Abstract
The effects of replacing 40% of dietary fish meal (FM) in a reference diet (REF) with either mussel meal (MM), zygomycete fungi (ZYG), extracted baker’s yeast (EY), or non-extracted baker’s yeast (NY) on the lipid and metabolic profile of Arctic charr (Salvelinus alpinus) were investigated. After a 14-week feeding trial, liver and muscle tissues were collected for lipid (lipid content, lipid class, fatty acid composition) and 1H NMR-based metabolomics analyses (aqueous and chloroform phases). Lipid analyses showed that fish fed ZYG diet had lower liver lipid content and thereby 10% higher level of docosahexaenoic acid compared with REF. Metabolomics analyses showed that on the one hand fish fed NY diet affected liver metabolites (2–3 fold higher concentrations of e.g., n,n-dimethylglycine and betaine) compared with REF, while, on the other hand, the muscle metabolic fingerprint was mainly affected by EY. In general, affected metabolites (e.g., alanine, anserine, betaine, hydroxyproline, isoleucine, malonate, n,n-dimethylglycine, proline, succinate, and valine) in fish fed test diets suggested that the test meal ingredients caused mainly a response in muscle metabolism. Fish metabolism was least affected by MM, which suggests that it may be suitable to replace fish meal in Arctic charr diets.
1. Introduction
Aquaculture is the fastest growing animal production sector in the world. During the period 2001–2016, world fish production expanded at an annual rate of about 5.8% [1]. Fish and fish products intended for human consumption have increased up to 151 MT per year, and approximately 47% of the seafood protein consumed worldwide originates from the aquaculture industry [1]. Thus, the demand for fish meal (FM), a traditional component and key source of feedstuffs for aquaculture, is steadily growing. Fish meal is the evolutionary base of formulated aquafeeds for carnivorous fish and thereby of good digestibility and amino acid (AA) profile, for maximal protein utilization and growth [2,3]. However, due to high demand and the fast growth of aquaculture worldwide, FM production for aquafeeds is becoming non-sustainable, with wild-caught FM products reaching their production limits and increasing feed costs [2]. Therefore, it is necessary to develop alternative proteins to feeds to achieve long-term sustainable fish production [4].
Alternative protein sources such as blue mussel (Mytilus edulis L.) and microorganisms are becoming attractive. Blue mussels feed on phytoplankton. The mussel flesh has a high protein content with suitable AA composition. The fat content is low, while the lipid composition satisfies the requirements for the essential fatty acids (FA) eicosapentaenoic acid (EPA, 20:5n-3) and docosahexaenoic acid (DHA, 22:6n-3) in e.g., rainbow trout (Oncorhynchus mykiss) [5]. Therefore, the use of blue mussel as a FM replacement has several advantages, such as lowering nutrient level in sea water by filtrating phytoplankton, and supplying a high-quality protein and lipid (DHA and EPA) source [6]. Furthermore, Pan [7] and Langeland et al. [8] showed that growth performance and digestibility in Arctic charr were unaffected when de-shelled blue mussels representing 520 g kg−1 and 320 g kg−1 of the diet (as is basis) were compared to fish meal-based diets.
Microorganisms such as yeast and other fungi, bacteria, and micro-algae can become a suitable source of proteins, vitamins, minerals, lipids, and carbohydrates [9] and are candidates for use in aquafeeds.
Zygomycete fungi (Rhizopus oryzae) are filamentous fungi which are cultivated on e.g., paper pulp wastewater (spent sulphite liquor). Previous studies have shown that the biomass obtained contains a high protein content (40–50% dry-matter basis depending on harvesting, dewatering, and drying method), vitamins, and an AA composition comparable to that of FM [10,11,12]. Partial replacement of FM with zygomycete fungi has been shown to have no effect on feed intake and growth performance in rainbow trout [13] as well as Atlantic salmon (Salmo salar) [14].
Another alternative protein source for aquafeeds could be baker’s yeast (Saccharomyces cerevisiae) [8]. The total nitrogen of baker’s yeast (40–65%) is slightly lower, than in FM [9,15]. Inclusion of yeast into fish diets can cause an AA imbalance compared with FM due to its lower levels of sulphur-containing AA such as methionine [9]. Several studies have shown that growth performance is affected depending on the amount of baker’s yeast used in fish diets. It has been shown that 50% replacement of total nitrogen with baker’s yeast in the diet of lake trout (Salvelinus namaycush) [16] and with brewer’s yeast in the diet of sea bass (Dicentrarchus labrax) [17] has no negative effect on growth performance. In contrast, lower final weight has been reported in Atlantic salmon fed a diet with baker’s yeast replacing 40% of crude protein from FM [18].
The replacement of feed ingredients with alternative sources may affect the metabolism of the fish. Therefore, it is of interest to analyse and compare the changes occurring in the fish body using a metabolomics approach. Metabolomics is defined as the comprehensive analysis of small metabolites in an organism, tissue, or biofluid under a given set of conditions [19]. Use of high-resolution 1H nuclear magnetic resonance (NMR) spectroscopy is a well-established technique and used in our previous studies, to explore metabolic changes in fish liver [12,14,20,21,22] and white muscle [20,23] in response to different diets. Metabolomics of white muscle can provide insights into changes in muscle metabolism and indicate possible effects on the final flesh quality of the fish dependent on the diet. The liver is the key organ in relation to biosynthesis and catabolism. Extracts from liver and white muscle can thus provide information regarding FA distribution (lipid phase) and small metabolites (aqueous phase).
We have performed several studies using alternative protein sources as replacement for fish meal, which are not usable as food for human. The focus was to use blue mussels, too small to be of interest for the food industry and microbes cultivated on waste streams (e.g., zygomycete fungi and baker’s yeast) as partial replacements.
The aim of the present study is to explore alternative protein sources to fish meal for feed of salmonid fish with focus on the metabolic profile.
2. Material and Methods
2.1. Fish Trial
A full description of the experimental design, feed, and growth of the fish can be found in Vidakovic et al. [24]. In brief, Arctic charr with an initial body weight of 47.8 ± 0.81 g (mean ± SEM) were individually tagged and distributed in triplicate groups of 50 fish into 15 experimental tanks (700 L) provided with flow-through freshwater (10 L/min) from Lake Ansjön at an average temperature of 7 °C. The growth trial was performed at the Aquaculture Center North, Kälarne (Vattenbrukscentrum Norr AB, Kälarne, Sweden) as described by Vidakovic et al. [24]. The fish were fed for 14 weeks (March–June) at a rate of 1% body weight (BW) per day using ‘automatic’ feeders. The feed ration was increased weekly based on a specific growth rate (SGR) of 0.7 at 6 °C [25]. Restricted feeding was used instead of feeding to satiation, to ensure that all fish eat the same amount of each diet to reduce the metabolic effects of different feed ratios. That all feed was consumed was checked by trained staff, before and during the experimental period. The experiment was carried out in accordance with EU legislation (i.e., Directive 2010/63/EU), and received the approval of the Ethical Committee for Animal Experiments in Umeå, Sweden.
