Abstract
Sexual growth dimorphism is a common phenomenon in teleost fish. However, the mechanism of this complex phenomenon remains unclear. The fine-patterned puffer (Takifugu poecilonotus; Temminck and Schlegel, 1850) exhibits female-biased sexual size dimorphism similar to other pufferfish. In this study, the transcriptomes of female and male T. poecilonotus were sequenced, 285.95 million raw read pairs were generated from sequence libraries. After identification and assembly, a total of 149,814 nonredundant unigenes were obtained with an N50 length of 3538 bp. Of these candidates, 122,719 unigenes (81.91% of the total) were successfully annotated with multiple public databases. The comparison analysis revealed 10,385 unigenes (2034 in females and 8351 in males) were differentially expressed between different sexes of T. poecilonotus. Then, we identified many candidate growth- and sex-related genes, including Dmrt1, Sox3, Spatas, Prl/Prlr, fabps, Ghr, and Igf1r. In addition to these well-known genes, Fabp4 was identified for the first time in fish. Furthermore, 68,281 simple sequence repeats (SSRs) loci were screened and identified in the transcriptome sequence of T. poecilonotus. The results of our study could provide valuable information on growth- and sex-associated genes and facilitate further exploration of the molecular mechanism of sexual growth dimorphism.
1. Introduction
Sexual growth dimorphism, growth rate differing between the genders, is common among animals, e.g., in mammals, birds, insects and fish [1,2,3]. In teleost fish, sexual growth dimorphism is widely encountered, and many species display different growth rates between the sexes. For some species, males grow faster than females to adult size, such as many tilapia species [4]; catfish such as channel catfish Ictalurus punctatus [5] and wels catfish Silurus glanis [6]; and salmonids, such as Oncorhynchus nerka [7] and Salmo trutta [8]. However, in most other fish species, females outgrow males. For instance, flatfish [9], sea bass [10], eel [11], cyprinids [12] and other fish [13,14]. The mechanism of sexual growth dimorphism is complex, and numerous factors are involved in gender growth differences [15].
Fish growth is a complex polygenic trait regulated by numerous factors, including nutrients, environment, reproductive activity and energy metabolism [16]. Fortunately, sexually dimorphic growth can be used to explore candidate networks and genes for enhancing body size or growth speed, which may offer rapid and significant economic gains [13]. An increasing number of growth-related genes were identified based on the analysis of differentially expressed genes between females and males [16,17,18]. Sex is one of the most intriguing propositions in life science. Its mechanisms in many animals (e.g., mammals and birds) are high conserved [19,20]. Facilitated by the conservation, sex-related genes seem to be also conserved (e.g., Sry for mammals and Dmrt1 for birds) [21]. However, the sex-related genes are highly variable in fish species. Previous researches have announced many genes related to sex in the teleost, including amhy in Odontesthes hatcheri [22], sdY in Oncorhynchus mykiss [23], pfpdz1 in Pelteobagrus fulvidraco [24]. Research on sex-related genes in teleost has aroused widespread interest in an increasing number of investigators. As teleost is an important protein resource for humans, it is important to understand the growth and sex of teleost to enable the prediction of potential impacts and effective management. Members of the genus Takifugu are mainly distributed in the north-western Pacific Ocean; they are extremely similar in external morphology, and natural hybrids within the genus have frequently been found in the natural environment [25,26]. Due to their excellent taste, abundant nutrients, and ability to serve as senior food ingredients in some cultures, these fish are important aquaculture species in East Asia [27,28]. Although the fine-patterned puffer (Takifugu poecilonotus Temminck and Schlegel, 1850) has not been developed as an aquaculture species, this species is commercially viable and highly traded [29]. Fine-patterned puffer exhibits sexual growth dimorphism; similar to other pufferfish; the female fish tend to be stronger and larger than the male fish [30,31,32]. These biological features make T. poecilonotus an ideal non-model marine fish species for growth-related and sex-related gene studies. In addition, as a result of taste and fast growth rate, particularly the sexual differences in growth, T. poecilonotus has tremendous future potential for future aquaculture and the gene breeding industry. However, genetic and molecular data of T. poecilonotus are limited. Thus, identifying growth- and sex-related genes and their functions may not only be valuable information of growth- and sex-associated genes but may also lead to an expanded genome database for T. poecilonotus and facilitate future study about molecular biology.
Comparative transcriptome analysis is useful for studying gene expression in many samples simultaneously [33,34]. This methodology is ideal for investigating the molecular mechanisms of gene or pathway responses to an experimental hypothesis [35]. Furthermore, for species whose genome is not yet available, comparing complex genomes is an attractive option to sequence the transcriptome for detecting genes. In addition, transcriptomics allows simultaneous analyses of multiple processes (metabolism, protein homeostasis and other bioprocesses) [36,37]. In this study, the muscle, liver, gonads and heart of females and males were used for transcriptome analysis. Then, the genes expression profiles of different T. poecilonotus individuals were generated, differentially expressed genes (DEGs) between T. poecilonotus males and females were detected. Subsequently, candidate important genes and pathways involved in growth-related and sex-related genes were identified. Moreover, abundant SSRs markers were detected in deep coverage sequence region reads. The present study’s objective was to detect the growth-related and sex-related genes and provide a foundation for discovering sexual growth dimorphism mechanisms of teleost fish. Furthermore, the transcriptomic resource in this study would support the future study of molecular regulatory mechanisms in teleost fish.
2. Materials and Methods
2.1. Ethics Statement
The procedures involving animals in this study were conducted in accordance with the Laboratory Animal Management Principles of China. All samples were anaesthetised and killed by severing the spinal cord. All the experimental procedures in the present study were approved by the ethics committee of Laboratory Animal Welfare and Ethics of South China Sea Fisheries Research Institute (project identification code: nhdf 2021-05, date of approval: 15 January 2021).
2.2. Sample Collection
Three males and three females T. poecilonotus were captured by using gill-net from the Beibu Gulf of China. Then, the six specimens were temporarily acclimated in aerated seawater for three days. After eliminating confounding effects, tissues (heart, liver, muscle and gonad) of each T. poecilonotus were collected and stored in liquid nitrogen. Age, standard size, body weight and maturation stage of gonad of the specimen were measured and shown in Table S1, gonadal maturation stages were measured according to Chen (2008) [38].
2.3. RNA Extraction, cDNA Library Building, and Transcriptome Sequencing
Total RNA of each tissue was extracted. Then, assessment of RNA integrity was performed using TRIzol reagent. RNA integrity was assessed by using an Agilent 2100 Bioanalyser (Agilent Technologies, Palo Alto, CA, USA), and samples with RNA integrity number ≥7 were used for subsequent cDNA library preparation [39]. We pooled each tissue RNA from every individual in equal amounts, and 3 μg mixed RNA of each individual was used for constructing the cDNA libraries.
Then, mRNA (RNA with a poly-A tail) was extracted from the total RNA using magnetic beads with Oligo (dT) probes and purified. Fragmentation buffer was applied to break the mRNA into fragments of suitable size, and the fragmented mRNA was reverse-transcribed into double-stranded cDNA with the N6 random primer. Then, the cDNA fragments were repaired with phosphate at the 5′end, and an “A” base was added to the 3′ end, after which adapters were ligated to the cDNA fragments. PCR was performed to amplify the ligation products. After heat denaturation, the single-stranded DNA was cyclised with a splint oligo and DNA ligase. Then, the six cDNA libraries were sequenced on the BGISEQ-500 platform.
