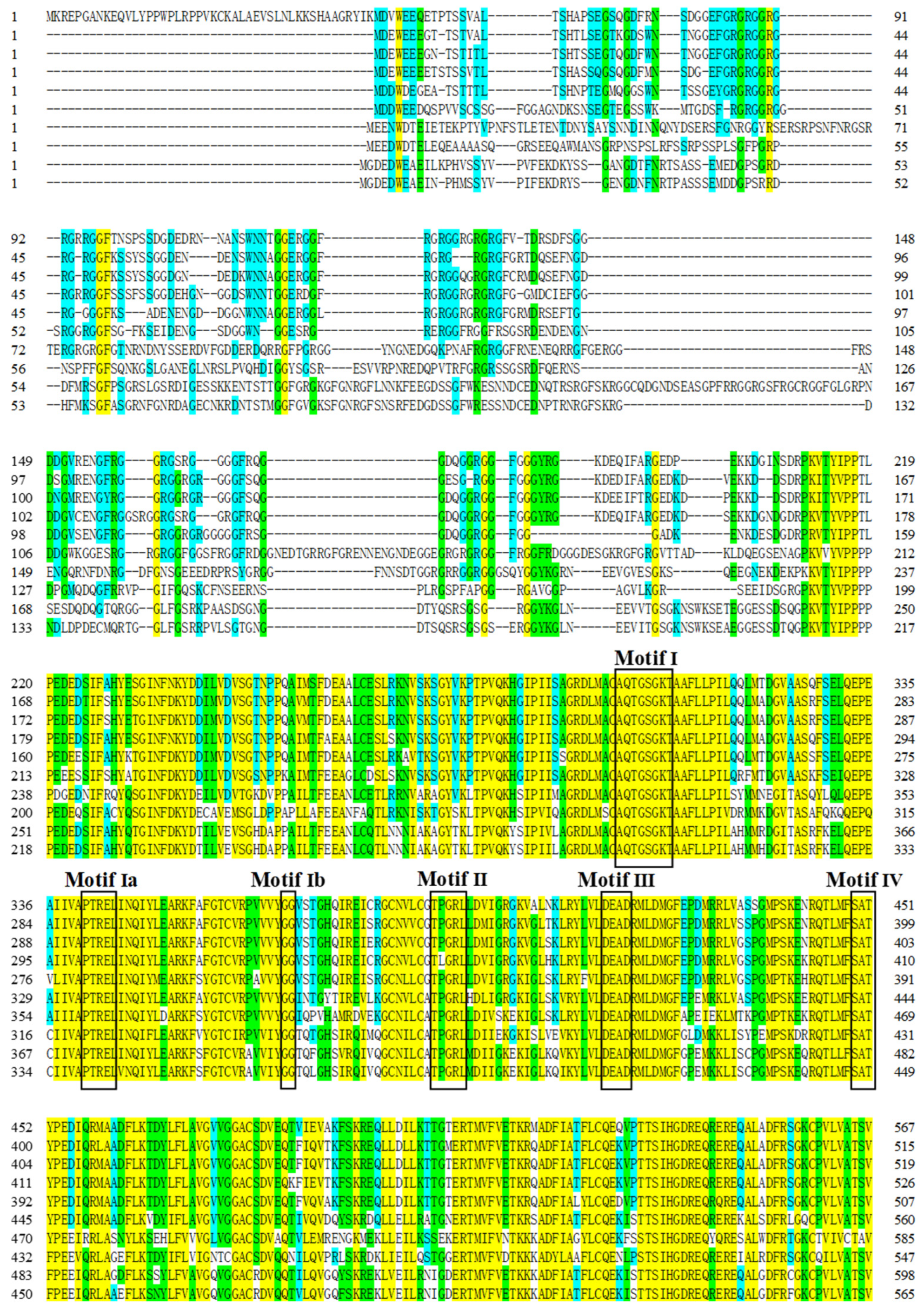
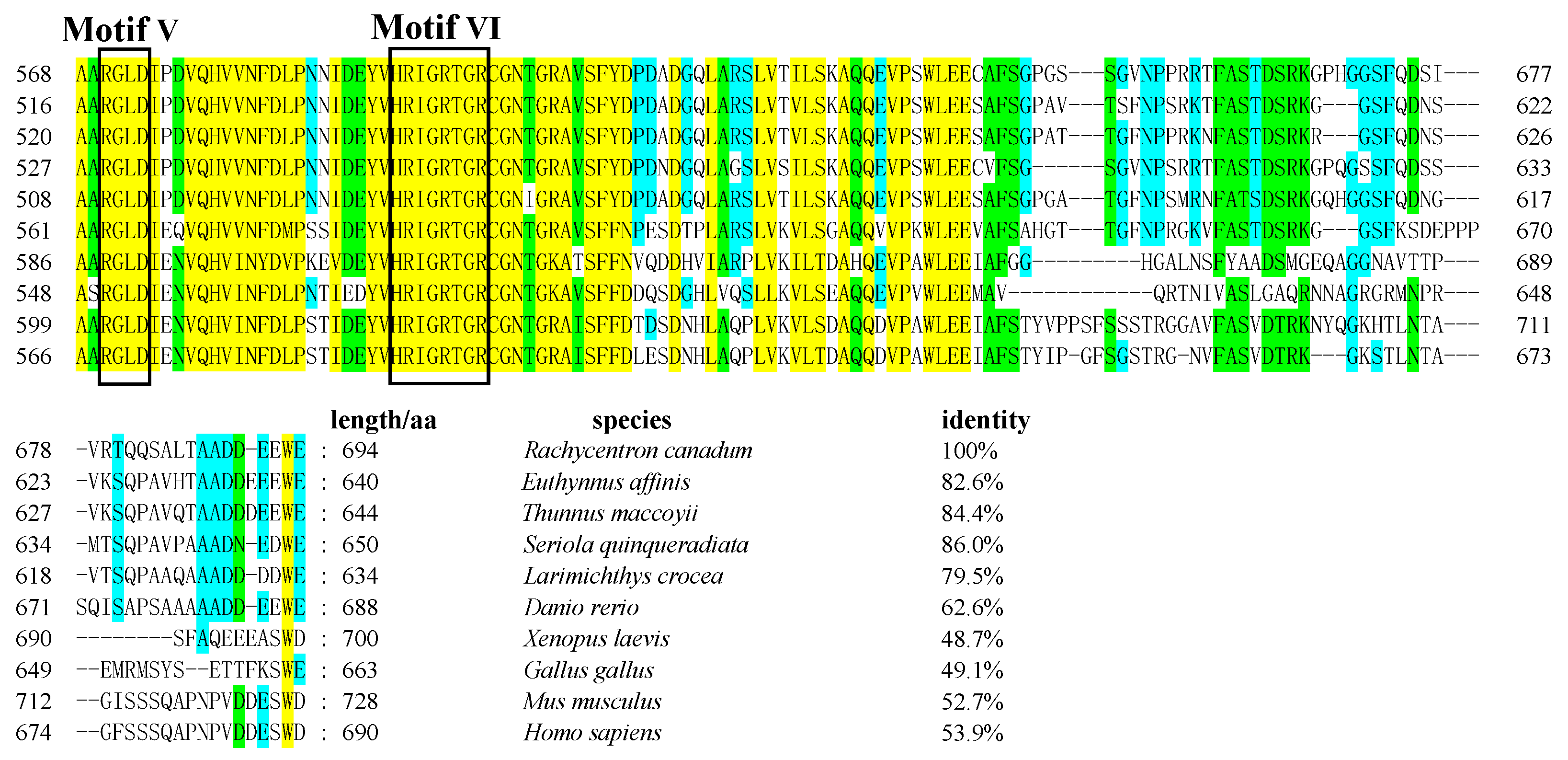
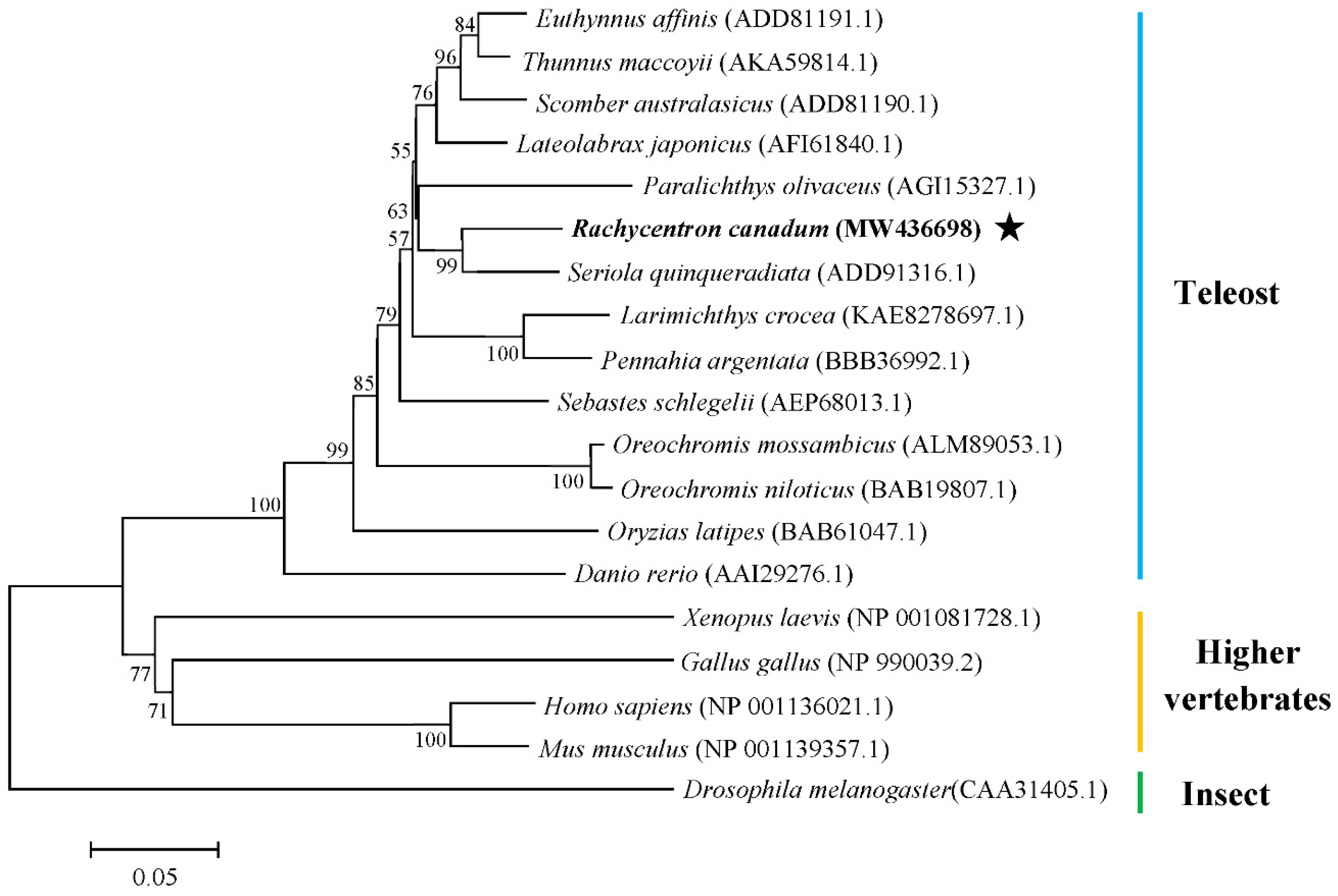
3.1. Cloning of Rcvasa cDNA and Phylogenetic Analysis
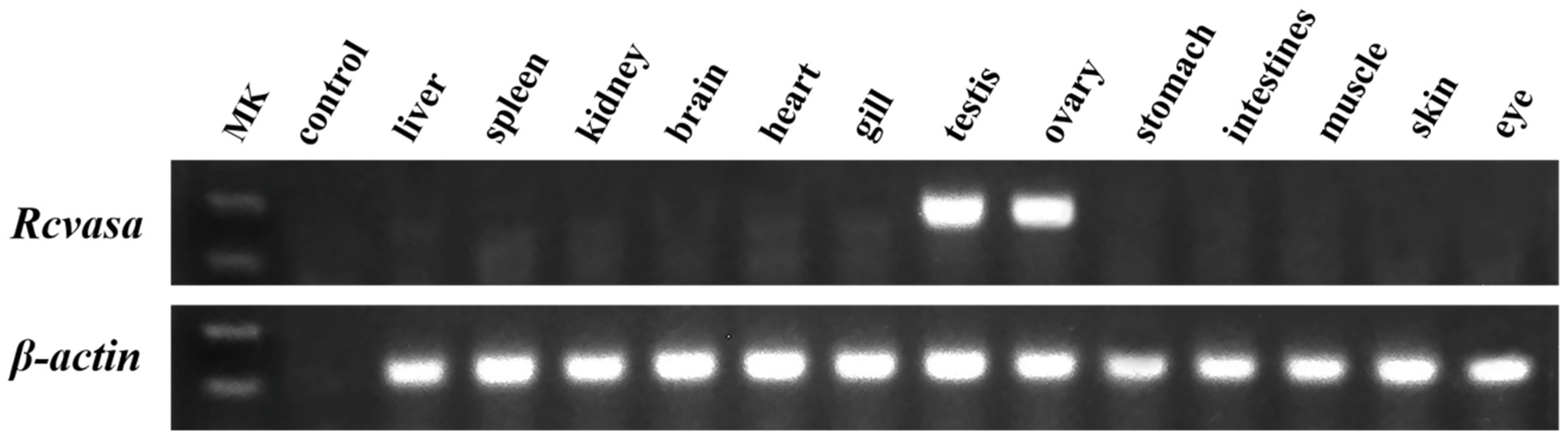
3.2. Tissue Distribution Patterns of Rcvasa mRNA
3.3. Expression Patterns of Rcvasa mRNA at Different Gonadal Development Stages
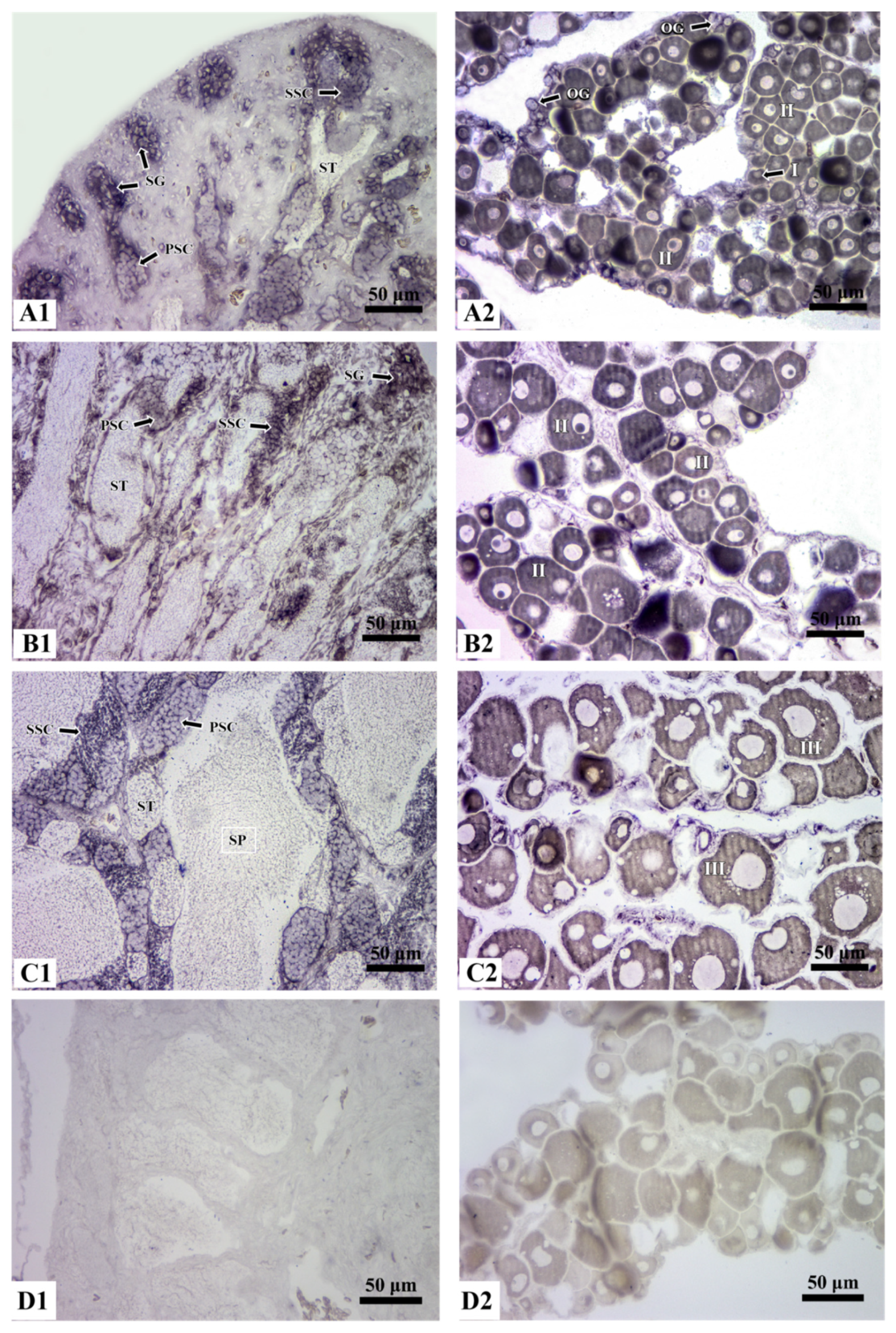
3.4. Localization of Rcvasa mRNA in Germ Cells during Gametogenesis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wylie, C. Germ cells. Cell 1999, 96, 165–174. [Google Scholar] [CrossRef]
- Barske, L.A.; Capel, B. Blurring the edges in vertebrate sex determination. Curr. Opin. Genet. Dev. 2008, 18, 499–505. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.Y.; Gui, J.F.; Hong, Y.H. Differential expression of vasa RNA and protein during spermatogenesis and oogenesis in the gibel carp (Carassius auratus gibelio), a bisexually and gynogenetically reproducing vertebrate. Dev. Dyn. 2005, 23, 872–882. [Google Scholar] [CrossRef]
- Liu, L.X.; Hong, N.; Xu, H.Y.; Li, M.Y.; Yan, Y.; Purwanti, Y.; Yi, M.S.; Li, Z.D.; Wang, L.; Hong, Y.H. Medaka dead end encodes a cytoplasmic protein and identifies embryonic and adult germ cells. Gene. Expr. Patterns 2009, 9, 541–548. [Google Scholar] [CrossRef]