2.2. Diet
The diet formulation and proximate composition of the feed ingredients and diets, including crude lipid and crude protein, are shown in Table 1 and the FA composition is shown in Table 2. The REF diet contained FM as the main protein source. In the four experimental diets, 40% of the crude protein content in FM was replaced: mussel meal (MM), non-extracted baker’s yeast (NY), extracted baker’s yeast (EY), or zygomycete fungi (ZYG), respectively (Table 1). The NY (Jästbolaget®, Stockholm, Sweden) was cultured on molasses with ammonia, phosphorus, magnesium, and vitamins and then dried on a fluidized bed. The EY (Alltech Serbia AB, Senta, Serbia) was produced by autolysis after removing the solid phase (cell walls) by centrifugation, and the liquid phase was then dried. The ZYG (Cewatech AB, Gothenburg, Sweden) was cultivated on spent sulphite liquor. The MM (Royal Frysk Muscheln GmbH, Emmelsbüll-Horsbüll, Germany) contained dried, de-shelled blue mussels of food grade [8]. All diets were extruded at the Finnish Game and Fisheries Research Institute (Laukaa Research Station, Laukaa, Finland) on a twin-screw extruder (3 mm die, BC-45 model; Clextral, Creusot Loir, France). During the extrusion process, 20% of additional moisture was added to the feed mash, which was heated to 120–130 °C for 30 s, dried overnight by warm air and then sprayed with lipids using a vacuum coater (Pegasus PG-10VC; Dinnissen, Sevenum, The Netherlands). Diets were formulated on a digestible iso-nitrogenous and iso-energetic basis in order to perform reliable comparisons between the diets.

Table 1.
Composition and nutrient content in experimental diets to Arctic charr. The dry matter (DM) of all feed ingredients are expressed as %, while crude protein, crude lipid, and ash are expressed as g/kg DM and energy content as (MJ kg−1 DM). Data from Vidakovic et al. [24].

Table 2.
Fatty acid composition in the experimental diets fed to Arctic charr. Expressed as % of total fatty acids.
2.3. Sampling
After 14 weeks, the fish were anesthetized (MS-222, 100 mg/L; Sigma Chemicals Co. St. Louis, MO, USA) and killed by cutting the spinal cord. White muscle samples (n = 9 per diet) were prepared by skinning and deboning the fillets. White muscle was taken from the region between the dorsal fin and lateral line. Liver (n = 9 per diet) was removed and dissected into two parts, for metabolomics (pars dexter) and lipid (pars sinister) analyses. The samples were frozen directly in liquid nitrogen and stored at −80 °C until further analyses by NMR spectroscopy and gas chromatography (GC).
2.4. 1H NMR-Based Metabolomics Assays
2.4.1. Sample Preparation for Metabolomics Study of Liver and White Muscle
The 1H-NMR analysis of liver and white muscle samples (n = 9 per group) was performed according to Wagner et al. [20] with a few modifications. In brief, frozen tissue sample (100 mg) was homogenized (Ultra Turrax T25, Ika Werke, Staufen, Germany) for 1 min with methanol-chloroform (2:1, v:v, 3 mL). After sonication (30 min) and then vortexing for 1 min with an additional 1 mL of ice-cold chloroform and water, the sample was centrifuged (Eppendorf centrifuge 5810 R, Eppendorf AB, Hamburg, Germany) at 1811 g for 35 min at 4 °C. After phase separation, the aqueous and chloroform supernatants were collected in separate tubes.
The supernatant of the aqueous phase (polar phase) was dried using an evacuated centrifuge (Savant, SVC 100H, Techtum Instrument AB, Umeå, Sweden) and re-dissolved in sodium phosphate buffer (280 µL, 0.25 M, pH = 7.0) and Millipore Water (240 µL). Nanosep centrifugal filters with 3 kDa cutoff (Pall Life Science, Port Washington, NY, USA) were used to filter the samples, after glycerol was removed from the filter membrane by washing 10 times with 0.5 mL Millipore water (1500 g, 36 °C, 15 min). Liver and white muscle samples (490 µL) were filtered at 12,000 g at 4 °C for 15 min. To the liver and white muscle filtrates (360 µL and 420 µL, respectively), deuterium oxide (D2O, 50 µL), sodium-3-(trimethylsilyl)-2,2,3,3-tetradeuteriopropionate solution (TSP-d4, 30 µL, 5.8 mmol/L, Cambridge Isotope Laboratories, Andover, MA, USA) as internal standard, Millipore water (82 µL and 55 µL, respectively), and sodium phosphate buffer (78 µL and 45 µL, respectively) were added, so that the total volume of each sample was 600 µL. The samples were then analysed with NMR spectroscopy.
The chloroform phase (organic phase) was evaporated under nitrogen, dissolved in 600 µL CDCL3 (99.96 atom% D) and analysed with NMR spectroscopy.
2.4.2. 1H NMR Spectroscopic Analyses
The NMR analyses were performed on the samples in 5 mm outer diameter NMR tubes using a Bruker spectrometer (Karlsruhe, Germany) operating at 600 MHz equipped with a cryogenically cooled probe and autosampler. Tuning and shimming was performed for each sample.
One-dimensional 1H-NMR analysis of the aqueous phase using zgesgp pulse sequence (Bruker BioSpin GmbH, Rheinstetten, Germany) was performed at 25 °C with 128 scans and 65,536 data points over a spectral width of 17,942.58 Hz. Acquisition time was 1.83 s and relaxation delay was 4.0 s. Two-dimensional correlation spectroscopy (COSY) and total correlation spectroscopy (TOCSY) with pre-saturation were performed with 32 scans and a spectral width of 7195 Hz for both F1 and F2. The mixing time for TOCSY was 80 ms. Heteronuclear single quantum correlation (HSQC) was performed using 32 scans and a spectral width of 7211 Hz and 25,002.09 Hz for proton and carbon, respectively. For the identification of signals a standard pulse sequence from the Bruker library was used. The 1H NMR spectra of the chloroform phase were recorded using zg30 pulse sequence (Bruker BioSpin GmbH, Rheinstetten, Germany) at 20 °C with 128 scans and 65,536 data points over a spectral width of 12,019.23 Hz. The acquisition time was 2.72 s and the relaxation delay 3 s.