2.4. Assembly and Annotation
Raw read quality control was performed using SOAPnuke v 1.6.0 (https://github.com/BGI-flexlab/SOAPnuke, accessed on 8 December 2016). Trimmomatic v0.35 [40] was used for raw read trimming. Clean data were obtained after removing low-quality reads and adaptors. Subsequently, the Trinity-v2.11.0 software package [41] was applied to assemble the high-quality clean reads de novo (-min_kmer_cov: 3). During this period, we set the k-mer value as 25. TGICL [42] was applied to remove the redundancy and acquire non-redundant unigenes with a minimum 70% similarity. Then, we generated two classed of unigenes: clusters (prefix CL) and singletons (prefix unigene). Then, the Benchmarking Universal Single-Copy Orthologs (BUSCO v5.0.0) was used to assess completeness of the assembled transcriptome [43]. All unigenes were then searched against the eukaryotic orthologous group (KOG) database, Kyoto Encyclopedia of Genes and Genomes (KEGG) database, NCBI nonredundant protein (NR) database, NCBI nucleotide database (NT), Gene Ontology (GO) and Swiss-Prot protein database.
2.5. Analysis of Potential Candidate Genes
To characterise gene expression variation between different sex, clean reads of the female and male T. poecilonotus were mapped back separately on the de novo assembled transcriptome by using Bowtie2 [44]. Then, unigenes expression levels were calculated by RSEM software [45]. The DEG-seq package was used to identify differentially expressed genes (DEGs) for each sample based on a Poisson distribution [46]. The thresholds of differentially expressed genes were defined as |log2 fold change| ≥ 2 and a Q-value ≤ 0.01. Then, DEGs were used for subsequent Go and KEGG pathways enrichment analyses under the threshold of p-value ≤ 0.05 with GOSeq [47] and KOBAS v2.0 based on over-representation analysis (ORA) [48].
2.6. Simple Sequence Repeat (SSR) Loci Detection
The MISA was used to identify the SSR loci in the assembled T. poecilonotus transcript reference. The threshold of the minimum repeat number for various unit types was twelve for mononucleotide microsatellites, six for dinucleotides, five for trinucleotide and tetranucleotide microsatellites, and four for pentanucleotide and hexanucleotide microsatellites. Then, SSR-containing unigenes were selected to design primers on flanking sequences of these loci with Primer 3 software [49].
2.7. Quantitative Real-Time PCR Validation
To validate the transcriptome data in this study, we selected 10 DEGs and analysed their expression level based on quantitative real-time PCR. These 10 randomly selected DEGs showed significantly different expressions between different sex (5 male-biased and 5 female-biased). Then, we used Primer Premier 6.0 to design the primers of 10 selected unigenes. In addition, β-actin gene and 18S gene were chosen to standardise the concentration of PCR products. Furthermore, standard curves were constructed to identify the ideal dilution times of cDNA samples and were used as calibrator. All cDNA samples were diluted 20-fold used nuclease-free water and were used as templates for PCR. Furthermore, the qRT-PCR analysis was designed following the manufacturer’s instructions for SYBR® Premix Ex Taq™ (Tli RNaseH Plus) RR420A. A reaction system of 25 μL was amplified using ABI PRISM 7300 Real-Time PCR System, including 2.5 μL of diluted cDNA template, 12.5 μL of SYBR Premix Ex Taq (2×), 0.5 μL of each of the forward and reverse primers, 0.5 μL of ROX Reference Dye (50×) and 8.5 μL of nuclease-free water. The amplification processes consisted of a holding stage of 2 min at 95 °C, followed by 40 cycles of 15 s at 95 °C and 30 s at 56 °C. Three parallel experiments of every cDNA template were performed to increase the veracity of the result. Then, ABI 7500 system software was applied to analyse the expression data. The relative transcript fold changes were calculated using 2−ΔΔCt method.
3. Results
3.1. Sequencing and Assembly
In the present study, 285.95 million raw read pairs were generated, and after preprocessing the raw data, 264.02 million clean paired-end sequence reads with a Q30 percentage of 86.46% were obtained. Statistical data of six T. poecilonotus cDNA libraries are shown in Table 1. Transcriptome assembly was completed after clustering by removing redundancy with Trinity and TGICL. A total of 149,814 unigenes were identified with an N50 length of 3538 bp and a mean length of 2066 bp (Table 2). The length distribution of the unigenes revealed that 23,132 (15.44%) were between 200 and 300 bp, and 38,528 (25.72%) were 3000 bp and longer (Figure S1).

Table 1.
Summary statistics of clean transcriptome sequencing data from each sample.

Table 2.
Statistics for the assembled unigenes.
The result of BUSCO revealed that 99.67% of the genome (30.36% as single genes and 69.31% as duplicated genes) was complete, and 0.03% of the genome was missing. In summary, the de novo assembly in this study was relatively successful (Figure S2). The Raw reads in this study are archived in the NCBI Short Read Archive (SRA) databases under BioProject PRJNA683736, with accession numbers SRR13236436-SRR13236441. This Transcriptome Shotgun Assembly project was deposited at DDBJ/EMBL/GenBank under the accession GIXS00000000. The version described in this paper is the first version, GIXS01000000.
3.2. Annotation of T. poecilonotus Transcriptome
The unigenes were functionally characterised based on the description of their similar sequences as obtained from databases. The results showed that 122,719 unigenes matched to various databases (Table 3). From the annotated information of six databases, 63,250 unigenes had significant hits in the NR database, and most unigenes were matched with genes from Takifugu rubripes (74.38%) (Figure S3). In summary, 110,737 unigenes (73.92%) were annotated with at least one of the five protein databases used in this study, while 52,989 unigenes (35.37%) were annotated with all five protein databases that were searched in this study, namely, KEGG, KOG, GO, NR and Swiss-Prot databases (Figure 1).

Table 3.
Summary of unigene annotations.

Figure 1.
Venn diagram of the functional annotations.
The results showed 63,518 unigenes were annotated to the three GO categories in total. The terms “binding” (31,409 unigenes), “catalytic activity” (24,082 unigenes) and “cellular process” (14,736 unigenes) were dominant in these GO categories, respectively (Figure S4). In addition, 90,961 unigenes were annotated with the KOG database and classified into 25 subcategories, and “general function prediction only” (15,183 unigenes) and “signal transduction mechanisms” (14,975 unigenes) were the top two subcategories (Figure 2). In total, blast search against the KEGG database showed 100,035 unigenes were assigned (Figure 3); the top three pathways were “human diseases” (51,859 unigenes), “organism system” (40,564 unigenes) and “metabolism” (29,797 unigenes). Furthermore, the predominant subcategories were “signal transduction” (16,907 unigenes), “global and overview maps” (11,183 unigenes), “immune system” (10,598 unigenes) and “cancers: overview” (10,177 unigenes).
