Abstract
Nuclear factor-E2-related factor (Nrf) belongs to the Cap ‘n’ collar basic leucine zipper (CNC-bZIP) family, which plays an important role in the resistance to oxidative stress in the body. In this study, 12 Nrf genes were identified in the common carp genome database. Comparative genomic analysis showed that the Nrf genes of common carp had significant amplification, confirming that the common carp had experienced four genome-wide replication events. Phylogenetic analysis showed that all common carp Nrf clustered with scleractinian fish Nrf, indicating that they were highly conserved during evolution. In addition, tissue distribution results showed that most Nrf genes had a broad tissue distribution but exhibited tissue-specific expression patterns, demonstrating functional differences after WGD events. At 30 and 60 days of Cd2+ stress, most of the Nrf genes showed an increase in expression compared with the control group, indicating that they played a key role in the organism’s response to oxidative stress. To find a suitable concentration of Bacillus coagulans to activate the Nrf genes, we added three different concentrations (2.0 × 107 CFU/g, 2.0 × 108 CFU/g, and 2.0 × 109 CFU/g) of B. coagulans into the feed and defined them as L1, L2, and L3 groups, respectively. We investigated the effect of different concentrations of B. coagulans in the feed on the expression level of Nrf genes in the intestine of common carp under Cd2+ stress at 30 and 60 days. The results showed that, compared with the control/stress group, the expression of different Nrf genes was improved to varying degrees at three concentrations, and the effect of the L2 group (2.0 × 108 CFU/g) was the best. This suggests that the L2 group is the optimum concentration for activating Nrf gene expression when subjected to heavy metal Cd2+ stress and may act as an activation switch with a prominent role in the body’s resistance to oxidative stress and immune response.
1. Introduction
Nuclear factor-E2-related factor (Nrf) belongs to the Cap ‘n’ collar basic leucine zipper (CNC-bZIP) family; it is an important class of regulatory factors that can be involved in oxidative stress response and detoxification of the body []. A total of four members have been recorded in mammals, including Nrf1 (Nfe2l1), Nrf2 (Nfe2l2), Nrf3 (Nfe2l3), and Nfe2, which have close homology to each other [].
Nrf1 was the first identified member of the Nrf family []. Nrf1 is often confused with nuclear respiratory factor 1 (NRF1) because the gene abbreviations are too similar; this led to statements such as “nothing is known about how Nrf1 is regulated” in 2010 []. Nrf1 was a gene identified in a cDNA library screen as binding to the erythrocyte-specific NF-E2/AP1 consensus sequence found in the 5′ β-globin locus control region 5′-GCTGAGTCA-3′, widely expressed in various tissues of animals. It participates in many cellular activities such as redox reactions, mitochondrial respiration, inflammatory responses, lipid metabolism, and cell differentiation [,]. Similarly, as a repressor of gene expression, Nrf1 plays a vital role in the antioxidant stress response to glutathione homeostasis.
Nrf2 was discovered in a cDNA library screen by Moi et al. and was named in 1994. It is widely distributed in the kidney, muscle, heart, liver, intestine, and other organs [,,]. Nrf2 is currently known to be regulated by more than 200 genes and is the most active transcription factor in the CNC family. Nrf2 plays an important role in protecting the body from oxidative damage. Nrf2 is also involved in regulating the expression of several antioxidant enzymes, phase II detoxification enzymes, which in turn exert cytoprotective effects []. Experiments performed in mice after knocking out Nrf2 have shown that it can be involved in the regulation of basal and non-inducible expression of genes in response to oxidative stress []. In addition, downregulation of Nrf2 and Nrf1 can lead to oxidative stress, apoptosiss, and embryonic death in mice. Thus, Nrf2 is very significant in mediating the oxidative stress response of organisms [].
There is little information regarding Nrf3 and Nfe2. Nrf3 is mainly distributed in the mammalian placenta and B-cell lineage []. Upon exposure to external stress, Nrf3 can translocate to the nucleus and exert its transcriptional activity through heterodimerization with small Maf proteins, allowing the antioxidant response element (ARE) or Maf recognition element (MARE) [,]. It is also essential in promoting phagocytosis of degenerated neurons via microglia and in cancer therapy. Nfe2 is expressed in the body’s bone marrow cells, erythrocytes and megakaryocyte cell lines, which show a high degree of conservation. Like other Nrf, Nfe2 can bind antioxidant response elements and thus regulate cellular protection against damage []. Nfe2 also increases the number of mature erythrocytes in the body and regulates the lesion of erythematosus disease. In summary, Nrf is inextricably linked to the antioxidant capacity of the species and the detoxification mechanism of the organism, and these pathways provide new therapeutic targets for the body’s resistance to oxidative damage and the detoxification of heavy metals [].
Environmental pollution and a sharp increase in farming density has resulted in serious exceedances of Cd2+ levels in water bodies. Among the various heavy metals, Cd2+ is the most difficult to excrete from the body and is one of the toxic metals that can cause serious damage to organisms []. When heavy metals enter the body of fish, they not only have toxic effects on them at the individual, tissue, and cellular levels [], but also the extension of exposure time also produces genotoxicity and indirectly causes DNA damage. In the study of zebrafish (Danio rerio), Cd2+ interferes with phoenix gene expression, delaying hair cell regeneration []. All tissues and organs in the body of fish absorb and accumulate heavy metals with a high degree of selectivity, and the intestine is not only the main area of nutrient digestion and absorption in the body but also a key part of intermediate metabolism, with a strong enrichment capacity. The damage to the intestine comes in two main forms. On the one hand, Cd2+ affects the physiological function and structure of the intestine, leading to increased permeability of the intestinal wall cells. On the other hand, Cd2+ causes changes in intestinal microbial population and flora diversity, increased lipopolysaccharide production, and impaired barrier function, leading to endotoxemia and systemic inflammation in the organism [].
In recent years, the application of probiotics in aquaculture has received increasing attention [,]. B. coagulans is one of the common Gram-positive bacteria, which is resistant to high temperatures, bile salts, and acids and maintains a very high survival rate after feed pelleting []. The addition of B. coagulans to feed in the aquatic animal gut has the following advantages: (1) The spores can produce a large number of hydrolytic enzymes to improve the utilization of feed, thus promoting the growth of animals []; (2) it can increase the reproduction of probiotics to resist the invasion of foreign pathogens []; (3) It can produce active substances with vibrio and have a strong inhibitory effect on Salmonella and Staphylococcus aureus []; (4) It can act as an immune stimulant to enhance the non-specific immune function of fish [].
Common carp is one of the most representative fish species, with a high proportion of the global freshwater aquaculture industry and has a very high commercial value [,]. As a major model species, common carp is widely used for research in developmental biology, immunology, environmental toxicology, physiology, and evolutionary genomics. This study aimed to investigate the phylogeny of the common carp Nrf gene family, its expression in healthy tissues, and the effect of B. coagulans in addition to feeding under 0.5 mg/L CdCl2 stress on its expression.