2.4.3. Data Processing and Signal Identification
The NMR spectra data were processed using Bruker Topspin 2.0 software (Bruker BioSpin GmbH, Rheinstetten, Germany), Fourier-transformed after multiplication by line broadening of 0.3 Hz and subsequently referenced to standard peak TSP-d4 at 0.0 ppm in the aqueous phase and to chloroform peak at 7.24 ppm in the chloroform phase. After the baseline and spectral phase were corrected manually, the spectra of 45 fish liver and white muscle sample extracts were integrated using AMIX 3.7.3 (Bruker BioSpin GmbH, Rheinstetten, Germany) into 0.01 ppm integral regions (buckets) between 0.3–9.5 ppm (without water signal: 4.7–5.0 ppm) for the aqueous phase and 0.5–5.6 ppm for the chloroform phase. For the aqueous phase, each spectral region was normalized to the intensity of internal standard (TSP-d4) to ensure quantitative measurements. For the chloroform phase, each spectral region was normalized to the total intensity. For the aqueous phase of liver and white muscle samples the ChenomX NMR Suite version 7.5 profiler (ChenomX Inc., Edmonton, AB, Canada) was used to identify and quantify compounds. 56 metabolites for liver and 48 metabolites for white muscle were identified by overlapping with standard spectra, and their concentrations were expressed in mmol/g tissue. The identification of 1H NMR signals was performed primarily using ChenomX NMR Suite 7.5 library (ChenomX Inc, Edmonton, AB, Canada), the Human Metabolome Database (www.hmdb.ca), and previous literature [14,20,26,27] and confirmed with 2D-NMR (COSY, TOCSY, and HSQC) in case of multiplicity.
2.5. Total Lipid and Fatty Acid Analysis
Total lipid analysis of liver, white muscle, and diets was performed according to Mraz et al. [28]. In brief, 1 g of sample was homogenized in hexane: isopropanol (HIP; 3:2, v:v) with an Ultra-Turrax (T25, IKA Werke, Staufen, Germany). For lipid and non-lipid phase separation, 6.67% of Na2SO4 was added to the homogenate and centrifuged. After gravimetrical identification of the total lipid content from dried samples, the lipids were stored in hexane at −80 °C until further analysis. All chemicals and solvents were purchased from Merck (Darmstadt, Germany) except chloroform (Sigma Chemicals Co. St. Louis, MO, USA). The solvents were used without further purification.
Fatty acid methyl esters (FAME) from total lipids in liver, white muscle, and diets were prepared with BF3 according to the method described by Appelqvist [29]. FAME were stored in hexane at −80 °C until further analysis.
FAME were analysed by GC using a CP 3800 instrument (Varian AB, Stockholm, Sweden) equipped with a flame ionization detector and a split injector, and separated on a 50 m length fused silica capillary column BPX 70 (SGE, Austin, TX, USA) (0.22 mm i.d. × 0.25 µm film thickness) [30]. The injector temperature was 230 °C and the detector temperature 250 °C. Helium was used as carrier gas, at a flow rate of 0.8 mL/min, and nitrogen was used as make-up gas. Peaks were identified by comparing their retention times with those of the standard mixture GLC 68A (Nu-check Prep, Elysian, MN, USA) and quantified using an internal standard (methyl-15-methylheptadecanoate; Larodan Fine Chemicals AB, Malmö, Sweden). Peak areas were integrated using Galaxie chromatography data system software version 1.9 (Varian AB, Stockholm, Sweden). For each experimental diet, liver (n = 9) and white muscle (n = 9) samples were analysed in duplicate.
2.6. Lipid Class Analyses
Lipid class composition was analysed according to Mraz and Pickova [28] with minor modifications. For the analysis of liver, samples with a concentration of 1 µg/µL hexane were prepared in duplicate 200 µL vials. The samples were applied on pre-coated silica gel 60 high performance thin layer chromatography (HPTLC) plates (20 cm × 10 cm; 0.2 mm layer; Merck, Darmstadt, Germany) using a Camag ATS 4 automatic TLC sampler. Lipid classes were separated using a Camag ADC 2 developing chamber and a hexane-diethyl ether-acetic acid (85:15:2, v:v:v) mobile phase for approximately 55 min, including drying. Afterwards, the plate was dipped in a phosphomolybdenic acid/ethanol (20 g in 200 mL) solution for derivatization and dried at 120 °C for 8 min. A detection scanner (Camag TLC scanner 3, Muttenz, Switzerland) at a wavelength of 650 nm was used for densitometric measurement of the proportions of different lipid classes. Lipid classes were identified by comparing the samples with an external standard (TLC 18-5A; Nu-Check Prep, Elysian, MN, USA), integrated with Wincats software package (version 1.4.8; Camag, Muttenz, Switzerland) and expressed as percentage of height.
2.7. Statistics
Multivariate data analysis for the absolute concentrations of the metabolites was performed using Simca-P software (version 13.0; Umetrics, Umeå, Sweden). All variables were “Unit Variance” (UV)-scaled. Principal component analysis (PCA) was used to get a first overview of the data and search for outliers. Outliers were determined using PCA-Hotelling T2 Ellipse (95% confidential interval (CI)) and DModX (95% CI). The multivariate data were checked for normal distribution using the normal probability plot of the PCA model. Partial least squares projection to latent structures-discriminant analysis (PLS-DA) and orthogonal partial least squares projection to latent structures-discriminant analysis (OPLS-DA), supervised techniques, were performed for classification of different treatments. OPLS-DA loading plot and variable influences in projection (VIP) plot for spectral regions with VIP > 1 and with jack-knife-based CI that did not include unity were considered to be discriminative between the different diets. Significance in the OPLS-DA model was tested using cross-validation (CV) analysis of variance (ANOVA) (p < 0.05), which is a diagnostic tool for assessing the reliability of OPLS-DA models [31]. Furthermore, the absolute concentrations of metabolites were tested for differences between diets and reference, using SAS 9.4 (SAS Institute Inc., Cary, NC, USA). Because of some non-normal distributions, the non-parametric Kruskal–Wallis test (PROC NPAR1WAY), was used to identify if there were any differences between groups. If the data showed a significant effect (p < 0.05), pairwise comparisons between REF and each of the four diets (REF vs. MM, REF vs. EY, REF vs. NY, REF vs. ZGY) were conducted by multiple Mann–Whitney tests with the DSCF option of PROC NPAR1WAY (see SAS online help). Furthermore, a false discovery rate (FDR) method was used with PROC MULTTEST to reduce the number of false positive results and obtain significant results, corrected for multiple testing, with an FDR alpha-level of 0.05.