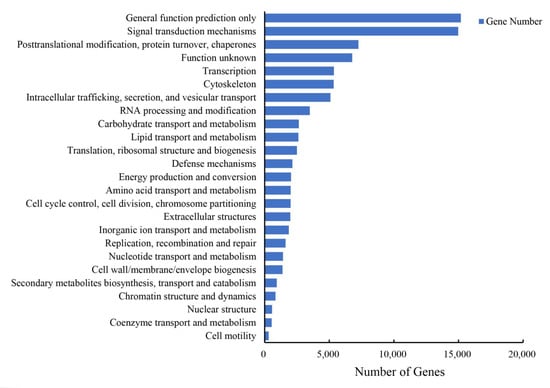
Figure 2.
Clusters of KOG functional classifications.
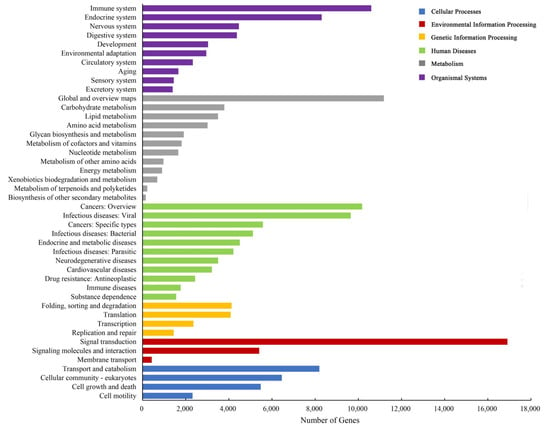
Figure 3.
KEGG pathway classifications of the unigenes.
3.3. Potential Candidate Genes and Pathways
To identify the potential growth-related genes, unigenes meeting the differential expression criteria, |log2 fold change| ≥ 2 and Q-values < 0.01, were determined to be DEGs between female T. poecilonotus and male T. poecilonotus. After filtering, 10,385 DEGs were obtained between the two sex samples; these genes might be associated with sex determination or be growth-related genes (Figure 4). Among these unigenes, 8351 were male-biased DEGs, and 2034 were female-biased DEGs.
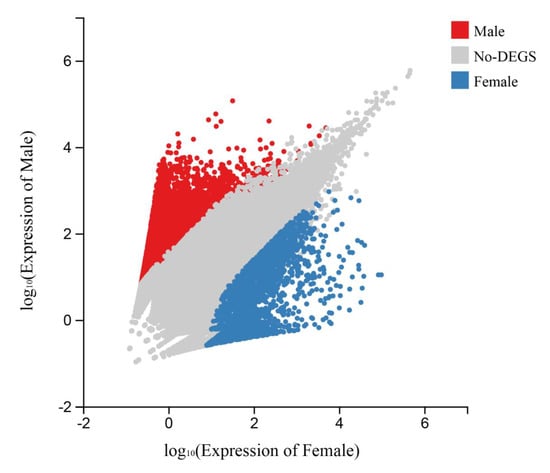
Figure 4.
Scatter plots showing gene expression profiles in female T. poecilonotus and male T. poecilonotus. Thresholds for inclusion were defined by Q-values < 0.01 and |log2 fold change| ≥ 2.
The 10,385 DEGs were then used as input to perform KEGG and GO enrichment analyses. A total of 3942 unigenes were assigned to 2146 GO terms. Figure 5 showed the top 20 statistically significant KEGG classifications. The results suggested that the three most-enriched KEGG classifications were “neuroactive ligand-receptor interaction” (ko04080), “PI3K-Akt signalling pathway” (ko04151) and “cellular senescence” (ko04218). The results of GO enrichment analyses showed that sex-biased genes were predominantly associated with “nucleus” (GO:0005634), “ATP binding” (GO:0005524), “integral component of membrane” (GO:0016021) and other terms (Table S2).
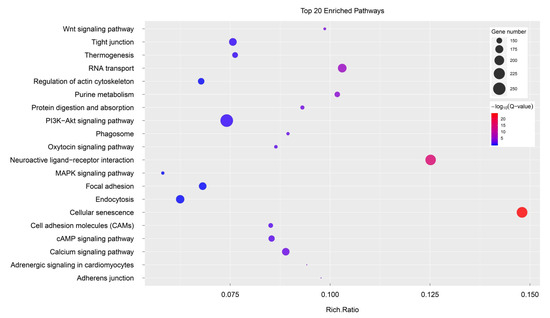
Figure 5.
KEGG pathway enrichment analyses of the DEGs.
The results of the sequence annotation showed that numerous differentially expressed genes were well-known as sex control and gonadal development genes, including the zona pellucida sperm-binding protein gene (Zps), sperm acrosome membrane-associated protein gene (Spacas), cytochrome P450 aromatase (Cyp19a), androgen-induced gene 1 protein, stAR-related lipid transfer protein gene (Start), double-sex and mab-3-related transcription factor 1 gene (Dmrt1) and other potential candidate protein-encoding genes (Table 4). As a complex trait, growth is controlled by numerous genes. In the present study, we identified multiple genes involved in regulating growth, such as genes related to controlling growth at the muscle tissue level, somatotropic axis growth and others (Table 5).

Table 4.
Genes related to the sex determination of Takifugu poecilonotus.

Table 5.
Growth-related genes in Takifugu poecilonotus.
3.4. Discovery of Molecular Markers
In the present study, we detected a large number of transcripts within which 68,281 potential SSRs were discovered. Dinucleotide SSRs represented the largest fraction (30,255 SSRs, 44.31%), followed by mononucleotide (20,211 SSRs, 29.60%) and trinucleotide (13,570 SSRs, 19.87%), as shown in Figure 6. Only a small fraction of quad-nucleotide (2382), penta-nucleotide (924) and hexa-nucleotide (936) SSR loci were screened. Furthermore, SSRs with six tandem repeats were the most common, followed by SSRs with five, twelve, seven, thirteen, fourteen and eight tandem repeats, whereas the remaining tandem repeats each accounted for fewer than 5% of the SSRs.
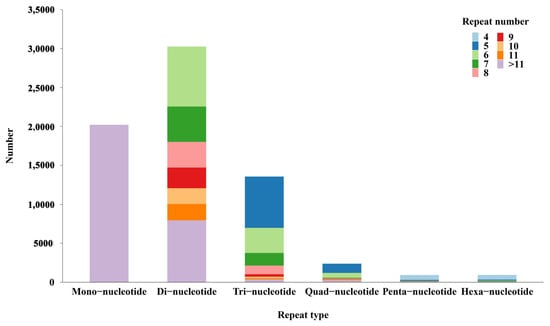
Figure 6.
The distribution of different SSRs.
3.5. Transcriptome Data Validation
We validated the expression patterns of 10 DEGs using qRT-PCR. Of these genes, five genes were predicted to be overexpressed in female specimens, and five genes were predicted to be overexpressed in male specimens. The specific primers for each gene are shown in Table S3. Similar expression patterns of these randomly selected genes were observed in the results of RNA-seq and qRT-PCR (Figure 7). Thus, the similar tendency in the expression patterns between qRT-PCR and RNA-seq data indicated the reliability and accuracy of the transcriptome expression analysis.