2. Materials and Method
2.1. Ethics Statement
This study was conducted under the permission of the Animal Conservation and Utilization Committee of Fisheries College of Henan Normal University (Approval code: 2021052501). Anesthetic (MS-222, Yufubao, China) was used to reduce common carp pain.
2.2. Identification and Naming of Nrf Genes
All available Nrf genes nucleotide and amino acid sequences of zebrafish (Danio rerio) were obtained from the public databases GenBank (http://www.ncbi.nlm.nih.gov/genbank/, accessed on 19 February 2021) or Ensembl (http://asia.ensembl.org/, accessed on 19 February 2021). These sequences were used as query sequences to search against all available common carp genome databases for orthologous genes by BLAST, with a cut off E-value of 1 × 10−5. The search results from the two histology databases were then compared backwards to ensure that no missing genes were present. The nomenclature of the common carp Nrf genes was based on the BLAST results of its homologue and the topological results of the phylogenetic evolutionary tree in zebrafish. If there were two or more genes in common carp corresponding to a gene in zebrafish, the Latin suffix (-1, -2, etc.) was added to the end of each gene to distinguish the different copies.
2.3. Structural Domain and Phylogenetic Analysis
The amino acid sequence of the common carp Nrf genes based on the Nrf secondary conserved structural domain was predicted by online functional software (SMART, http://smart.embl-heidelberg.de/, accessed on 20 February 2021) and validated by NCBI conserved domain structure prediction. The 3D structural domain of the Nrf genes was predicted using a simple modular research tool (https://swissmodel.expasy.org/, accessed on 22 February 2021).
To better understand the phylogeny and denomination of Nrf genes, a total of 54 amino acid sequences of Nrf genes were retrieved from common carp, human (Homo sapiens), mouse (Mus musculus), clawed frog (Xenopus laevis), coelacanth (Latimeria chalumnae), zebrafish, tilapia (Oreochromis nioticus), medaka (Oryzias latipes), channel catfish (Ictalurus punctatus), stickleback (Gasterosteus aculeatus), and fugu (Takifug rubripes). The species sequences were aligned by ClustalW2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/, accessed on 22 February 2021). The phylogenetic tree was constructed by the neighbor-joining (NJ) method in the MEGA6 software, and the reliability in the topology was tested by 1000 bootstrap replications. To further demonstrate the validity of the NJ phylogenetic tree, we also constructed maximum likelihood (ML) and maximum parsimony (MP) phylogenetic trees using the online software Evolview (https://www.evolgenius.info/evolview/#login, accessed on 25 February 2021) for visualization.
2.4. Tissue Expression of Nrf Genes
Healthy common carp with a body mass of (35.0 ± 1.5) g were taken from the aquatic base of Henan Normal University, and three tails were randomly selected after 2 weeks of domestication. Twelve tissues (heart, liver, spleen, head and kidney, kidney, muscle, skin, blood, gonads, brain, gills, and intestine) were extracted, and the organizational samples were placed in QIAzol Lysis Reagent (Qiagen). A Qiagen TissueRuptor (Qiagen) was used to cut up the tissue samples, and RNA was extracted from each of these pools using the Qiagen miRNeasy Mini Kit (Qiagen) according to the manufacturer’s instructions. Finally, RNA quality was detected by gel electrophoresis and NanoDrop2000. Strand cDNA was obtained according to cDNA Reverse Transcription Kit (TaKaRa) instructions for the subsequent qRT-PCR amplification template. Fluorescence quantification was performed with a ChamQ Universal SYBR qPCR kit from Vazyme. The total reaction system was 10 μL, including 2ChamQ Universal SYBR qPCR Master Mix 5 μL, 0.3 μL each of upstream and downstream primers (Table 1), 1 μL of cDNA, and 3.4 μL of ddH2O. The running reaction procedure was 95 °C pre-denaturation for 30 s, followed by denaturation at 95 °C for 5 s, annealing at 60 °C for 30 s, and extension at 72 °C for 30 s for 40 cycles. The assay was repeated three times for all samples to ensure the accuracy of the data.

Table 1.
Primers used for the qRT-PCR.
2.5. Experimental Water Environment and Feed Preparation
Heavy metals such as Cd2+ are not easily soluble in hard water bodies, and their toxicity is also susceptible to water hardness (Ca2+). Therefore, deionized water was used as the experimental water, and soft water with a water hardness of 28 mg CaCO3/L was prepared as the experimental water with reference to the Chinese method for determination of acute toxicity of water quality substances to freshwater fish (zebrafish) (GB/T 13,267.91). CdCl2-5H2O (Chongqing Kelong Chemical Reagent Factory (AR)) was used as the experimental water Cd2+ ion source reagent, which was first prepared in a stock solution with a concentration of 2500 mg/L for storage and then diluted to 0.5 mg/L according to experimental needs. The bacterium was isolated from the intestinal contents of common carp and identified as B. coagulans by the Institute of Microbiology, Henan Normal University. Different concentration groups (2.0 × 107 CFU/g, 2.0 × 108 CFU/g and 2.0 × 109 CFU/g) of B. coagulans, defined as groups L1, L2, and L3, were mixed evenly with the common carp basal feed, and the mixture was kept at 20 °C for 24 h before feeding.
2.6. Experimental Sample Preparation
The experimental fish were randomly divided into five groups, and each 30 common carp with a weight of (35 ± 2 g) were kept in a 200 L tank with three parallel replicates. Water quality parameters: DO (7.9 ± 0.2) mg·L−1, temperature (20.0 ± 2) °C, PH 7.4 ± 0.1, nitrite (0.07 ± 0.01) mg·L−1, total ammonia nitrogen (0.45 ± 0.05) mg·L−1, free CO2 (7.5 ± 0.5) mg·L−1. Control and stress groups were given basal diets without B. coagulans, while the remaining three groups were given different concentrations of B. coagulans. The feed was fed at 3% of the fish mass four times a day, and one-third of the water was changed every day. After the water change, the Cd2+ content of the water was measured by a portable ion detector, and the Cd2+ chloride content in the water was adjusted to 0.5 mg/L in the other four groups, except for the control group. The water temperature was maintained at (20.0 ± 2) °C and sufficient oxygen was provided during the culture. Tissue from the intestine (anterior, middle, and hindgut) of nine common carp was randomly selected from each group at 30 d and 60 d, and stored in a −80 °C refrigerator for further tissue RNA extraction. The RNA extraction methods as well as real-time quantitative PCR methods, are referred to in Section 2.4. After real-time quantitative PCR assays were performed for each subgroup, the data were analyzed for expression levels using the 2−ΔΔCt method [].
2.7. Data Statistics
The data were analyzed statistically with the independent sample T-test and one-way analysis of variance (ANOVA) method using SPSS 17.0 software (IBM Corp., Armonk, NY, USA). p values of less than 0.05 were regarded as being of statistical significance.