- Bhat, N.; Hong, Y.H. Cloning and expression of boule and dazl in the Nile tilapia (Oreochromis niloticus). Gene 2014, 540, 140–145. [Google Scholar] [CrossRef]
- Sun, Z.H.; Wang, Y.; Lu, W.J.; Li, Z.; Liu, X.C.; Li, S.; Zhou, L.; Gui, J.F.; Lin, L. Divergent expression patterns and function implications of four nanos genes in a hermaphroditic fish, Epinephelus coioides. Int. J. Mol. Sci. 2017, 18, 685. [Google Scholar] [CrossRef]
- Duangkaew, R.; Jangprai, A.; Ichida, K.; Yoshizaki, G.; Boonanuntanasarn, S. Characterization and expression of a vasa homolog in the gonads and primordial germ cells of the striped catfish (Pangasianodon hypophthalmus). Theriogenology 2019, 131, 61–71. [Google Scholar] [CrossRef]
- Linder, P.; Lasko, P.F.; Ashburner, M.; Leroy, P.; Nielsen, P.J.; Nishi, K.; Schnier, J.; Slonimski, P.P. Birth of the D-E-A-D box. Nature 1989, 337, 121–122. [Google Scholar] [CrossRef]
- Liang, L.; Diehl-Jones, W.; Lasko, P. Localization of vasa protein to the Drosophila pole plasm is independent of its RNA-binding and helicase activities. Development 1994, 120, 1201–1211. [Google Scholar] [CrossRef]
- Deshpande, G.; Calhoun, G.; Yanowitz, J.L.; Schedl, P.D. Novel functions of nanos in downregulating mitosis and transcription during the development of the Drosophila Germline. Cell 1999, 99, 271–281. [Google Scholar] [CrossRef]