For the lipid analysis, percentage concentration of individual FA, FA groups, and lipid classes were arcsine-transformed for normal distribution (Anderson-Darling test) approximation, followed by a test of equal variance (Bartlett or Levene test). The data were further analysed by the Kruskal–Wallis test, to check for any differences between groups. In case of significant differences (p < 0.05) pairwise comparisons between REF and each of the other four groups were conducted by multiple Mann–Whitney tests with the DSCF option using software SAS.
All data presented are mean ± SEM and differences were regarded as significant at p < 0.05.
3. Results
3.1. Growth Performance
Minor mortalities (0.67% in REF, NY, EY and ZYG) were detected during the feeding trial. The average fish body start weight was 47.7 ± 0.32 g (p = 0.652) and the final weights for the different fish groups were 133.3 ± 2.56 g (REF), 126.4 ± 2.30 g (MM), 117.9 ± 2.74 *** g (EY), 125.6 ± 2.34 g (NY) and 118.5 ± 2.21 *** g (ZYG) (mean ± SEM, *** = p < 0.001) after a 14-week feeding trial [24]. A similar pattern was found for SGR; fish fed REF diet had highest SGR (1.08 ± 0.01%/day), while EY (0.95 ± 0.01%/day) and ZYG (0.97 ± 0.01%/day) (p ≤ 0.001) showed the lowest SGR value. The SGR did not significantly differ in fish fed MM (1.02 ± 0.01%/day) and NY (1.04 ± 0.01%/day) diet (for more details see [24]) compared with REF.
3.2. 1H NMR-Based Metabolomics Analysis
3.2.1. Aqueous Extracts from Liver Tissue
A representative 1H NMR spectrum of the aqueous phase from liver extracts is shown in Figure 1A. In order to identify metabolic changes in fish, the absolute concentrations of 56 metabolites were quantified through a profiling approach from 1H NMR spectra. One outlier was observed in the MM group and one in the ZYG group by PCA model and therefore, excluded from the dataset. In the PCA model, the first and second components explained 29.4% and 8.96% of variation, respectively, and the total amount of X variation was 68.5% using seven principal components (model parameters: R2X = 68.5%, Q2 = 21.9%) (Figure 2A). The investigation with the PLS-DA model showed no valid results, therefore only OPLS-DA model was used for further analyses. An OPLS-DA model with four predictive (24.5%) and two orthogonal components (33.6%) was generated (model parameters: R2X = 58.1%, R2 = 76.2%, Q2 = 47.0%, R2Y = 100%, CV-ANOVA = 0.0023) from the dataset (Figure 2B). The OPLS-DA score plot showed clear separations between REF and ZYG along vertical axis and REF and NY as well as REF and MM along the horizontal axis. For identification of the metabolites in liver causing the differences between REF and the other four groups, the S-plot, VIP-plot and loading column plot (Supplementary, Figure S1A) of OPLS-DA were used. The concentrations of metabolites were further investigated using Kruskal–Wallis test. The analyses of aqueous liver extract showed an effect in four metabolites. Compared with REF fish, 3-aminoisobutyrate was higher in MM fish, o-phosphocholine in ZYG fish, and betaine and n,n-dimethylglycine in NY fish (Table 3).
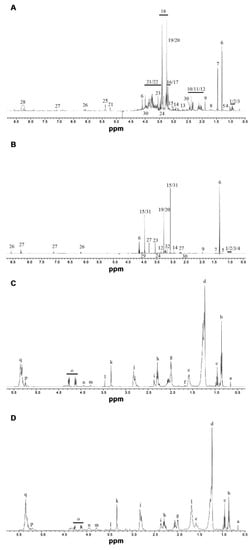
Figure 1.
Representative 600 MHz 1H NMR spectra of Arctic charr liver and muscle extracts fed REF diet. (A) aqueous liver; (B) aqueous muscle; (C) chloroform liver and (D) chloroform muscle. Assignments (the relevant proton(s) for specific assignments are indicated by bold letter): (1/2/3) isoleucine, leucine, 2-aminobutyrate; (4) valine; (5) 3-aminoisobutyrate; (6) lactate; (7) alanine; (8) lysine; (9) acetate; (10/11/12) glutamine/glutamate/proline; (13) sarcosine; (14) n,n-dimethylglycine; (15) creatine; (16/17) choline/phosphocholine; (18) taurine; (19/20) betaine/trimethylamine n-oxide; (21/22) glucose/glycerol; (23) glycine; (24) hydroxyproline; (25) glycogen; (26) nucleosides; (27) anserine; (28) inosine; (29) serine; (30) succinate; (31) creatine phosphate; (32) malonate. (a) cholesterol; (b) all FA, –CH3 (except n-3 FA); (c) n-3 FA –CH3; (d) all FA –(CH2)n– (except EPA and DHA); (e) all FA –CH2-CH2-COOH (except DHA); (f) EPA (20:5n-3) =CH-CH2-CH2-CH2-COOH; (g) unsaturated FA –CH2-CH=CH; (h) all FA –CH2-COOH (except DHA); (i) DHA (22:6n-3) =CH-CH2-CH2-COOH; (j) polyunsaturated FA =CH-CH2-CH=; (k) phosphatidylcholine –N(CH3)3; (l) unassigned resonance; (m) phospholipid; (n) phosphatidylcholine, phosphatidylethanolamine glyceryl moiety; (o) glyceryl moiety (C1,3 proton); (p) glyceryl moiety (C2); (q) unsaturated FA –CH=CH–.