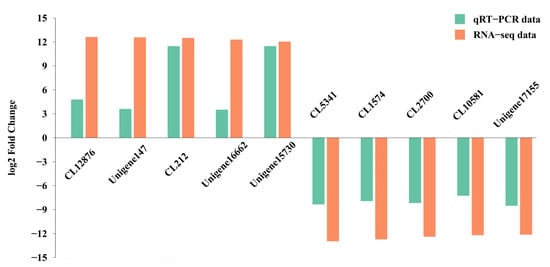
Figure 7.
The expression levels of 10 selected unigenes by RNA-seq and qRT-PCR.
4. Discussion
As a valuable commercial fish, Takifugu poecilonotus has high exploitability and aquaculture prospects. However, there were few studies regarding growth-related and sex-related genes of T. poecilonotus. In order to explore the mechanisms of sexual growth dimorphism and sex determination, we identified sex-related and growth-related genes and their biological pathways. It is the first time that the transcriptome data of fine-patterned puffer (Takifugu poecilonotus) have been reported. The result of annotation revealed that numerous sequences of fine-patterned puffer could be annotated to well-known genes and biological pathways regarding growth and sex. In the present study, 149,814 unigenes were sequenced and assembled. In total, 110,737 (73.92%) unigenes were significantly matched to protein databases. In the present study, 10,385 DEGs were identified between different sexes of T. poecilonotus. Some of these DEGs play important roles in sex growth and determination. Our results may provide fundamental resources for further exploration of the molecular mechanism of the biological processes of T. poecilonotus.
In the present study, numerous growth- and sex-related genes were detected from the transcriptome data of T. poecilonotus. Obviously, it is widely accepted that genes encoding components of the somatotropic axis play critical roles in regulating the formation of skeletal muscles in finfish, including the growth hormone gene (Gh), insulin-like growth factors (Igf), somatostatin and their carrier proteins and receptors [50]. Jia et al. [51,52] showed that specific growth rate, body weight and plasma growth hormone of female tiger puffers (Takifugu rubripes) reared in an offshore sea cage aquaculture system (OSCS) were significantly higher than those of fish reared in other aquaculture systems. The mRNA expression results indicated that female individuals reared in the OSCS showed higher somatic growth axis-related genes (Igf1, Igf2, Igf1r, Igf2r, Ghr1, and Ghr2) expression levels. In the present study, compared to male T. poecilonotus, female T. poecilonotus showed significantly higher Igf1r, Igfbp1, Igfbp3 and Ghr expression levels.
Fatty acid-binding proteins (FABPs) belong to the protein superfamily of lipid-binding proteins [53]. It was reported that the length of FABPs usually ranged from 126 to 137 amino acids, and the average molecular mass was 14–16 kDa [54,55,56]. Fabps have been well studied for decades [53,57]. The first teleost FABPs were identified in the hearts of sea ravens (Hemitripterus americanus) and ocean pouts (Macrozoarces americanus) [58]. Subsequently, teleost FABPs/Fabps were identified in different tissues of many teleosts, including striped bass (Morone saxatilis) [59], rainbow trout (Oncorhynchus mykiss) [60], Atlantic salmon (Salmo salar L.) [61]. Fatty acid-binding protein genes are involved in several processes of cell physiology, including development, growth, and cell differentiation [62]. In many respects, Fabps participates in the binding, sequestration and metabolism of long-chain fatty acids, eicosanoids, bile salts, and other hydrophobic ligands [63,64,65]. Fabps may also be possible carriers of certain hydrophobic reactants in their passage from the cytosol to chromatin, and thus, they may have a direct or indirect effect on cell growth [66]. Previous studies identified and characterised the Fabps genes of pufferfish (Tetraodon nigroviridis), including Fabp1–3, Fabp6, Fabp7, Fabp10, and Fabp11 [67,68]. In the present study, compared to female T. poecilonotus, male T. poecilonotus exhibited higher Fabp1, Fabp6 and Fabp7 expression levels, indicating that these genes may play important roles in reproduction and growth.
More importantly, we also found Fabp4 in the transcriptome of T. poecilonotus. This is the first time that the expression of Fabp4 was found in fish. It was reported that Fabp4 could affect the esterification, transportation, b-oxidation and uptake of fatty acids, with binding with long-chain fatty acids, and regulate the energy balance and lipid signalling within cells [69,70]. In mammals, fabp4 was also found to be associated with carcass traits, fat deposition and growth. It was reported that Fabp4 is a potential candidate gene for obesity, as it is located within a quantitative trait locus (QTL) region for serum leptin levels in mice [71]. In addition, FABP4 protein content may be a marker of intramuscular fat accretion in the longissimus thoracis muscle in pigs [72,73]. Obviously, Fabp4 may be a strong candidate gene for fat metabolism. However, the biological function of the Fabp4 gene in fish is unknown, and further work is required to fully characterise the Fabp4 gene in fish.
Prolactin (PRL) plays a critical role in multiple biological functions by binding to its receptor (PRLR) in fish [74]. In adult fish, the major action of PRL is freshwater osmoregulation. In addition, it was also reported that PRL is associated with reproduction, behaviour, growth, and immunoregulation [74,75,76]. Prolactin and PRLR are also present in embryos and exhibit widespread tissue distribution in fish larvae. Yang et al. [77] identified transcripts of PRL and PRLR in the early embryo of rainbow trout (O. mykiss) and indicated PRL and PRLR were associated with the post-hatching development of larvae. In the early development and metamorphosis of amphibians and mammals, the role of PRL has been well understood. The potential roles of PRL in fish embryos and larvae are considered in relation to their physiological status, and the spectrum of activities differs by species [74]. Furthermore, researchers have detected the expression of the PRL gene in the pituitary but also in the intestine, gill, liver, ovary and testes in teleost [78,79]. However, the role of PRL in fish is not clearly established. Some researchers indicated that PRL influences the growth of Mozambique tilapia by stimulating liver IGF-I production [80]; however, it is still unclear that the way in which PRL stimulates IGF-I production. In the present study, the expression level of PRL was significantly higher in females than that in males. The possible source of PRL transcripts may be the liver or ovary. There is relatively little information about the gene for PRL and its regulation in teleost. Hence, further work is required to fully characterise the biological function of PRL and PRLR in fish. Sex determination is the process of establishing individual gender and regulating the differentiation of sex characteristics [19,20]. In contrast to other vertebrates (birds or humans), the sex determination of teleost fish is diverse [36,81,82]. According to the annotations, many sex-related genes were identified in the T. poecilonotus transcriptome. As a sex-determining gene, doublesex and mab-3-related transcription factor 1 (Dmrt1) is highly conserved and was identified in many teleost fish [81,83,84]. The dmrt1 gene plays an important role in the testis differentiation and maintenance of male-specified germ cells by encoding a transcription factor [85]. In the present study, Dmrt1 showed higher expression levels in male T. poecilonotus than in female T. poecilonotus, indicating that Dmrt1 may be crucial for the development and maturation of gonads in male T. poecilonotus.