- Lasko, P. The DEAD-box helicase Vasa: Evidence for a multiplicity of functions in RNA processes and developmental biology. Biochim. Biophys. Acta. 2013, 1829, 810–816. [Google Scholar] [CrossRef]
- Schüpbach, T.; Wieschaus, E. Maternal-effect mutations altering the anterior-posterior pattern of the Drosophila embryo. Roux’s Arch. Dev. Biol. 1986, 195, 302–317. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.Y.; Li, M.Y.; Gui, J.F.; Hong, Y.H. Fish germ cells. Sci. China Life Sci. 2010, 53, 435–446. [Google Scholar] [CrossRef] [PubMed]
- Braat, A.K.; Zandbergen, M.A.; Water, S.V.D.; Goos, H.J.; Zivkovic, D. Characterization of zebrafish primordial germ cells: Morphology and early distribution of vasa RNA. Dev. Dyn. 1999, 216, 153–167. [Google Scholar] [CrossRef]
- Kobayashi, T.; Kajiura-Kobayashi, H.; Nagahama, Y. Differential expression of vasa homologue gene inthe germ cells during oogenesis and spermatogenesisin a teleost fish, tilapia, Oreochromis niloticus. Mech. Dev. 2000, 99, 139–142. [Google Scholar] [CrossRef]
- Wang, Z.K.; Gao, J.N.; Song, H.Y.; Wu, X.M.; Sun, Y.; Qi, J.; Yu, H.Y.; Wang, Z.G.; Zhang, Q.Q. Sexually dimorphic expression of vasa isoforms in the tongue sole (Cynoglossus semilaevis). PLoS ONE 2014, 9, e93380. [Google Scholar] [CrossRef]