3. Results
3.1. Identification and Naming of Nrf Genes
A total of 12 Nrf genes were identified in the common carp genome database, distributed on 11 chromosomes (Table 2), significantly more than the number of Nrf genes in other vertebrates. Among them, human, mouse, clawed frog, coelacanth, tilapia, medaka, channel catfish, stickleback, and fugu had the same number of genes with four Nrf genes, but common carp and zebrafish showed significant amplification of Nrf genes with 12 and 6 Nrf genes, respectively. For gene annotation and nomenclature, 12 common carp Nrf genes were classified based on the similarity of Nrf genes to six known zebrafish Nrf homologs. Duplicated genes were assigned latin word suffixes (-1, -2, etc.). Therefore, the common carp Nrf genes were named as Nrf1a-1, Nrf1a-2, Nrf1b-1, Nrf1b-2, etc. All Nrf gene sequences were uploaded to the NCBI database with MW838195-206 consecutive landing numbers, and the detailed coding sequences, loci, and landing numbers are shown in Table 3.

Table 2.
Summary of Nrf family in common carp.

Table 3.
Comparison of the exon-intron organization of Nrf genes in common carp, zebrafish, and human.
3.2. Exon-Intron, Domain Structures and 3D Structure Prediction
A comparison of Nrf gene structures in common carp, zebrafish, and humans revealed that most genes have exon-intron structural differences. The common carp has one gene containing 9 exons, and the remaining 11 genes have between 3 and 6 exons. In contrast to common carp, zebrafish have one gene containing 10 exons and the remaining 6 genes have 3 genes with 3 exons and 3 genes with 4 exons, respectively. However, in humans there are 4 Nrf genes with 11, 7, 5, and 3 exons, respectively (Table 4). Protein structure prediction was performed based on the protein sequence pairs of Nrf genes, all of which contain different Nrf structural domains, transmembrane structural domains, and low complexity (Figure 1). In addition, there are 4 Nrf genes with coiled helixes and all belong to different copies of the Nrf2 gene. In three-dimensional structures, for example, Nrf1a-1 mainly consists of 4 α-helices, a β-turn angle, and a Nrf structural domain (Figure 2 and Supplementary Figure S1).

Table 4.
Comparative analysis of Nrf genes from common carp with other species.
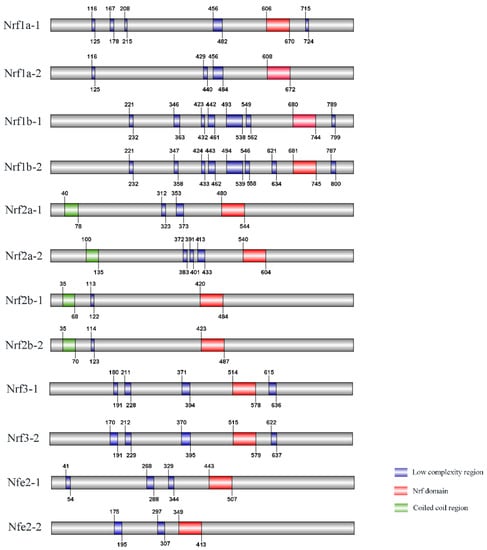
Figure 1.
Schematic representation of the Nrf gene domain architecture in common carp.
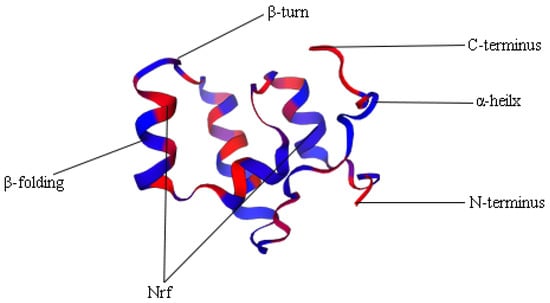
Figure 2.
3D structure domain of Nrf1a-1.
3.3. Phylogenetic Analysis of the Nrf Gene Family
To examine the phylogenetic relationships of Nrf genes in common carp and other vertebrates, we extracted the amino acid sequences of 54 Nrf genes from different representative species and used them to construct the phylogenetic topology. The NJ topology was divided into four subfamilies: subfamily I (contains Nrf1), subfamily II (contains Nrf2), subfamily III (contains Nrf3), and subfamily IV (contains Nrf4) (Figure 3). Based on topological features, common carp Nrf (blue pentagram) is homologous to other vertebrates except for the clawed frog and coelacanth, which are on a branch with tetrapods and are particularly close to the homologue of the model organism of vertebrates, zebrafish (green pentagram). In the tetrapod branch, mammals converge first (gray circles) followed by other tetrapods (red circles). Finally, the homologous genes of tetrapods converge with those of vertebrates onto the main branch (blue part). The Nrf1b-1/2 gene of common carp was first clustered with the Nrf1b of zebrafish, then clustered with the homologous genes of other bony fish, and finally clustered with the homologous genes of quadrupeds to form a branch. Additionally, we constructed ML evolutionary trees to support the NJ evolutionary tree, and their topologies were in high agreement with bootstrap values, indicating the reliability of the phylogenetic topology (Supplementary Figure S2).
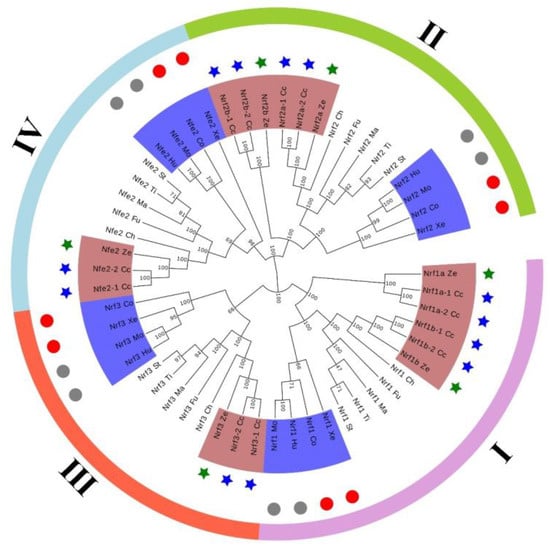
Figure 3.
Neighbor-joining-based phylogenetic tree of 54 Nrf protein sequences. Cc represents common carp, Hu represents human, Mo represents mouse, Xe represents clawed frog, Co represents coelacanth, Ze represents zebrafish, Ti represents tilapia, Ma represents medaka, Ch represents channel catfish, St represents stickleback, and Fu represents fugu.
3.4. Tissue Expression Pattern of Nrf Genes
Analysis of Nrf gene family expression in the healthy tissues of common carp based on qRT-PCR using gene-specific primers showed that most Nrf genes were widely distributed in tissues, with higher expression in muscle, blood, brain, liver, and intestine, but with conversely lower expression in kidney, spleen, and heart. In addition, Nrf1a-1 is expressed at low levels in gills, blood, spleen, gonads, head, and kidneys, but Nrf1a-2 has low expression levels only in gills and spleen. In Nfe2-2, the expression levels were low in skin, heart, and muscle, while Nfe2-1 only had low expression levels in muscle and relatively high expression levels in skin and heart. We not only observed some differences in expression between Nrf gene copies, but also found similarities such as a largely consistent tissue expression pattern between Nrf1b-1 and Nrf1b-2. Based on the overall tissue expression levels, Nrf2b-1 of the Nrf genes had extremely low expression levels in most tissues, while in contrast Nrf1a-2 had high expression levels in most tissues (Figure 4).