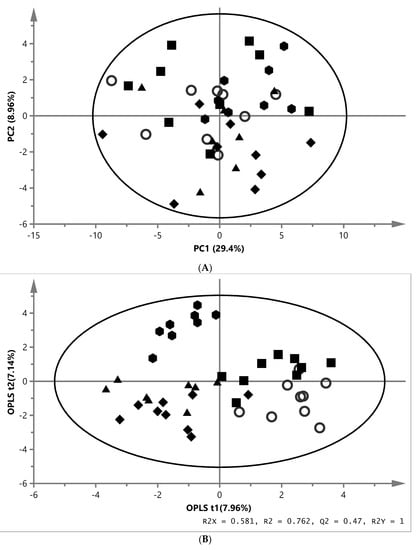
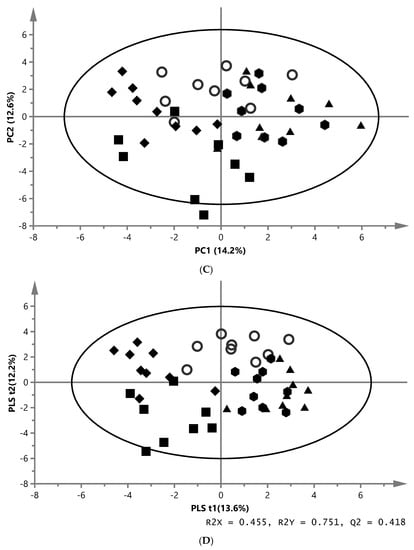
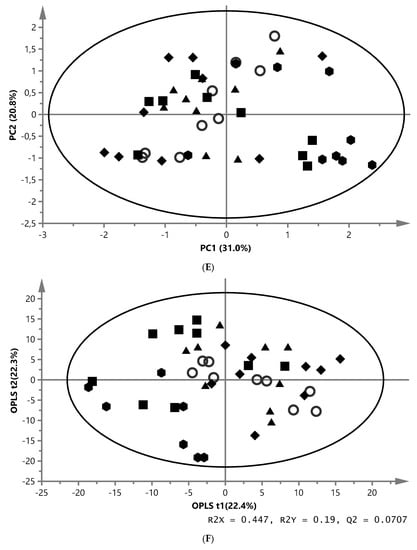
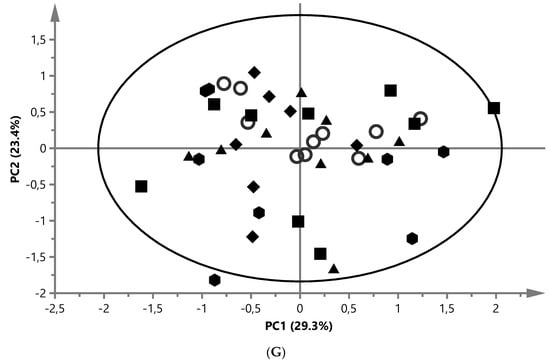
Figure 2.
Principal component analysis (PCA), partial least squares (PLS-DA) and orthogonal partial least squares (OPLS-DA) score plots based on the metabolic fingerprint from liver and white muscle extracts of fish fed with. REF ( ), MM (
), MM ( ), EY (
), EY ( ), NY (
), NY ( ) and ZYG (
) and ZYG ( ). (A) PCA score plot of aqueous liver extract. The first component explained 29.4% and the second 8.96% of variation (model parameters: R2X = 68.5%, Q2 = 21.9%, 7 components). (B) OPLS-DA score plot of aqueous liver extract. The total explained X variation was 58.1%. Of this, 24.5% was predictive and 33.6% was structured (model parameters: R2X = 58.1%, R2 = 76.2%, Q2 = 47.0%, R2Y = 100%, CV-ANOVA = 0.0023, 4 + 2 + 0 components). (C) PCA score plot of aqueous white muscle extracts. The first two components explained 14.2% and 12.6% of variation, respectively (R2X = 45.6%, Q2 = 4.06%, 4 components). (D) PLS-DA score plot of aqueous white muscle extract, using five components (R2X = 45.5%, Q2 = 41.8%, R2Y = 75.1%, CV-ANOVA = 0.000033). (E) PCA score plot of chloroform liver extract. The first component explained 31.0% of variation and the second 20.8% (model parameters: R2X = 61.5%, Q2 = 47.7%, 3 components). (F) OPLS-DA score plot of chloroform liver extract using two predictive components (model parameters: R2X = 44.7%, Q2 = 7.07%, R2Y = 19.0%, CV-ANOVA = 0.018). (G) PCA score plot of chloroform muscle extract. The first and second components explained 29.3% and 23.4% of variation, respectively (model parameters: R2X = 94.1%, Q2 = 70.8%, 10 components).
). (A) PCA score plot of aqueous liver extract. The first component explained 29.4% and the second 8.96% of variation (model parameters: R2X = 68.5%, Q2 = 21.9%, 7 components). (B) OPLS-DA score plot of aqueous liver extract. The total explained X variation was 58.1%. Of this, 24.5% was predictive and 33.6% was structured (model parameters: R2X = 58.1%, R2 = 76.2%, Q2 = 47.0%, R2Y = 100%, CV-ANOVA = 0.0023, 4 + 2 + 0 components). (C) PCA score plot of aqueous white muscle extracts. The first two components explained 14.2% and 12.6% of variation, respectively (R2X = 45.6%, Q2 = 4.06%, 4 components). (D) PLS-DA score plot of aqueous white muscle extract, using five components (R2X = 45.5%, Q2 = 41.8%, R2Y = 75.1%, CV-ANOVA = 0.000033). (E) PCA score plot of chloroform liver extract. The first component explained 31.0% of variation and the second 20.8% (model parameters: R2X = 61.5%, Q2 = 47.7%, 3 components). (F) OPLS-DA score plot of chloroform liver extract using two predictive components (model parameters: R2X = 44.7%, Q2 = 7.07%, R2Y = 19.0%, CV-ANOVA = 0.018). (G) PCA score plot of chloroform muscle extract. The first and second components explained 29.3% and 23.4% of variation, respectively (model parameters: R2X = 94.1%, Q2 = 70.8%, 10 components).
 ), MM (
), MM ( ), EY (
), EY ( ), NY (
), NY ( ) and ZYG (
) and ZYG ( ). (A) PCA score plot of aqueous liver extract. The first component explained 29.4% and the second 8.96% of variation (model parameters: R2X = 68.5%, Q2 = 21.9%, 7 components). (B) OPLS-DA score plot of aqueous liver extract. The total explained X variation was 58.1%. Of this, 24.5% was predictive and 33.6% was structured (model parameters: R2X = 58.1%, R2 = 76.2%, Q2 = 47.0%, R2Y = 100%, CV-ANOVA = 0.0023, 4 + 2 + 0 components). (C) PCA score plot of aqueous white muscle extracts. The first two components explained 14.2% and 12.6% of variation, respectively (R2X = 45.6%, Q2 = 4.06%, 4 components). (D) PLS-DA score plot of aqueous white muscle extract, using five components (R2X = 45.5%, Q2 = 41.8%, R2Y = 75.1%, CV-ANOVA = 0.000033). (E) PCA score plot of chloroform liver extract. The first component explained 31.0% of variation and the second 20.8% (model parameters: R2X = 61.5%, Q2 = 47.7%, 3 components). (F) OPLS-DA score plot of chloroform liver extract using two predictive components (model parameters: R2X = 44.7%, Q2 = 7.07%, R2Y = 19.0%, CV-ANOVA = 0.018). (G) PCA score plot of chloroform muscle extract. The first and second components explained 29.3% and 23.4% of variation, respectively (model parameters: R2X = 94.1%, Q2 = 70.8%, 10 components).