- Xu, H.Y.; Lim, M.; Dwarakanath, M.; Hong, Y.H. Vasa identifies germ cells and critical stages of oogenesis in the Asian seabass. Int. J. Biol. Sci. 2014, 10, 225–235. [Google Scholar] [CrossRef]
- Yoshizaki, G.; Sakatani, S.; Tominaga, H.; Takeuchi, T. Cloning and characterization of a vasa-like gene in rainbow trout and its expression in the germ cell lineage. Mol. Reprod. Dev. 2015, 55, 364–371. [Google Scholar] [CrossRef]
- Nagasawa, K.; Takeuchi, Y.; Miwa, M.; Higuchi, K.; Morita, T.; Mitsuboshi, T.; Miyaki, K.; Kadomura, K.; Yoshizaki, G. cDNA cloning and expression analysis of a vasa-like gene in Pacific bluefin tuna Thunnus orientalis. Fish. Sci. 2009, 75, 71–79. [Google Scholar] [CrossRef]
- Kobayashi, T.; Kajiura-Kobayashi, H.; Nagahama, Y. Two isoforms of vasa homologs in a teleost fish: Their differential expression during germ cell differentiation. Mech. Dev. 2002, 111, 167–171. [Google Scholar] [CrossRef]
- Castellanos-Galindo, G.; Baos, R.; Zapata, L. Mariculture-induced introduction of cobia Rachycentron canadum (Linnaeus, 1766), a large predatory fish, in the Tropical Eastern Pacific. BioInvasions Rec. 2016, 5, 55–58. [Google Scholar] [CrossRef]