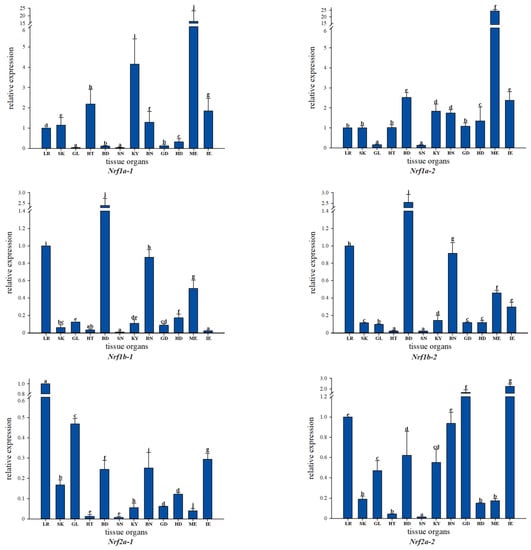
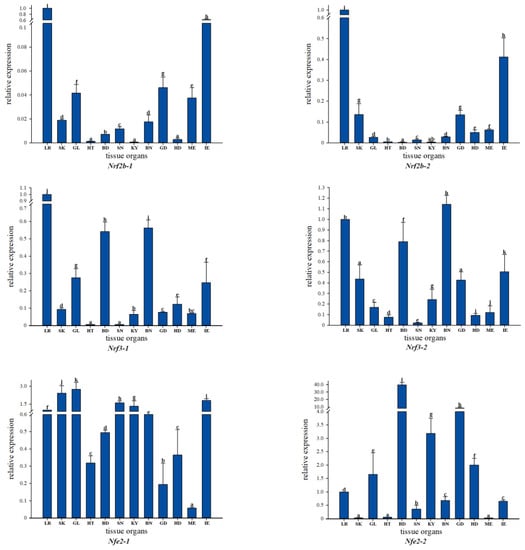
Figure 4.
qRT-PCR-based expression analysis of Nrf genes in different tissues of healthy common carp: LR (liver), SK (skin), GL (gill), HT (heart), BD (blood), SN (spleen), KY (kidney), BN (brain), GD (gonad), HD (head kidney), ME (muscle), and IE (intestine).
3.5. Effect of Adding B. coagulans to Feed under Cd2+ Stress on the Expression Level of Nrf Genes
To better understand the changes in the detoxification levels of Nrf genes under Cd2+ stress, we chose to add different concentrations of B. coagulans to the feed for qRT-PCR assay. The results showed that the addition of different concentrations of B. coagulans to the feed after 30 and 60 days of feeding resulted in differences in Nrf gene expression (Figure 5). All Nrf1 genes had the same expression pattern at 30 days, all Nrf1 gene expression appeared to be up-regulated when Cd2+ stress was applied, and additional feeding of L1 and L2 to B. coagulans also differentially reactivated the increase in Nrf1 gene expression, but L3 did not significantly upregulate Nrf1 expression. L2 can significantly increase Nrf gene expression and reached the highest level between both copies of Nrf2b-1/2. In Nrf2a-2, L1 had the highest level of gene expression at 30 and 60 days (Figure 6). There was a similar expression pattern between the two copies of Nrf3-1/2, with L2 of B. coagulans provoking the highest expression levels of both copies of the gene at 30 and 60 days, while L1 showed a down-regulation of expression at 30 days in comparison to the stress group in both cases (Figure 7). Interestingly, at 60 days, both copies again showed a simultaneous increase in expression after L1 induction compared to the stress group. Nfe2-1/2 had different expression trends between the two copies. At 60 days, Nfe2-1 showed a decrease in its expression level when subjected to Cd2+ stress (Figure 8). However, different levels of increase were observed under different concentrations of B. coagulans induction, and the highest expression level was reached under B. coagulans stimulation at L2, while Nfe2-2 differed, with all groups showing different levels of decrease in expression levels when subjected to Cd2+ stress.
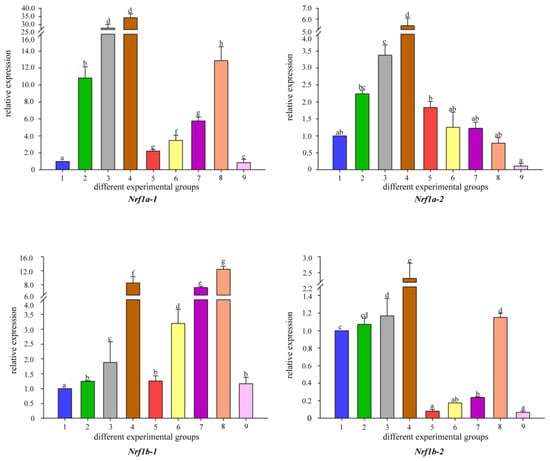
Figure 5.
Temporal expression of Nrf1 mRNA relative to control and normalized by qRT-PCR for changes in β-actin in the gut of common carp, challenged for 30 and 60 days. Significant differences between the challenge, infection, and control groups are indicated by letters, p < 0.05: 1 is the blank control group; 2 is the heavy metal Cd2+-stress group at 30 days; 35 is the B. coagulans group with L1, L2, and L3 concentrations in the feed at 30 days; 6 is the heavy metal Cd2+-stress group at 60 days; 79 is the B. coagulans group with L1, L2, and L3 concentrations in the feed at 60 days group.
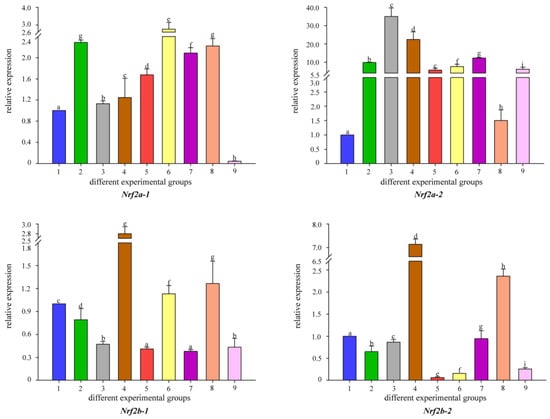
Figure 6.
Temporal expression of Nrf2 mRNA relative to control and normalized by qRT-PCR for changes in β-actin in the gut of common carp, challenged for 30 and 60 days. Significant differences between the challenge, infection, and control groups are indicated by letters, p < 0.05: 1 is the blank control group; 2 is the heavy metal Cd2+-stress group at 30 days; 35 is the B. coagulans group with L1, L2, and L3 concentrations in the feed at 30 days; 6 is the heavy metal Cd2+-stress group at 60 days; 79 is the B. coagulans group with L1, L2, and L3 concentrations in the feed at 60 days group.