The sex-determining region Y (SRY) box, or SOX, is characterised as a conserved signature sequence in the high-mobility group (HMG) DNA-binding domain. SOX genes were identified in most vertebrates (mammals, birds and fish) [86]. In teleost fish, the sox superfamily plays crucial roles in many biological processes, including gonad development [87]. The sox3 plays a significant role in gametogenesis, sex determination, and gonad differentiation in vertebrates [88]. Yao et al. (2007) found that the expression of sox3 was dynamic along with the process of sex reversal in protogynous hermaphroditic grouper [89]. Li et al. (2020) detected sox3 was mainly expressed in the ovary of yellowfin seabream [90]. Nevertheless, Takehana et al. (2014) announced sox3 was the male-determining factor on the Y chromosome in the fish Oryzias dancena [88]. In the present study, the sox3 showed significantly higher expression in male T. poecilonotus than that in females, indicating the gene may play critical roles in sex differentiation in T. poecilonotus.
As an extracellular matrix surrounding oocytes, zona pellucida plays a protective role in fish oocytes, and it is important in sperm binding [91]. Zona pellucida comprises four kinds of glycoproteins (ZP1–4), they are incorporated into long filaments [92,93]. The zona pellucida proteins were originally detected in the egg envelope of mammals and were also found in the inner layer of the fish chorion [94]. A previous study has shown that Zp2 plays an important role in the early formation of oocyte envelope, and Zp3 could be treated as a major class of female-specific reproductive molecules [95]. In the present study, Zp1, Zp2, and Zp3 showed higher expression levels in females than that in males, indicating these genes may also play critical roles in folliculogenesis and reproduction in T. poecilonotus.
5. Conclusions
In the present study, we sequenced the transcriptome of T. poecilonotus. Through comparison of female and male T. poecilonotus transcriptome, we detected numerous candidate genes associated with growth and sex differentiation. Furthermore, amount of SSR loci were detected in the transcriptome of T. poecilonotus. This informative transcriptome analysis provides valuable data to increase the genomic resources of T. poecilonotus. The results could provide valuable information on growth- and sex-associated genes and facilitate further exploration of the molecular mechanism of sexual growth dimorphism in teleost.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/fishes6040079/s1, Figure S1: The length distribution of the unigenes, Figure S2: Completeness of the assembly and annotations, Figure S3: Distribution of the unigene hits by species in the NR database, Figure S4: GO terms for the DEGs in the biological process, cellular component, and molecular function categories, Table S1: Characters of six specimens used in the present study, Table S2: Top 20 GO pathways, Table S3: Specific primers for the selected unigenes and reference genes.
Author Contributions
Conceptualization, B.S. and Y.L. (Yan Liu); methodology, B.S. and L.W.; software, B.S. and C.Y.; validation, B.S., Y.L. (Yan Liu) and C.Y.; formal analysis, B.S. and Y.L. (Yan Liu); investigation, B.S. and L.W.; resources, B.S. and Y.L. (Yuan Li); data curation, Y.L. (Yan Liu) and C.Y.; writing—original draft preparation, B.S.; writing—review and editing, C.Y. and D.S.; visualization, B.S.; supervision, Y.L. (Yuan Li) and D.S.; project administration, L.W. and D.S.; funding acquisition, D.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Science and Technology Project of Guangdong Province, grant number 2019B121201001; Hainan Natural Science Foundation, grant number 319QN337; Central Public-interest Scientific Institution Basal Research Fund, South China Sea Fisheries Research Institute, CAFS, grant number 2021SD14; the National Programme on Global Change and Air-Sea Interaction, grant number GASI-02-SCS-YDsum.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the ethics committee of Laboratory Animal Welfare and Ethics of South China Sea Fisheries Research Institute (code: nhdf 2021-05 and date of approval: 15 January 2021).
Data Availability Statement
All raw reads in the present study are archived in the National Center for Biotechnology Information (NCBI) Short Read Archive (SRA) databases under BioProject PRJNA683736, with accession numbers SRR13236436-SRR13236441. This Transcriptome Shotgun Assembly project was deposited at DDBJ/EMBL/GenBank under the accession GIXS00000000. The version described in this paper is the first version, GIXS01000000.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Isaac, J.L. Potential causes and life-history consequences of sexual size dimorphism in mammals. Mammal. Rev. 2005, 35, 101–115. [Google Scholar] [CrossRef]
- Loonstra, A.J.; Verhoeven, M.A.; Piersma, T. Sex-specific growth in chicks of the sexually dimorphic Black-tailed Godwit. Ibis 2018, 160, 89–100. [Google Scholar] [CrossRef]
- Pauly, D. Female fish grow bigger-let’s deal with it. Trends Ecol. Evol. 2019, 34, 181–182. [Google Scholar] [CrossRef]
- Toguyeni, A.; Fauconneau, B.; Boujard, T.; Fostier, A.; Kuhn, E.R.; Mol, K.A.; Baroiller, J.F. Feeding behaviour and food utilisation in tilapia, Oreochromis niloticus: Effect of sex ratio and relationship with the endocrine status. Physiol. Behav. 1997, 62, 273–279. [Google Scholar] [CrossRef]
- Simco, B.A.; Goudie, C.A.; Klar, G.T.; Parker, N.C.; Davies, K.B. Influence of sex on growth of channel catfish. Trans. Am. Fish. Soc. 1989, 118, 427–434. [Google Scholar] [CrossRef]
- Haffray, P.; Vauchez, C.; Vandeputte, M.; Linhart, O. Different growth and processing traits in males and females of European catfish, Silurus glanis. Aquat. Living Resour. 1998, 11, 341–345. [Google Scholar] [CrossRef]
- Quinn, T.P.; Foote, C.J. The effects of body size and sexual dimorphism on the reproductive behavior of sockeye salmon, Oncorhynchus nerka. Anim. Behav. 1994, 48, 751–761. [Google Scholar] [CrossRef]