- Velde, T.; Griffiths, S.P.; Fry, G.C. Reproductive biology of the commercially and recreationally important cobia Rachycentron canadum in northeastern Australia. Fish. Sci. 2010, 76, 33–43. [Google Scholar] [CrossRef]
- Hamilton, S.; Severi, W.; Cavalli, R.O. Biology and aquaculture of cobia: A review. Bol. Inst. Pesca 2013, 39, 461–477. [Google Scholar]
- Schmittgen, T.D.; Livak, K.J. Analyzing realtime PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Kuang, J.; Chen, G.; Ma, Q.; Mao, F.; Zhou, Q.; Huang, J.; Shi, G.; Zhang, J. Histological observation on gonadal differentiation and first annual gonadal development of cobia (Rachycentron canadum). Haiyang Xuebao 2021, 43, 1–11. [Google Scholar]
- Liu, Y. Propagation Physiology of Main Cultivated Fish in China; Agricultural Publishing Press: Beijing, China, 1993; pp. 20–42. [Google Scholar]
- Rocak, S.; Linder, P. DEAD-box proteins: The driving forces behind RNA metabolism. Nat. Rev. Mol. Cell Biol. 2004, 5, 232–241. [Google Scholar] [CrossRef] [PubMed]
- Bartfai, R.; Orban, L. The vasa locus in zebrafish: Multiple RGG boxes from duplications. DNA Cell Biol. 2003, 22, 47–54. [Google Scholar] [CrossRef]
- Tanner, N.K. The newly identified Q motif of DEAD-box helicases is involved in adenine recognition. Cell Cycle 2003, 2, 18–19. [Google Scholar] [CrossRef]