). (A) PCA score plot of aqueous liver extract. The first component explained 29.4% and the second 8.96% of variation (model parameters: R2X = 68.5%, Q2 = 21.9%, 7 components). (B) OPLS-DA score plot of aqueous liver extract. The total explained X variation was 58.1%. Of this, 24.5% was predictive and 33.6% was structured (model parameters: R2X = 58.1%, R2 = 76.2%, Q2 = 47.0%, R2Y = 100%, CV-ANOVA = 0.0023, 4 + 2 + 0 components). (C) PCA score plot of aqueous white muscle extracts. The first two components explained 14.2% and 12.6% of variation, respectively (R2X = 45.6%, Q2 = 4.06%, 4 components). (D) PLS-DA score plot of aqueous white muscle extract, using five components (R2X = 45.5%, Q2 = 41.8%, R2Y = 75.1%, CV-ANOVA = 0.000033). (E) PCA score plot of chloroform liver extract. The first component explained 31.0% of variation and the second 20.8% (model parameters: R2X = 61.5%, Q2 = 47.7%, 3 components). (F) OPLS-DA score plot of chloroform liver extract using two predictive components (model parameters: R2X = 44.7%, Q2 = 7.07%, R2Y = 19.0%, CV-ANOVA = 0.018). (G) PCA score plot of chloroform muscle extract. The first and second components explained 29.3% and 23.4% of variation, respectively (model parameters: R2X = 94.1%, Q2 = 70.8%, 10 components).

Table 3.
Significantly different absolute concentrations of metabolites (mmol/g) in aqueous liver and white muscle extracts of Arctic charr fed the experimental diets.
3.2.2. Aqueous Extracts from White Muscle Tissue
A representative 1H NMR spectrum of the aqueous phase from white muscle extracts is shown in Figure 1B. Multivariate data analyses were similar to those performed for aqueous liver extracts. The absolute concentrations of 48 metabolites were quantified through the profiling approach in order to identify metabolic changes in the muscle. One outlier from the EY group was removed. The fitted PCA model was explained by four PC, whereof PC1 explained 14.2% and PC2 explained 12.6% of variation (model parameters: R2X = 45.6%, Q2 = 4.06%, 4 components) (Figure 2C). The score plot of the PLS-DA model using five components achieved separation between the groups (model parameters: R2X = 45.5%, Q2 = 41.8%, R2Y = 75.1%, CV-ANOVA = 0.000033) (Figure 2D). The loading plot was used to identify the metabolites which cause the differences between the REF and the four experimental groups (Supplementary, Figure S1B). The analyses of aqueous muscle extracts showed an effect on eleven metabolites. In comparison with REF, higher concentrations of alanine, betaine, isoleucine, proline, and valine and a lower level of hydroxyproline were observed in EY, while higher concentrations of 3-aminoisobutyrate and malonate were found in MM. Furthermore, betaine, n,n-dimethylglycine, and succinate increased in NY and 3-aminoisobutyrate was higher, while hydroxyproline was lower in ZYG compared with REF (Table 3).
3.2.3. Chloroform Extracts from Liver Tissue
A representative 1H NMR spectrum showing the metabolic profile of hepatic lipids extract using chloroform is shown in Figure 1C. The data analysis of chloroform liver samples gave analogous results to those obtained for extracts from the aqueous phase, with the difference that the spectral data were scaled to the total integrated area of each spectrum. In the PCA score plot, the first component described 31.0% of the variation, while the second component explained 20.8% of spectral variation (model parameters: R2X = 61.5%, Q2 = 47.7%, three components). The data on the diets showed a tendency for grouping in the score plot, separating ZYG (right) from the REF group (left) (Figure 2E). Further analysis with the OPLS-DA model (two outliers in ZYG group were excluded) resulted in two predictive components, the first and second component explained 22.4% and 22.3% of variation, respectively (model parameters: R2X = 44.7%, R2Y = 19.0%, Q2 = 7.07%, CV-ANOVA = 0.018). The model revealed that ZYG fish exhibited discriminating metabolites that tended to differ in comparison with the other four diets (Figure 2F). The 1H NMR-based metabolomics analysis of chloroform extracts showed a higher percentage of n-3 FA, EPA, DHA, PUFA, and phosphatidylcholine/-ethanolamine in ZYG fish compared with the REF. In addition, signals assigned to all FA except EPA and DHA, unsaturated FA, and glyceryl of lipids were lower in ZYG fish than the other four groups (Table 4).

Table 4.
Metabolites along the first predictive component of the OPLS-DA model in the chloroform liver extract of Arctic charr fed the experimental diets.
3.2.4. Chloroform Extracts from White Muscle Tissue
A representative 1H NMR spectrum showing the metabolic profile of white muscle chloroform extract is shown in Figure 1D. Multivariate data analysis gave analogous results to those obtained for extracts from the hepatic chloroform phase. The first and second principal components explained 29.3% and 23.4%, respectively, of spectral variation (R2X) in the PCA model (model parameters: R2X = 94.1% and Q2 = 70.8%) after removing three outliers (2xWY and 1xZYG) (Figure 2G). Overall, there was no clear clustering separation between the REF and the other four diets. Further investigations with the PLS-DA and OPLS-DA models showed no significant differences between the REF and four experimental groups in terms of their R2Y, Q2Y, and CV-ANOVA (p > 0.05). Therefore, no further data analysis was performed.
3.3. Lipid Class, Lipid Content, and Fatty Acid Analysis
Lipid class composition in the liver of fish fed the five different experimental diets showed that ZYG fish had the highest percentage of PL and cholesterol, and the lowest percentage of TAG (Table 5). Also, the lipid content was significantly lower for ZYG fish (4.85%) in comparison of REF vs. the other four groups (ranging from 7.89 to 9.11%, p = 0.0186). Muscle lipid content did not differ between the five groups, ranging from 1.60 to 1.89%. The FA composition of liver and white muscle are shown in Table 6 and Table 7, respectively.

Table 5.
Composition of lipid classes (% of total lipids) in the liver of Arctic charr fed the experimental diets (mean ± SEM, n = 9).

Table 6.
Hepatic lipid content (%) and fatty acid composition (% of total lipids) of Arctic charr fed the experimental diets (mean ± SEM, n = 9).

Table 7.