- Bonnet, S.; Haffray, P.; Blanc, J.M.; Vallée, F.; Vauchez, C.; Fauré, A.; Fauconneau, B. Genetic variation in growth parameters until commercial size in diploid and triploid freshwater rainbow trout (Oncorhynchus mykiss) and seawater brown trout (Salmo trutta). Aquaculture 1999, 173, 359–375. [Google Scholar] [CrossRef]
- Imsland, K.; Folkvord, A.; Grung, G.L.; Stefansson, S.O.; Taranger, G.L. Sexual dimorphism in growth and maturation of turbot, Scophthalmus maximus (Rafinesque, 1810). Aquac. Res. 1997, 28, 101–114. [Google Scholar] [CrossRef]
- Saillant, E.; Fostier, A.; Menu, B.; Haffray, P.; Chatain, B. Sexual growth dimorphism in sea bass Dicentrarchus labrax. Aquaculture 2001, 202, 371–387. [Google Scholar] [CrossRef]
- Roncarati, P.; Melotti, O.; Mordenti, L.; Gennari, L. Influence of stocking density of European eel (Anguilla anguilla, L) elvers on sex differentiation and zootechnical performances. J. Appl. Ichtyol. 1997, 13, 131–136. [Google Scholar] [CrossRef]
- Tyus, H.M. Ecology and management of Colorado squawfish. In Battle Against Extinction; Minckley, W.L., Ed.; Native Fish Management in the American West: Tucson, AZ, USA, 1993; pp. 379–402. [Google Scholar]
- Dutney, L.; Elizur, A.; Lee, P. Analysis of sexually dimorphic growth in captive reared cobia (Rachycentron canadum) and the occurrence of intersex individuals. Aquaculture 2017, 468, 348–355. [Google Scholar] [CrossRef]
- Pongthana, N.; Penman, D.J.; Baoprasertkul, P.; Hussain, M.G.; Islam, M.S.; Powell, S.F.; McAndrew, B.J. Monosex female production in the silver barb (Puntius gonionotus Bleeker). Aquaculture 1999, 173, 247–256. [Google Scholar] [CrossRef]
- Rennie, M.D.; Purchase, C.F.; Lester, N.; Collins, N.C.; Shuter, B.J.; Abrams, P.A. Lazy males? Bioenergetic differences in energy acquisition and metabolism help to explain sexual size dimorphism in percids. J. Anim. Ecol. 2008, 77, 916–926. [Google Scholar] [CrossRef]
- Wang, N.; Wang, R.; Wang, R.; Chen, S. Transcriptomics analysis revealing candidate networks and genes for the body size sexual dimorphism of Chinese tongue sole (Cynoglossus semilaevis). Funct. Integr. Genom. 2018, 18, 327–339. [Google Scholar] [CrossRef] [PubMed]
- Lou, F.; Yang, T.; Han, Z.; Gao, T. Transcriptome analysis for identification of candidate genes related to sex determination and growth in Charybdis japonica. Gene 2018, 677, 10–16. [Google Scholar] [CrossRef]
- Ma, D.; Ma, A.; Huang, Z.; Wang, G.; Wang, T.; Xia, D.; Ma, B. Transcriptome analysis for identification of genes related to gonad differentiation, growth, immune response and marker discovery in the turbot (Scophthalmus maximus). PLoS ONE 2016, 11, e0149414. [Google Scholar] [CrossRef]
- Yang, L.; Wang, Y.; Zhang, Z.; He, S. Comprehensive transcriptome analysis reveals accelerated genic evolution in a Tibet Fish Gymnodiptychus pachycheilus. Genome Biol. Evol. 2015, 7, 251–261. [Google Scholar] [CrossRef]
- Chen, H.; Xiao, G.; Chai, X.; Lin, X.; Fang, J.; Teng, S. Transcriptome analysis of sex-related genes in the blood clam Tegillarca granosa. PLoS ONE 2017, 12, e0184584. [Google Scholar] [CrossRef]
- Koopman, P.; Gubbay, J.; Vivian, N.; Goodfellow, P.; Lovell-Badge, R. Male development of chromosomally female mice transgenic for Sry. Nature 1991, 351, 117–121. [Google Scholar] [CrossRef]
- Hattori, R.S.; Murai, Y.; Oura, M.; Masuda, S.; Majhi, S.K.; Sakamoto, T.; Fernandino, J.I.; Somoza, G.M.; Yokota, M.; Strüssmann, C.A. AY-linked anti-Mullerian hormone duplication takes over a critical role in sex determination. Proc. Natl. Acad. Sci. USA 2012, 109, 2955–2959. [Google Scholar] [CrossRef]
- Yano, A.; Guyomard, R.; Nicol, B.; Jouanno, E.; Quillet, E.; Klopp, C.; Cabau, C.; Bouchez, O.; Fostier, A.; Guiguen, Y. An immune-related gene evolved into the master sex-determining gene in rainbow trout, Oncorhynchus mykiss. Curr. Biol. 2012, 22, 1423–1428. [Google Scholar] [CrossRef] [PubMed]
- Dan, C.; Lin, Q.; Gong, G.; Yang, T.; Xiong, S.; Xiong, Y.; Huang, P.; Gui, J.-F.; Mei, J. A novel PDZ domain-containing gene is essential for male sex differentiation and maintenance in yellow catfish (Pelteobagrus fulvidraco). Sci. Bull. 2018, 63, 1420–1430. [Google Scholar] [CrossRef]
- Masuda, Y.; Shinohara, N.; Takahashi, Y.; Tabeta, O.; Matsuura, K. Occurrence of natural hybrid between pufferfishes, Takifugu xanthopterus and T. vermicularis, in Ariake Bay, Kyushu, Japan. Nippon. Suisan Gakkaishi 1991, 57, 1247–1255. [Google Scholar] [CrossRef][Green Version]
- Miyaki, K.; Tabeta, O.; Kayano, H. Karyotypes in six species of pufferfishes genus Takifugu (Tetraodontidae, Tetraodontiformes). Fish. Sci. 1995, 61, 594–598. [Google Scholar] [CrossRef]
- Kikuchi, K.; Iwata, N.; Furuta, T.; Kawabata, T.; Yanagawa, T. Growth of tiger puffer Takifugu rubripes in closed recirculating culture system. Fish. Sci. 2006, 72, 1042–1047. [Google Scholar] [CrossRef]
- Wang, Q.L.; Zhang, H.T.; Ren, Y.Q.; Zhou, Q. Comparison of growth parameters of tiger puffer Takifugu rubripes from two culture systems in China. Aquaculture 2016, 453, 49–53. [Google Scholar] [CrossRef]
- Masuda, H.; Amaoka, K.; Muzik, C.K.; Uyeno, T.T.; Yoshimo, T. The Fishes of the Japanese Archipelago; Tokai University Press: Tokyo, Japan, 1984; p. 437. [Google Scholar]
- Yang, Z.; Chen, Y. Length-weight relationship of obscure puffer (Takifugu obscurus) during spawning migration in the Yangtze River, China. J. Freshw. Ecol. 2003, 18, 349–352. [Google Scholar] [CrossRef]
- Ueda, Y.; Sano, J.; Uchida, H.; Amano, C.; Matsumura, Y.; Katayama, T. Growth and age-length key of the tiger puffer Takifugu rubripes in the East China Sea, Sea of Japan, and Seto Inland Sea, Japan. Nippon. Suisan Gakkaishi 2010, 76, 803–811. [Google Scholar]
- Zhou, H.; Zhuang, Z.; Zhang, R. Temperature-control-induced masculinization in tiger puffer Takifugu rubripes. J. Oceanol. Limnol. 2019, 37, 1125–1135. [Google Scholar] [CrossRef]
- Chatchaiphan, S.; Srisapoome, P.; Kim, J.H.; Devlin, R.H.; Na-Nakorn, U. De novo transcriptome characterization and growth-related gene expression profiling of diploid and triploid bighead catfish (Clarias macrocephalus Günther 1864). Mar. Biotechnol. 2017, 19, 36–48. [Google Scholar] [CrossRef]