- Lin, F.; Xu, S.H.; Ma, D.Y.; Xiao, Z.Z.; Zhao, C.Y.; Xiao, Y.S.; Chi, L.; Liu, Q.H.; Li, J. Germ line specific expression of a vasa homologue gene in turbot (Scophthalmus maximus): Evidence for vasa localization at cleavage furrows in euteleostei. Mol. Reprod. Dev. 2012, 79, 803–813. [Google Scholar] [CrossRef]
- Mu, W.J.; Wen, H.H.; He, F.; Li, J.F.; Liu, M.; Ma, R.Q.; Zhang, Y.Q.; Hu, J.; Qi, B.X. Cloning and expression analysis of vasa during the reproductive cycle of Korean rockfish, Sebastes schlegeli. J. Ocean Univ. China 2013, 12, 115–124. [Google Scholar] [CrossRef]
- Nishimura, T.; Tanaka, M. Gonadal development in fish. Sex. Dev. 2014, 8, 252–261. [Google Scholar] [CrossRef] [PubMed]
- Raghuveer, K.; Senthilkumaran, B. Cloning and differential expression pattern of vasa in the developing and recrudescing gonads of catfish, Clarias gariepinus. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2010, 157, 79–85. [Google Scholar] [CrossRef] [PubMed]
- Xiao, J.; Luo, Y.J.; Chen, L.B.; Yang, L.; Huang, Y.L.; Guo, Z.B.; Guo, E.Y.; Tang, Z.Y.; Zhang, M.; Gan, X. Molecular cloning of vasa gene and the effects of LHRH-A on its expression in blue tilapia Oreochromis aureus. Fish Physiol. Biochem. 2013, 39, 931–940. [Google Scholar] [CrossRef]