White muscle lipid content (%) and fatty acid composition (% of total lipids) of Arctic charr fed the experimental diets (mean ± SEM, n = 9).
3.3.1. Liver
Liver FA composition differed considerably between REF and dietary groups, whereas ZYG fish showed the highest effect (Table 6). The liver of ZYG fish had the lowest percentage of 17:0, 18:4n-3 and total MUFA, mainly reflected in lower proportion of 16:1n-7, 18:1n-9, 18:1n-7, 20:1n-9, 22:1n-9 and 24:1 compared with REF. Furthermore, ZYG showed higher proportion of 15:0, PUFA, total n-6 PUFA (e.g., 18:2n-6, 20:2n-6, 20:3n-6, 20:4n-6) and total n-3 PUFA (e.g., 22:6n-3) compared with REF. The FA 17:0 and 22:1n-9 were lower and 20:3n-6 higher in fish fed EY diet compared with REF. In comparison to REF fish fed NY showed lower levels of 17:0, 22:1n-9, 24:1, 18:2n-6, 18:3n-3 and 22:5n-3 and higher levels of 18:0. The lowest effect was observed when comparing REF vs. MM, there only the FA 22:1n-9 and 24:1 were found to be lower in MM fish.
3.3.2. White Muscle
White muscle FA profile was not as affected as liver, although some differences were apparent, particularly in ZYG fish compared with REF (Table 7). Total n-6 PUFA was higher in ZYG and EY fish, which was mainly reflected in a higher level of 18:2n-6 (not significant in EY), 18:3n-6, and 20:3n-6. The FA 18:3n-6 and 20:3n-6 were also higher in NY compared to REF. The n-3/n-6 ratio was lower in ZYG fish compared with REF, which agreed with higher levels of total n-6, but also the lower percentage of 22:5n-3 compared with REF fish. Although the percentages of total SAFA, MUFA, and PUFA were unaffected, some single FA showed differences, namely 14:0, which was higher in EY fish than in REF fish. Percentage of 15:0 was higher in MM, EY and ZYG compared with REF fish. The NY fish had higher percentage of 16:1n-7 compared with REF. Lower percentage of 22:1n-9 and 24:1 were observed in NY and ZYG fish compared with REF fish. MM had a lower percentage of 24:1 compared to REF.
4. Discussion
Changes in the lipid and metabolic profiles of Arctic charr muscle and liver were observed when 40% of dietary FM was replaced either by mussel, zygomycete fungi or yeast meal.
Replacement of FM with ZYG had a negative effect on final body weight and SGR, in agreement with results presented by Abro et al. [12] and Langeland et al. [8]. The contradictory results reported by Bankefors et al. [14] may be explained by much lower replacement of FM with 19% zygomycete biomass compared with 40% replacement in the present experiment. Furthermore, we observed decreased growth performance in Arctic charr fed extracted yeast, while non-extracted yeast showed no effect [24]. Contrary to these results, Øverland et al. [18] found reduced growth in Atlantic salmon fed 34.5% non-extracted yeast. Possible underlying reasons for these conflicting results may be the feed production method. Most previous studies were testing intact baker’s yeast in fish diets, including that of Øverland et al. [18], used cold pelleting, while diets in the present study were produced by extrusion. As suggested by Vidakovic et al. [24] extrusion may have a positive effect on protein digestibility from intact baker’s yeast to fish.
It is well known that changes in the tissue FA profile are caused by dietary FAs [32]. Thus, changes in FA profile such as decrease in 20:1n-9, 22:1n-9 and increase in 18:2n-6 in the ZYG diet compared with the REF diet were reflected in the liver of fish fed those diets. The difference in 15:0 is an effect of the microbial origin of the ingredients, causing this FA incorporation into the fish tissues (REF). The lipid class analyses of the liver showed an increase of 29% in phospholipids (PL) and 10% in cholesterol content, and a decrease of 42% in triacylglycerol (TAG) content in ZYG fish compared with REF fish. It is known that PL serves as cell membrane constituents, whereas TAG are mainly used for energy purposes [33]. Therefore, the lower lipid content in the liver and the lower final body weight of ZYG fish seem to agree with the lower TAG percentage found in the liver of these fish, suggesting either lower TAG deposition or greater use of energy stores. Additionally, a major effect was found on the FA composition of ZYG fish, resulting in a 54% increase in DHA level compared with REF fish. This is in agreement with the almost 50% lower lipid content in the liver of fish fed this diet, as this difference is most likely due to lower DHA content commonly found in storage lipids in comparison with PL [33,34]. In addition, the ZYG diet had the lowest amount of dietary oil (10.9%). The reason for the increased DHA content may be the low lipid content, which is supported by the high PL level found in the liver of ZYG fish, which results in conservation of DHA for membrane use. These findings are consistent with those in a previous study by Pan [7], who showed that an experimental diet containing 23% R. oryzae as DHA being high in PL fraction of white muscle in Arctic charr. In addition, DHA is also commonly preserved in PL when fish are fed diets based on oils of vegetable origin [35]. The white muscle shows no difference in either lipid or DHA numbers, giving the same relation between PL and TAG in all groups [33].
1H NMR-based metabolomics is a tool in nutrition for understanding the metabolic changes caused by dietary ingredients and, in the present study, to evaluate the replacement of FM. The metabolomics analysis showed changes in several metabolites in muscle and liver of fish when FM was replaced by mussel or microorganism meal.
Fish fed the NY diet showed the highest levels of betaine and n,n-dimethylglycine in the liver and muscle. Betaine, which comes from the diet or by oxidation of choline, is an organic osmolyte and methyl donor which is essential for proper liver function and is important in protein and energy metabolism. It can be de-methylated to produce n,n-dimethylglycine and simultaneously homocysteine is converted via single carbon metabolisms to methionine [36,37]. Increased levels of betaine cause increased levels of n,n-dimethylglycine and methionine. In line with this, an increase in n,n-dimethylglycine and methionine (NY: 0.26 ± 0.05 g compared with REF: 0.19 ± 0.03 g, p = 0.153) was observed in NY fish.
We observed higher concentrations of phosphocholine and phosphatidylcholine in the liver of ZYG fish. The higher levels of phosphatidylcholine and its precursor phosphocholine are in agreement with the higher level of PL in ZYG fish compared with REF fish, since phosphatidylcholine has been regarded as the major phospholipid [38].
White muscle, which is a dominant muscle in fish, is known for its high amount of AA and species-dependent concentration of lipids [39]. The 1H NMR analysis showed that several AA were affected (alanine, isoleucine, proline, and valine) when the protein source was altered, with the highest effect observed in EY fish. These AA were also higher in the EY diet compared with the REF diet [24].