- Morozova, O.; Hirst, M.; Marra, M.A. Applications of new sequencing technologies for transcriptome analysis. Annu. Rev. Genom. Hum. G. 2009, 10, 135–151. [Google Scholar] [CrossRef]
- Wolf, J.B. Principles of transcriptome analysis and gene expression quantification: An RNA-seq tutorial. Mol. Ecol. Resour. 2013, 13, 559–572. [Google Scholar] [CrossRef]
- Casas, L.; Saborido-Rey, F.; Ryu, T.; Michell, C.; Ravasi, T.; Irigoien, X. Sex change in clownfish: Molecular insights from transcriptome analysis. Sci. Rep. 2016, 6, 35461. [Google Scholar] [CrossRef]
- Yang, X.; Ikhwanuddin, M.; Li, X.; Lin, F.; Wu, Q.; Zhang, Y.; You, C.; Liu, W.; Cheng, Y.; Shi, X.; et al. Comparative transcriptome analysis provides insights into differentially expressed genes and long non-coding RNAs between ovary and testis of the mud crab (Scylla paramamosain). Mar. Biotechnol. 2018, 20, 20–34. [Google Scholar] [CrossRef] [PubMed]
- Chen, X. Fisheries Resources and Fisheries Science; China Ocean Press: Beijing, China, 2008; pp. 57–58. [Google Scholar]
- Schroeder, A.; Mueller, O.; Stocker, S.; Salowsky, R.; Leiber, M.; Gassmann, M.; Lightfoot, S.; Menzel, W.; Granzow, M.; Ragg, T. The RIN: An RNA integrity number for assigning integrity values to RNA measurements. BMC Mol. Biol. 2006, 7, 3. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Pertea, G.; Huang, X.; Liang, F.; Antonescu, V.; Sultana, R.; Karamycheva, S.; Lee, Y.; White, J.; Cheung, F.; Parvizi, B.; et al. TIGR Gene Indices clustering tools (TGICL): A software system for fast clustering of large EST datasets. Bioinformatics 2003, 19, 651–652. [Google Scholar] [CrossRef] [PubMed]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Feng, Z.; Wang, X.; Wang, X.; Zhang, X. DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 2010, 26, 136–138. [Google Scholar] [CrossRef] [PubMed]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Mao, X.; Huang, J.; Ding, Y.; Wu, J.; Dong, S.; Kong, L.; Gao, G.; Li, C.-Y.; Wei, L. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011, 39, W316–W322. [Google Scholar] [CrossRef]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3-new capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef]
- De-Santis, C.; Jerry, D.R. Candidate growth genes in finfish-Where should we be looking? Aquaculture 2007, 272, 22–38. [Google Scholar] [CrossRef]
- Jia, Y.; Jing, Q.; Xing, Z.; Gao, X.; Zhai, J.; Guan, C.; Huang, B. Effects of two different culture systems on the growth performance and physiological metabolism of tiger pufferfish (Takifugu rubripes). Aquaculture 2018, 495, 267–272. [Google Scholar] [CrossRef]
- Jia, Y.; Jing, Q.; Zhai, J.; Guan, C.; Huang, B. Alternations in oxidative stress, apoptosis, and innate-immune gene expression at mRNA levels in subadult tiger puffer (Takifugu rubripes) under two different rearing systems. Fish. Shellfish. Immunol. 2019, 92, 756–764. [Google Scholar] [CrossRef] [PubMed]
- Hanhoff, T.; Lucke, C.; Spener, F. Insights into binding of fatty acids by fatty-acid binding proteins. Mol. Cell. Biochem. 2002, 239, 45–54. [Google Scholar] [CrossRef]
- Kanda, T.; Iseki, S.; Hitomi, M.; Kimura, H.; Odani, S.; Kondo, H.; Matsubara, Y.; Muto, T.; Ono, T. Purification and characterization of a fatty-acid-binding protein from the gastric mucosa of rats: Possible identity with heart fatty-acid-binding protein and its parietal cell localization. Eur. J. Biochem. 1989, 185, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Börchers, T.; Spener, F. Fatty acid binding proteins. Curr. Top. Membr. 1994, 40, 261–294. [Google Scholar]
- Pelsers, M.M.; Hermens, W.T.; Glatz, J.F. Fatty acid-binding proteins as plasma markers of tissue injury. Clin. Chim. Acta 2005, 352, 15–35. [Google Scholar] [CrossRef]
- Ockner, R.K.; Manning, J.A.; Poppenhausen, R.B.; Ho, W.K. A binding protein for fatty acids in cytosol of intestinal mucosa, liver, myocardium, and other tissues. Science 1972, 177, 56–58. [Google Scholar] [CrossRef]
- Stewart, J.M.; Driedzic, W.R. Fatty acid binding proteins in teleost fish. Can. J. Zool. 1988, 66, 2671–2675. [Google Scholar] [CrossRef]
- Londraville, R.L.; Sidell, B.L. Cold acclimation increases fatty acidbinding protein concentration in aerobic muscle of striped bass, Morone saxatilis. J. Exp. Zool. 1996, 275, 36–44. [Google Scholar] [CrossRef]
- Ando, S.; Xue, X.H.; Tibbits, G.F.; Haunerland, N.H. Cloning and sequencing of complementary DNA for fatty acid binding protein from rainbow trout heart. Comp. Biochem. Physiol. B 1998, 119, 213–217. [Google Scholar] [CrossRef]
- Jordal, A.E.O.; Hordvik, I.; Pelsers, M.; Bernlohr, D.A.; Torstensen, B.E. FABP3 and FABP10 in Atlantic salmon (Salmo salar L.)-General effects of dietary fatty acid composition and life cycle variations. Comp. Biochem. Physiol. B 2006, 145, 147–158. [Google Scholar] [CrossRef] [PubMed]
- Lücke, C.; Gutiérrez-González, L.H.; Hamilton, J.A. Intracellular lipid binding proteins: Evolution, structure, and ligand binding. In Cellular Proteins and Their Fatty Acids in Health and Disease; Duttaroy, A.S., Ed.; Wiley VCH: Weinheim, Germany, 2003; pp. 95–118. [Google Scholar]
- Bass, N.M. The cellular fatty acid binding proteins: Aspects of structure, regulation, and function. Int. Rev. Cytol. 1988, 3, 143–184. [Google Scholar]
- Binas, B.; Danneberg, H.; McWhir, J.; Mullins, L.; Clark, A.J. Requirement for the heart-type fatty acid binding protein in cardiac fatty acid utilization. FASEB J. 1999, 13, 805–812. [Google Scholar] [CrossRef]
- Leaver, M.J.; Boukouvala, E.; Antonopoulou, E.; Diez, A.; Favre-Krey, L.; Ezaz, M.T.; Bautista, J.M.; Tocher, D.R.; Krey, G. Three peroxisome proliferators activated receptor isotypes from each of two species of marine fish. Endocrinology 2005, 146, 3150–3162. [Google Scholar] [CrossRef]
- Wolfrum, C.; Borchers, T.; Sacchettini, J.C.; Spener, F. Binding of fatty acids and peroxisome proliferators to orthologous fatty acid binding proteins from human, murine, and bovine liver. Biochemistry 2000, 39, 1469–1474. [Google Scholar] [CrossRef] [PubMed]