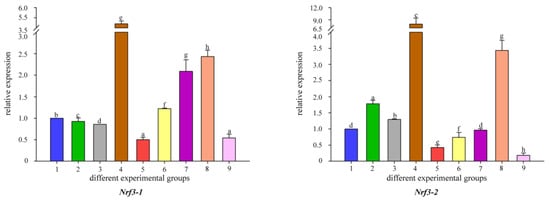
Figure 7.
Temporal expression of Nrf3 mRNA relative to control and normalized by qRT-PCR for changes in β-actin in the gut of common carp, challenged for 30 and 60 days. Significant differences between the challenge, infection, and control groups are indicated by letters, p < 0.05: 1 is the blank control group; 2 is the heavy metal Cd2+-stress group at 30 days; 35 is the B. coagulans group with L1, L2, and L3 concentrations in the feed at 30 days; 6 is the heavy metal Cd2+-stress group at 60 days; 79 is the B. coagulans group with L1, L2, and L3 concentrations in the feed at 60 days group.
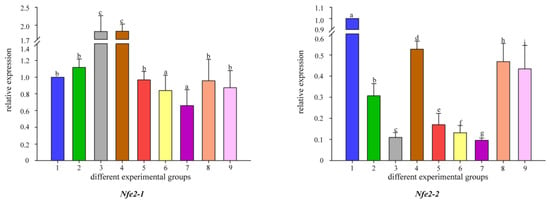
Figure 8.
Temporal expression of Nfe2 mRNA relative to control and normalized by qRT-PCR for changes in β-actin in the gut of common carp, challenged for 30 and 60 days. Significant differences between the challenge, infection and control groups are indicated by letters, p < 0.05: 1 is the blank control group; 2 is the heavy metal Cd2+-stress group at 30 days; 35 is the B. coagulans group with L1, L2, and L3 concentrations in the feed at 30 days; 6 is the heavy metal Cd2+-stress group at 60 days; 79 is the B. coagulans group with L1, L2, and L3 concentrations in the feed at 60 days group.
Overall, compared with the control/stress group, B. coagulans at L1, L2, and L3 concentrations increased Nrf genes expression, which enhanced the body’s detoxification capacity.
4. Discussion
In recent years, environmental pollution and indiscriminate discharge of wastewater have led to a serious excess of heavy metals in water bodies, and heavy metal toxicity can lead to oxidative stress in organisms, which is considered a key factor in the development of various diseases []. Many studies have shown that Nrf genes have a major role in regulating oxidative stress and detoxification mechanisms in the body []. For example, Nrf1 can mediate the body’s antioxidant response to elemental responses, and when the Nrf1 gene is deficient in mice, it causes the body’s fibroblasts to respond to various oxidant toxicities. When subjected to HgCl2 stress, activation of Nrf2 alleviates oxidative stress and mitochondrial damage and reduces toxic injuries. Therefore, it is essential to investigate the mechanism of oxidative stress and detoxification of common carp Nrf genes in the body.
In the present study, we found that common carp and zebrafish have 12 and 6 Nrf genes, respectively, significantly more than mammals after database matching. This is due to the significant increase in the number of genes in early teleosts after experiencing additional genome-wide replication events, so that most teleost have several times more copies of genes than homologous copies in the mammalian genome [,]. However, in some species, such as medaka, tilapia, and channel catfish, no significant amplification of the Nrf gene was found, which may be the result of the species’ evolutionary process leading to gene loss and is consistent with three forms of copy gene duplication, i.e., one of them forming a nonfunctional pseudogene lost during evolution, the formation of a new gene, and two copies complementing each other to form subfunctionalization []. Through further phylogenetic analysis, we confirmed that the common carp Nrf genes all clustered with their counterparts, indicating that the gene was highly conserved during evolution. In addition, teleosts are mainly composed of total finfishes, which can represent a deep lineage containing evolutionary information related to terrestrial vertebrates (tetrapods or coelacanths are one) and spoked finfishes (which evolved into extant teleost) []. The coelacanth Nrf genes are homologous to mammals, which is consistent with the evolutionary history of fish.
Expression analysis in healthy tissues showed that most Nrf genes were widely expressed and exhibited tissue-specific expression patterns. We observed high expression levels in muscle, blood, brain, liver, and intestine, which are the main accumulation sites of heavy metals in these tissues, such as the intestines as the main organ of digestion and absorption, which is the first interface of the organism to the aqueous environment []. Some of the genes with similar tissue expression patterns, e.g., Nrf1b-1 and Nrf1b-2, have essentially the same tissue expression pattern, which may indicate a similar gene function between the two copies. In addition, we also found differences in expression patterns in different copies of Nrf genes, such as Nrf1a-1/2 and Nfe2-1/2. This may be diverse and assume additional functions through various forms of regulatory evolution, such as divergence in expression patterns []. This also provides evidence for functional differentiation after WGD events.
After WGD, duplicated genes are subject to subsequent divergent processes, including gene loss, functionalization, and expression differences. However, the WGD of most vertebrates is too old for such analysis and has not been adequately studied to date []. Common carp WGD is relatively recent, so here we determined the expression levels of Nrf genes in intestinal tissues after feeding with different concentrations of B. coagulans in diets under heavy metal stress by qRT-PCR and performed a comparative analysis of the gene fate in recent replicates. Under the same treatment conditions, common carp Nrf1a-1/2, Nrf2a-1/2, and Nfe2-1/2 had significant differences in expression trends, and similar differences in expression levels were found for Nrf1b-1/2, Nrf2b-1/2, and Nrf3-1/2. Differences in expression patterns between most gene copies occur after WGD events, although copies can still be coexpressioned, but nearly half of the gene copies are no longer closely related, gradually evolving to divergent expressions and are thus subjected to one of the three fates []. Similar phenomena have been observed in other studies, such as the study of the common carp HIFs gene family, where functional differences in replicated genes can avoid potential adaptive conflicts and have important implications for the evolution of the species, and differences in expression levels between gene copies when infected by Aeromonas hydrophila []. In turbot (Scophthalmus maximus) studies, different copies of TLRs genes have an irreplaceable role in the immune response, and when infected by Vibrio eeleri, both TLR5a and TLR5b gene copies showed different expression levels after bacterial infection [].