- Zhu, W.X.; Wang, T.; Zhao, C.; Wang, D.; Zhang, X.Y.; Zhang, H.Y.; Chi, M.L.; Yin, S.W.; Jia, Y.Y. Evolutionary conservation and divergence of Vasa, Dazl and Nanos1 during embryogenesis and gametogenesis in dark sleeper (Odontobutis potamophila). Gene 2018, 672, 21–33. [Google Scholar] [CrossRef]
- Qu, L.; Wu, X.; Liu, M.F.; Zhong, C.Y.; Xu, H.Y.; Li, S.S.; Lin, H.R.; Liu, X.C. Identification and characterization of germ cell genes vasa and dazl in a protogynous hermaphrodite fish, orange-spotted grouper (Epinephelus coioides). Gene Expr. Patterns 2020, 35, 11905. [Google Scholar] [CrossRef]
- Cardinali, M.; Gioacchini, G.; Candiani, S.; Pestarino, M.; Yoshizaki, G.; Carnevali, O. Hormonal regulation of vasa-like messenger RNA expression in the ovary of the marine teleost Sparus aurata. Biol. Reprod. 2004, 70, 737–743. [Google Scholar] [CrossRef]

| Primer | Sequence (5′–3′) | Usage |
|---|---|---|
| Rcvasa-F | ATGAAAAGAGAACCAGGCGCGA | CDS sequence cloning |
| Rcvasa-R | CTACTCCCATTCTTCATCATCAGCTG | |
| Rcvasa3′-F1 | TGACCTCCCCAACAACATAGACG | 3′ RACE |
| Rcvasa3′-F2 | TGGGAGGGCGGTGTCTTTC | |
| Rcvasa5′-R1 | GCCAGCAGAAATGATGGGGAT | 5′ RACE |
| Rcvasa5′-R2 | CTCCCTGATCTCCACCTTGTCTG | |
| Long-UPM | CTAATACGACTCACTATAGGGCAAGCAGTGGTATCAACGCAGAGT | RACE universal primer |
| Short-UPM | CTAATACGACTCACTATAGGGC | |
| Rcvasa-F1 | GGTTGGCAGGAGCAGTAACT | PCR amplification |