- Parmar, M.B.; Wright, J.M. Comparative genomic organisation and tissue-specific transcription of the duplicated fabp7 and fabp10 genes in teleost fishes. Genome 2013, 56, 691–701. [Google Scholar] [CrossRef] [PubMed]
- Thirumaran, A.; Wright, J.M. Fatty acid-binding protein (fabp) genes of spotted green pufferfish (Tetraodon nigroviridis): Comparative genomics and spatial transcriptional regulation. Genome 2014, 57, 289–301. [Google Scholar] [CrossRef] [PubMed]
- Furuhashi, M.; Hotamisligil, G.S. Fatty acid-binding proteins: Role in metabolic diseases and potential as drug targets. Nat. Rev. Drug Discov. 2008, 7, 489–503. [Google Scholar] [CrossRef]
- Hotamisligil, G.S.; Bernlohr, D.A. Metabolic functions of FABPs-mechanisms and therapeutic implications. Nat. Rev. Endocrinol. 2015, 11, 592. [Google Scholar] [CrossRef]
- Ogino, T.; Moralejo, D.H.; Kose, H.; Yamada, T.; Matsumoto, K. Serum leptin concentration is linked to chromosomes 2 and 6 in the OLETF rat, an animal model of type 2 diabetes with mild obesity. Mamm. Genome 2003, 14, 239–244. [Google Scholar] [CrossRef]
- Gerbens, F.; Verburg, F.J.; van Moerkerk, H.T.B.; Engel, B.; Buist, W.; Veerkamp, J.H.; Te Pas, M.F.W. Associations of heart and adipocyte fatty acid-binding protein gene expression with intramuscular fat content in pigs. J. Anim. Sci. 2001, 79, 347–354. [Google Scholar] [CrossRef]
- Mercadé, A.; Pérez-Enciso, M.; Varona, L.; Alves, E.; Noguera, J.L.; Sánchez, A.; Folch, J.M. Adipocyte fatty-acid binding protein is closely associated to the porcine FAT1 locus on chromosome 4. J. Anim. Sci. 2006, 84, 2907–2913. [Google Scholar] [CrossRef]
- Power, D.M. Developmental ontogeny of prolactin and its receptor in fish. Gen. Comp. Endocr. 2005, 142, 25–33. [Google Scholar] [CrossRef]
- Bern, H.A. Functional evolution of prolactin and growth hormone in lower vertebrates. Am. Zool. 1983, 23, 663–671. [Google Scholar] [CrossRef]
- WClarke, C.; Bern, H.A. Comparative endocrinology of prolactin. In Hormonal Proteins and Peptides; Li, C.H., Ed.; Academic Press: New York, NY, USA, 1980; pp. 105–197. [Google Scholar]
- Yang, B.Y.; Greene, M.; Chen, T.T. Early embryonic expression of the growth hormone family protein genes in the developing rainbow trout, Oncorhynchus mykiss. Mol. Reprod. Dev. 1999, 53, 127–134. [Google Scholar] [CrossRef]
- Santos, C.R.A.; Brinca, L.; Ingleton, P.M.; Power, D.M. Cloning, expression, and tissue localisation of prolactin in adult sea bream (Sparus aurata). Gen. Comp. Endocr. 1999, 114, 57–66. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.M.; Kaneko, T.; Aida, K. Prolactin and prolactin receptor expressions in a marine teleost, pufferfish Takifugu rubripes. Gen. Comp. Endocr. 2006, 146, 318–328. [Google Scholar] [CrossRef]
- Shepherd, B.S.; Sakamoto, T.; Nishioka, R.S.; Richman, N.H.; Mori, I.; Madsen, S.S.; Chen, T.T.; Hirano, T.; Bern, H.A.; Grau, E.G. Somatotropic actions of the homologous growth hormone and prolactins in the euryhaline teleost, the tilapia, Oreochromis mossambicus. Proc. Natl. Acad. Sci. USA 1997, 94, 2068–2072. [Google Scholar] [CrossRef]
- Ribas, L.; Robledo, D.; Gómez-Tato, A.; Viñas, A.; Martínez, P.; Piferrer, F. Comprehensive transcriptomic analysis of the process of gonadal sex differentiation in the turbot (Scophthalmus maximus). Mol. Cell Endocrinol. 2016, 422, 132–149. [Google Scholar] [CrossRef]
- Kobayashi, Y.; Nagahama, Y.; Nakamura, M. Diversity and plasticity of sex determination and differentiation in fishes. Sex. Dev. 2013, 7, 115–125. [Google Scholar] [CrossRef]
- Du, X.; Wang, B.; Liu, X.; Liu, X.; He, Y.; Zhang, Q.; Wang, X. Comparative transcriptome analysis of ovary and testis reveals potential sex-related genes and pathways in spotted knifejaw Oplegnathus punctatus. Gene 2017, 637, 203–210. [Google Scholar] [CrossRef] [PubMed]
- Cui, Z.; Liu, Y.; Wang, W.; Wang, Q.; Zhang, N.; Lin, F.; Wang, N.; Shao, C.; Dong, Z.; Li, Y.; et al. Genome editing reveals dmrt1 as an essential male sex-determining gene in Chinese tongue sole (Cynoglossus semilaevis). Sci. Rep. 2017, 7, 42213. [Google Scholar] [CrossRef]
- Webster, K.A.; Schach, U.; Ordaz, A.; Steinfeld, J.S.; Draper, B.W.; Siegfried, K.R. Dmrt1 is necessary for male sexual development in zebrafish. Dev. Biol. 2017, 422, 33–46. [Google Scholar] [CrossRef]
- Bowles, J.; Schepers, G.; Koopman, P. Phylogeny of the SOX family of developmental transcription factors based on sequence and structural indicators. Dev. Biol. 2000, 227, 239–255. [Google Scholar] [CrossRef]
- Kamachi, Y.; Kondoh, H. Sox proteins: Regulators of cell fate specification and differentiation. Development 2013, 140, 4129–4144. [Google Scholar] [CrossRef] [PubMed]
- Takehana, Y.; Matsuda, M.; Myosho, T.; Suster, M.L.; Kawakami, K.; Shin-I, T.; Kohara, Y.; Kuroki, Y.; Toyoda, A.; Fujiyama, A.; et al. Co-option of Sox3 as the male-determining factor on the Y chromosome in the fish Oryzias dancena. Nat. Commun. 2014, 5, 4157. [Google Scholar] [CrossRef] [PubMed]
- Yao, B.; Zhou, L.; Wang, Y.; Xia, W.; Gui, J. Differential expression and dynamic changes of SOX3 during gametogenesis and sex reversal in protogynous hermaphroditic fish. J. Exp. Zool. A Ecol. Genet. Physiol. 2007, 307, 207–219. [Google Scholar] [CrossRef]
- Li, S.; Lin, G.; Fang, W.; Huang, P.; Gao, D.; Huang, J.; Xie, J.; Lu, J. Gonadal transcriptome analysis of sex-related genes in the protandrous yellowfin seabream (Acanthopagrus latus). Front. Genet. 2020, 11, 709. [Google Scholar] [CrossRef] [PubMed]
- Dumont, J.N.; Brummett, A.R. Egg envelopes in vertebrates. Dev. Biol. 1985, 1, 235–288. [Google Scholar]
- Wassarman, P.M. Zona pellucida glycoproteins. Annu. Rev. Biochem. 1988, 57, 415–442. [Google Scholar] [CrossRef]
- Litscher, E.S.; Wassarman, P.M. The fish Egg’s zona Pellucida. Curr. Top. Dev. Biol. 2018, 130, 275–305. [Google Scholar]
- Litscher, E.S.; Wassarman, P.M. Egg extracellular coat proteins: From fish to mammals. Histol. Histopathol. 2007, 22, 337–347. [Google Scholar]
- Liu, X.; Wang, H.; Gong, Z. Tandem-repeated zebrafish zp3 genes possess oocyte-specific promoters and are insensitive to estrogen induction1. Biol. Reprod. 2006, 74, 1016–1025. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).