When the heavy metal level in the water environment exceeds the standard, there will be a high accumulation in the intestinal tissue of fish exposed to water and diet []. However, the accumulation of metal nanomaterials needs to be excluded, and nanomaterials will help drug targeted transportation []. However, the toxic effects of heavy metal Cd2+ on fish are regulated by intestinal excretion, cellular metabolism, and the detoxification mechanisms of the organism. Nrf genes are an important class of regulators in the organism and can play an important role in the regulation of oxidative stress and induced defense in vertebrates when they are subjected to heavy metal stress []. It also works with genes such as HO-1, AHR, and NFκB to reduce oxidative stress in the body [,,]. In zebrafish studies, Nrf genes were found to bind antioxidant response elements to reduce organismal damage; gene expression levels were upregulated in response to oxidative stress in the organism, again with the regulation of primitive lupus erythematosus disease []. In the present study, most of the Nrf genes showed a tendency to increase in expression during the initial Cd2+ stress, suggesting that they are very considerable in the body’s response to oxidative stress. Often a single copy of a gene is sufficient for its function, while other daughter copies of the gene may accumulate deleterious mutations []. However, the expression of Nrf2b-1/2, Nrf3-1, and Nfe2-2 was not up-regulated during the initial Cd2+ stress, which may indicate that different copies of the genes gradually diverged due to molecular convergence after the WGD event, resulting in differential gene expression levels, but the functions may not have been fully differentiated. This is also consistent with Timme’s findings in the zebrafish Nrf genes expression study [].
Probiotics not only have strong antioxidant and immunomodulatory abilities, but also have certain regulatory effects on gene expression levels at different concentrations []. In studies on tilapia, when feed was supplemented with different concentrations (0.5%, 1%, and 2%) of the additive Salvadora persica, a concentration of 1% increased the antioxidant status and nonspecific immunity of tilapia, resulting in a significant increase in the expression levels of IL-1β and INF-γ genes in the kidney after feeding []. In studies on largemouth bass (Micropterus nigricans), after adding 0%, 1.5%, 3.0%, and 4.5% hydrolyzed yeast to the feed, the yeast hydrolysate at 1.5% or 3.0% significantly improved the antioxidant capacity of the organism and enhanced the immune response, resulting in significantly higher expression levels of IL-8, SOD, and CAT genes []. In the study of Takifugu obscurus, when three different concentrations of B. coagulans were used as feed additives, the 0.1% addition group had the best effect and was able to induce the rapid growth of fish, while the other high concentration groups had no significant difference compared with the control group []. The effect of probiotics on cytokine production is often related to the concentration of added strains. The appropriate strain concentration has a positive regulatory function, which not only regulates all genes but also promotes a higher levels of gene expression, while a too-high strain concentration has the ability to inhibit gene expression. In the present study, most Nrf genes were upregulated by L1, L2, or L3 at different infection time points compared with the control/stress groups, except for the differential expression of two gene copies, and L2 had a significantly stronger induction of Nrf genes than L1 and L3. This result suggests that the L2 group is the optimum concentration for activating Nrf gene expression when subjected to heavy metal Cd2+ stress and immune responses. These findings may provide new ideas for further research on the mechanisms related to the body’s resistance to oxidative stress, but the specific regulatory mechanisms are still unclear and need to be further investigated.
5. Conclusions
This study presented the first identification of the Nrf gene family in common carp. Comparative genomics and analysis showed considerable expansion of the Nrf genes and verified the four round whole genome duplication events in common carp. The spatial and temporal expression measured by qRT-PCR showed that expression divergences were observed between these copy genes, proving functional divergence after the WGD. At 30 and 60 days of cadmium stress, most of the Nrf genes showed an increase in expression compared with the control group, indicating that they played important roles in the organism’s response to oxidative stress. We then investigated the effect of different concentrations of B. coagulans in the feed on the expression levels of Nrf genes in the intestine of common carp under Cd2+ stress at 30 and 60 days. Three concentrations improved the expression of different Nrf genes variously compared with the control/stress groups. However, the L2 group was the optimum concentration for activating Nrf gene expression when subjected to Cd2+ stress and may act as an activation switch with a prominent role in the body’s resistance to oxidative stress and immune response.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/fishes7010048/s1, Figure S1: 3D structure domains; Figure S2: Maximum likelihood phylogenetic tree of 54 Nrf protein sequences.
Author Contributions
C.D. conceived the project and designed the scientific objectives. Z.J. collected and prepared the fish samples. Z.J. and C.D. conducted the experiment and finished bioinformatics analysis. C.D. and Z.J. prepared the manuscript. C.D. and X.L. revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by National Natural Science Foundation of China (31801032) and The APC was funded by Xuejun Li and Chuanju Dong.
Institutional Review Board Statement
This study was conducted under the permission of the Animal Conservation and Utilization Committee of Fisheries College of Henan Normal University (Approval code: 2021052501). Common carp were treated appropriately to minimize suffering.
Data Availability Statement
All Nrf gene sequences were deposited in the NCBI database with the continuous accession numbers MW838195–MW838206.
Acknowledgments