| Rcvasa-R1 | TGTGGTTGTGATCTCCGGTG | |
| β-actin-F | AGGGAAATTGTGCGTGAC | internal control gene |
| β-actin-R | AGGCAGCTCGTAGCTCTT | |
| Rcvasa-Pro | CAGGACGTTACACCCCCGGCATATTTCTC | probe of CISH |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, Q.; Kuang, J.; Chen, G.; Zhang, J.; Huang, J.; Mao, F.; Zhou, Q. Cloning and Expression Profiling of the Gene vasa during First Annual Gonadal Development of Cobia (Rachycentron canadum). Fishes 2022, 7, 60. https://doi.org/10.3390/fishes7020060
Ma Q, Kuang J, Chen G, Zhang J, Huang J, Mao F, Zhou Q. Cloning and Expression Profiling of the Gene vasa during First Annual Gonadal Development of Cobia (Rachycentron canadum). Fishes. 2022; 7(2):60. https://doi.org/10.3390/fishes7020060
Chicago/Turabian StyleMa, Qian, Jiehua Kuang, Gang Chen, Jiandong Zhang, Jiansheng Huang, Feifan Mao, and Qiling Zhou. 2022. "Cloning and Expression Profiling of the Gene vasa during First Annual Gonadal Development of Cobia (Rachycentron canadum)" Fishes 7, no. 2: 60. https://doi.org/10.3390/fishes7020060
APA StyleMa, Q., Kuang, J., Chen, G., Zhang, J., Huang, J., Mao, F., & Zhou, Q. (2022). Cloning and Expression Profiling of the Gene vasa during First Annual Gonadal Development of Cobia (Rachycentron canadum). Fishes, 7(2), 60. https://doi.org/10.3390/fishes7020060