The authors are grateful to the National Natural Science Foundation of China (31801032) for financial support.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Pi, J.; Leung, L.; Xue, P.; Wang, W.; Hou, Y.; Liu, D.; Yehu, D.E.; Lee, C.; Lau, J.; Kurtz, T.W. Deficiency in the nuclear factor E2-related factor-2 transcription factor results in impaired adipogenesis and protects against diet-induced obesity. J. Biol. Chem. 2010, 285, 9292–9300. [Google Scholar] [CrossRef]
- Mark, E.H.; Alicia, R.T.; Sibel, I.K.; John, J.S. Nrf2 and Nrf2-related proteins in development and developmental toxicity: Insights from studies in zebrafish (Danio rerio). Free Radic. Biol. Med. 2015, 88, 275–289. [Google Scholar]
- Chan, Y.J.; Han, L.X.; Kan, W.Y. Cloning of Nrf1, an NFE2-related transcription factor, by genetic selection in yeast. Proc. Natl. Acad. Sci. USA 1993, 90, 11371–11375. [Google Scholar] [CrossRef] [PubMed]
- Biswas, M.; Chan, J.Y. Role of Nrf1 in antioxidant response element-mediated gene expression and beyond. Toxicol. Appl. Pharmacol. 2010, 244, 16–20. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.Y.; Kwong, M.; Lu, R.; Chang, J.; Wang, B.; Kan, Y.W. Targeted disruption of the ubiquitous CNC-bZIP transcription factor, Nrf-1, results in anemia and embryonic lethality in mice. EMBO J. 2014, 17, 1779–1787. [Google Scholar] [CrossRef]
- Leung, L.; Kwong, M.; Hou, S.; Lee, C.; Chan, J.Y. Deficiency of the Nrf1 and Nrf2 Transcription Factors Results in Early Embryonic Lethality and Severe Oxidative Stress. J. Biol. Chem. 2003, 278, 48021–48029. [Google Scholar] [CrossRef]
- Moi, P.; Chan, K.; Asunis, I.; Cao, A.; Kan, Y. Isolation of NF-E2-related factor 2 (Nrf2), a NF-E2-like basic leucine zipper transcriptional activator that binds to the tandem NF-E2/AP1 repeat of the beta-globin locus control region. Proc. Natl. Acad. Sci. USA 1994, 91, 9926–9930. [Google Scholar] [CrossRef]
- Xiang, Y.; Kensler, T. Nrf2 as a target for cancer chemoprevention. Mutat. Res.-Fundam. Mol. Mech. Mutagenesis 2005, 591, 93–102. [Google Scholar]
- Lv, E.; Deng, J.; Yu, Y.; Wang, Y.; Gong, X.; Jia, J.; Wang, X. Nrf2-ARE signals mediated the anti-oxidative action of electroacupuncture in an MPTP mouse model of Parkinson’s disease. Free Radic. Res. 2015, 49, 1296–1307. [Google Scholar] [CrossRef]
- Hmlinen, M.; Teppo, H.R.; Skarp, S.; Haapasaari, K.M.; Karihtala, P. NRF1 and NRF2 mRNA and Protein Expression Decrease Early during Melanoma Carcinogenesis: An Insight into Survival and MicroRNAs. Oxidat. Med. Cell. Longev. 2019, 4, 1–15. [Google Scholar] [CrossRef]
- Ramos, M.; Dolan, P.M.; Itoh, K.; Yamamoto, M.; Kensler, T.W. Interactive effects of nrf2 genotype and oltipraz on benzo pyrene-DNA adducts and tumor yield in mice. Carcinogenesis 2003, 24, 461–467. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, A.; Ito, E.; Toki, T.; Kogame, K.; Takahashi, S.; Igarashi, K.; Hayashi, N.; Yamamoto, M. Molecular Cloning and Functional Characterization of a New Cap‘n’ Collar Family Transcription Factor Nrf3. J. Biol. Chem. 1999, 274, 6443–6452. [Google Scholar] [CrossRef] [PubMed]
- BChowdhury, A.; Katoh, H.; Hatanaka, A.; Iwanari, H.; Nakamura, N.; Hamakubo, T.; Natsume, T.; Waku, T.; Kobayashi, A. Multiple regulatory mechanisms of the biological function of NRF3 (NFE2L3) control cancer cell proliferation. Sci. Rep. 2017, 7, e12494. [Google Scholar] [CrossRef] [PubMed]
- Rost, M.S.; Shestopalov, I.; Yang, L.; Vo, A.H.; Richter, C.E. Nfe2 is dispensable for early, but required for adult thrombocyte formation and function in zebrafish. Blood 2016, 128, 2534. [Google Scholar] [CrossRef]
- Motohashi, H.; Katsuoka, F.; Shavit, J.A.; Yamamomt, M. Positive or negative mare-dependent transcriptional regulation is determined by the abundance of small maf proteins. Cell 2000, 103, 865–875. [Google Scholar] [CrossRef][Green Version]
- Williams, L.M.; Lago, B.A.; Mcarthur, A.G.; Raphenya, A.R.; Pray, N.; Saleem, N.; Salas, S.; Paulson, K.; Mangar, R.S.; Liu, Y. The transcription factor, Nuclear factor, erythroid 2 (Nfe2), is a regulator of the oxidative stress response during Danio rerio development. Aquat. Toxicol. 2016, 180, 141–154. [Google Scholar] [CrossRef]
- Liu, G.J.; Wu, Q.H.; Feng, R.E.; Guo, J.K.; Wang, R.G.; Mo, L.Y. Efficiency evaluation for remediating paddy soil contaminated with cadmium and arsenic using water management, variety screening and foliage dressing technologies. J. Environ. Manag. 2016, 170, 116–122. [Google Scholar] [CrossRef]
- Zhao, X.; Yao, L.A.; Ma, Q.L.; Zhou, G.; Xu, Z.C. Distribution and ecological risk assessment of cadmium in water and sediment in Longjiang River, China: Implication on water quality management after pollution accident. Chemosphere 2017, 194, 107–116. [Google Scholar] [CrossRef]
- Giuseppe, M.; Ike, O.; Antonino, G.; Basilio, R. Evaluation of the hair cell regeneration and claudin b and phoenix gene expression during exposure to low concentrations of cadmium and zinc in early developing zebrafish larvae, Comparative Biochemistry and Physiology Part C: Toxicol. Pharmacol. 2021, 248, 109116. [Google Scholar]
- Liu, J.; Pang, J.J.; Yan, H. The accumulation, histopathology, and intestinal microorganism effects of waterborne cadmium on Carassius auratus gibelio. Fish Physiol. Biochem. 2018, 45, 231–243. [Google Scholar] [CrossRef]
- Sevdan, Y.; Ebru, Y.; Mahmoud, D.; Einar, R.; Ehsan, A.; Hany, M.R.A. Probiotics, prebiotics, and synbiotics used to control vibriosis in fish: A review. Aquaculture 2022, 547, 737514. [Google Scholar]
- El-Saadony, M.T.; Alagawany, M.; Patra, A.K.; Kar, I.; Tiwari, R.; Dawood, M.A.; Dhama, K.; Abdel-Latif, H.M. The functionality of probiotics in aquaculture: An overview. Fish Shellfish Immunol. 2021, 117, 36–52. [Google Scholar] [CrossRef] [PubMed]
- Niu, Z.K.; Wu, H.Z.; Fu, C.; Zhang, Z.Y. Application status of Bacillus condensate. China Feed 2019, 17, 22–24. [Google Scholar] [CrossRef]
- Xu, Y.F.; Hu, H.; Zhou, J.D.; Wang, Z.G. The biological characteristics of Bacillus Coagulans and its application in aquaculture. J. Yancheng Inst. Technol. (Nat. Sci. Ed.) 2017, 30, 52–55. [Google Scholar] [CrossRef]
- Mu, Y.; Cong, Y. Bacillus coagulans and its applications in medicine. Benef. Microbes 2019, 10, 679–688. [Google Scholar] [CrossRef]
- Han, L.; Wang, X.H.; Yang, B. Analysis on pathogen in an outbreakdeath of Marsupenaeus japonicas andeffect of multi-probiotics combinationon enhancement of shrimp immunity. J. Fish. China 2017, 42, 431–441. [Google Scholar]
- Aguilar, J.; Hall, F.G.; Urbizo, U.C.; Garcia, H.S.; Vallejo, B.; González, A.; Hernández, A.; Liceaga, A.M. In silico prediction and in vitro assessment of multifunctional properties of postbiotics obtained from two probiotic bacteria. Probiotics Antimicrob. Proteins 2019, 12, 608–622. [Google Scholar] [CrossRef]
- Dong, C.; Lin, C.; Feng, J.; Jian, X.; Peng, X. Genome Wide Identification, Phylogeny, and Expression of Aquaporin Genes in Common Carp (Cyprinus carpio). PLoS ONE 2016, 11, e0166160. [Google Scholar] [CrossRef]
- Dong, C.; Jiang, L.; Peng, W.; Xu, J.; Shahid, M.; Al-Ghanim, K.A.; Sun, X.; Xu, P.; Cui, Z. Phylogenetic and Evolutionary Analyses of the Frizzled Gene Family in Common Carp (Cyprinus carpio) Provide Insights into Gene Expansion from Whole-Genome Duplications. PLoS ONE 2015, 10, e0144037. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T. Analysis of relative gene expression data using real-time quantitative pcr and the 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Ramos, E.; Pablo, M. Free radicals, antioxidants, nuclear factor-E2-related factor-2 and liver damage. Wiley Anal. Sci. 2020, 40, 151–168. [Google Scholar]
- Kwong, M.; Wai, Y.K.; Jefferson, Y. The CNC Basic Leucine Zipper Factor, Nrf1, Is Essential for Cell Survival in Response to Oxidative Stress-inducing Agents. J. Biol. Chem. 1999, 274, 37491–37498. [Google Scholar] [CrossRef]
- Meyer, M.; Schartl, M. Gene and genome duplications in vertebrates: The one-to-four rule and the evolution of novel gene functions. Curr. Opin. Cell Biol. 1999, 11, 699–704. [Google Scholar] [CrossRef]
- Xu, P.; Zhang, X.F.; Wang, X.; Li, J.T.; Liu, J.; Kuang, Y.; Xu, J.; Zheng, X.H. Genome sequence and genetic diversity of the common carp, Cyprinus carpio. Nat. Genet. 2014, 46, 1212–1219. [Google Scholar] [CrossRef] [PubMed]
- Sigbjørn, L.; Ben, F.K.; Simen, R.S.; Jason, R.M.; Matthew, P.K.; Torfinn, N.; Torgeir, R.H. The Atlantic salmon genome provides insights into rediploidization. Nature 2016, 533, 200–205. [Google Scholar]
- RytkNen, K.T.; Prokkola, J.M.; Salonen, V.; Nikinmaa, M. Transcriptional divergence of the duplicated hypoxia-inducible factor alpha genes in zebrafish. Gene 2014, 541, 60–66. [Google Scholar] [CrossRef] [PubMed]
- Dane, H.; Sisman, T. A morpho-histopathological study in the digestive tract of three fish species influenced with heavy metal pollution. Chemosphere 2020, 242, 125211–125218. [Google Scholar] [CrossRef]
- Li, J.T.; Hou, G.Y.; Kong, X.F.; Li, C.Y.; Zeng, J.M.; Li, H.D.; Xiao, G.B.; Li, X.M.; Sun, X.W. The fate of recent duplicated genes following a fourth-round whole genome duplication in a tetraploid fish, common carp (Cyprinus carpio). Sci. Rep. 2015, 5, 8199–8208. [Google Scholar] [CrossRef] [PubMed]
- Rastogi, S.; Liberles, D.A. Subfunctionalization of duplicated genes as a transition state to neofunctionalization. BMC Evol. Biol. 2005, 5, 1–7. [Google Scholar] [CrossRef]
- Zhang, J.; Dong, C.; Feng, J.; Li, J.; Li, X. Effects of dietary supplementation of three strains of Lactococcus lactis on HIFs genes family expression of the common carp following Aeromonas hydrophila infection. Fish Shellfish Immunol. 2019, 92, 590–599. [Google Scholar] [CrossRef]
- Liu, F.; Su, B.; Fu, Q.; Shang, M.; Gao, C.; Tan, F.; Li, C. Identification, characterization and expression analysis of TLR5 in the mucosal tissues of turbot (Scophthalmus maximus L.) following bacterial challenge. Fish Shellfish Immunol. 2017, 68, 272–279. [Google Scholar] [CrossRef] [PubMed]
- Shaikh, Z.A.; Vu, T.T.; Zaman, K. Oxidative stress as a mechanism of chronic cadmium-induced hepatotoxicity and renal toxicity and protection by antioxidants. Toxicol. Appl. Pharmacol. 1999, 54, 256–263. [Google Scholar] [CrossRef] [PubMed]
- Magro, M.; Baratella, D.; Bonaiuto, E.; Jessica, D.A.R.; Vianello, F. Stealth iron oxide nanoparticles for organotropic drug targeting. Biomacromolecules 2019, 20, 1375–1384. [Google Scholar] [CrossRef] [PubMed]
- Espada, S.; Ortega, F.; Molina-Jijón, E.; Rojo, A.I.; Cuadrado, A. The purinergic P2Y13 receptor activates the Nrf2/HO-1 axis and protects against oxidative stress-induced neuronal death. Free Radic. Biol. Med. 2010, 49, 416–426. [Google Scholar] [CrossRef]
- Ulin, A.; Henderson, J.; Pham, M.; Meyo, J.; Chen, Y.; Karchner, S.I.; Goldstone, J.V.; Hahn, M.E.; Williams, L.M. Developmental Regulation of Nuclear Factor Erythroid-2 Related Factors (Nrfs) by AHR1b in zebrafish (Danio rerio). Toxicol. Sci. Off. J. Soc. Toxicol. 2018, 167, 536–545. [Google Scholar] [CrossRef]
- Nourbakhsh, M.; Hauser, H. The Transcriptional Silencer Protein NRF: A Repressor of NF-κB Enhancers. Immunobiology 1997, 198, 65–72. [Google Scholar] [CrossRef]
- Dong, C.; Jiang, Z.; Zhang, X.; Feng, J.; Li, X. Phylogeny of Slc15 family and response to Aeromonas hydrophila infection following Lactococcus lactis dietary supplementation in Cyprinus carpio. Fish Shellfish Immunol. 2020, 106, 705–714. [Google Scholar] [CrossRef]
- Williams, L.M.; Timme-Laragy, A.R.; Goldstone, J.V.; Mcarthur, A.G.; Stegeman, J.J.; Smolowitz, R.M.; Hahn, M.E.; Yann, G. Developmental Expression of the Nfe2-Related Factor (Nrf) Transcription Factor Family in the Zebrafish, Danio rerio. PLoS ONE 2013, 8, e79574. [Google Scholar] [CrossRef]
- Kimmel, C.A.; Judy, B. Toxicologic Pathology. In Reproductive and Developmental Toxicology; Pralhad, W., Tausif, A., Eds.; Academic Press: Cambridge, MA, USA, 2011; Volume 76, pp. 1003–1026. [Google Scholar]
- Mohamed, A.L.; Waleed, N.E.; Ramy, M.; Shourbela, A.H.; ElFar, R.S.; Shewita, A.M. Miswak (Salvadora persica) dietary supplementation improves antioxidant status and nonspecific immunity in Nile tilapia (Oreochromis niloticus). Fish Shellfish Immunol. 2019, 88, 619–626. [Google Scholar]
- Gong, Y.; Yang, F.; Hu, J.; Liu, C.; Xie, S. Effects of dietary yeast hydrolysate on the growth, antioxidant response, immune response and disease resistance of largemouth bass (Micropterus salmoides). Fish Shellfish Immunol. 2019, 94, 548–557. [Google Scholar] [CrossRef]
- Hua, X.M.; Zhou, H.Q.; Zhang, Y.F.; Zhou, H. Effect of dietary supplemental chitosan and probiotics on growth and some digestive enzyme activities in juvenile Fugu Obscurus. Acta Hydrobiol. Sin. 2005, 29, 299–305. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).