Hydroxyproline, an abundant AA in FM, is produced in mammals via hydroxylation of proline by the enzyme prolyl hydroxylase, while in fish biosynthesis is largely unknown [40]. In previous studies, supplementation of the diet with hydroxyproline significantly increased the hydroxyproline levels in muscle, indicating that this AA is absorbed and transported in tissues without dihydroxylation [40]. In the present study, replacement of FM with EY (without dietary hydroxyproline) decreased the level of hydroxyproline in the muscle while proline levels increased, indicating that the capacity of proline hydroxylation in fish was affected. Sunde et al. [41] reported that free hydroxyproline in muscle seems to be positively correlated to the growth rate of juvenile salmon. Thus, in the present study, the decreased level of hydroxyproline might explain the reduced growth rate found in fish fed the EY diet. This indicates that one possibility to improve fish growth could be to add hydroxyproline to EY diets. In addition, proline, which can act as an antioxidant, is a substrate for glucose synthesis. According to previous studies, stimulated degradation of proline and increased activity of proline oxidase (a mitochondrial inner membrane enzyme which can generate ATP when proline is further metabolized via the tricarboxylic acid) was observed in response to stress, including nutritional stress [42]. Therefore, increase in proline in the present study might be also an indication of nutritional stress due to a potential lack of essential nutrients when FM was replaced, an effect reflected in lower final body weight of EY fish compared with REF fish.
ZYG fish showed different regulation of several metabolites in the muscle, such as anserine and hydroxyproline, compared with REF fish. Skeletal muscles of fish usually contain large amounts of histidine and histidine-derived dipeptides, such as carnosine and anserine. The physiological functions of these dipeptides have not been completely clarified, although they have been suggested to act as a muscle buffer and may have antioxidative activity [23]. A few studies have reported that content of histidine and, to some extent, anserine can be influenced by dietary composition. A previous study showed that high inclusion of protein (55%) increases the content of histidine in the muscle of masu salmon (Oncorhynchus masou masou). It was stated that the anserine level increased by dietary histidine level, as anserine seems to be more metabolically stable than histidine [43]. The ZYG diet used in our study had a higher level of histidine than the other diets, which might explain the higher amount of anserine in the muscle of ZYG fish. However, contrary to our results, Bankefors et al. [14] found lower amounts of anserine in Atlantic salmon fed a diet with 20% zygomycetes fungi.
The occurrence of 3-aminoisobutyrate in Arctic charr fed a diet containing MM might indicate a metabolic response to increased utilization of pyrimidine. Thymine, a derivate of pyrimidine, can be catabolized to 3-aminoisobutyrate, which is involved in valine, leucine, and isoleucine metabolism [44]. Awapara and Allen [45] reported the presence of 3-aminoisobutyrate in Mytilus edulis, which might also explain the higher levels found in MM fish compared with fish fed the REF diet. Malonate, a competitive inhibitor of succinate dehydrogenase in the respiratory electron transport chain [46] and also involved in pyrimidine metabolism, was present at three-fold higher concentrations in the muscle of MM fish than REF fish. Malonate is also involved in FA synthesis in the form of malonyl-CoA. However, no effect on the liver and muscle FA profile due to dietary MM was observed. Therefore, the increased malonate concentration with the replacement of FM by mussel meal in Arctic charr needs further investigation.
In summary, the lipid analyses showed that FM replacement generally affected the liver more than muscle tissue. The liver of ZYG fish had the lowest lipid content and lowest percentage of MUFA, and the highest level of n-3, n-6 PUFA, and DHA, mostly due to the high proportion of PL. The microbial signature FA 15:0 was on the other hand highest in ZYG liver. The metabolomics analyses showed that replacing FM with NY affected liver of Arctic charr most, while replacing FM with EY the muscle was more affected. The increase in some metabolites (e.g., alanine, betaine, isoleucine, proline, and valine) suggests a change in muscle metabolism in Arctic charr in response to replacement of FM by different feed ingredients. The smallest changes in liver and muscle metabolism were found in fish fed the MM diet, suggesting that it is a suitable feed source.
Supplementary Materials
The following are available online at https://www.mdpi.com/2410-3888/4/3/46/s1, Figure S1: Loading plot of OPLS-DA and PLS-DA based on metabolites from aqueous (A) liver and (B) muscle extract, respectively, of fish fed reference diet and four experimental diets.
Author Contributions
T.L., A.K., A.V. and M.L. conceived and designed the feeding trial. L.W., P.G.-R., T.L., A.V. and M.L. sampled the fish. L.W., P.G.-R., A.A.M. and J.P. planned the lipid and metabolomics analysis. L.W. performed the metabolomics and lipid study, analysed the experimental data, interpreted the results and wrote the first draft of the manuscript. P.G.-R. assisted in the performance of the lipid and metabolomics analyses and A.A.M. in the performance of the metabolomics study. P.G.-R., A.A.M. and J.P. contributed in the interpretation of the results. J.P., P.G.-R., A.A.M. and T.L. contributed in the writing and critical reading of the manuscript. A.V., A.K. and M.L. read the manuscript critically.
Funding
This work was supported by Formas (Swedish Research Council for Environment, Agricultural Sciences and Spatial Planning, grant number 2009-526). In addition, the study was financially supported by the Ministry of Education, Youth and Sports of the Czech Republic—projects, “CENAKVA“ (No. CZ.1.05/2.1.00/01.0024), “CENAKVA II“ (No. LO1205 under the NPU I program).
Acknowledgments
The authors thank Hanna Carlberg for her assistance during the muscle lipid analyses and the technical staff at Aquaculture Center North, Kälarne (Vattenbrukscentrum Norr AB). The authors thank also Michael Judas for assistance with statistical analysis.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| AA | amino acid |
| BW | body weight |
| CI | confidential interval |
| COSY | correlation spectroscopy |
| CV-ANOVA | cross validation analysis of variance |
| DHA | docosahexaenoic acid (22:6n-3) |
| EPA | eicosapentaenoic acid (20:5n-3) |
| EY | extracted baker’s yeast (Saccharomyces cerevisiae) |
| FA | fatty acid(s) |
| FAME | fatty acid methyl esters |
| FDR | false discovery rate |
| FM | fish meal |
| GC | gas chromatography |
| HPTLC | high performance thin layer chromatography |
| HSQC | heteronuclear single quantum correlation |
| MM | mussel meal |
| MT | million tonnes |
| MUFA | monounsaturated fatty acid(s) |
| n-3 | omega-3 |
| n-6 | omega-6 |
| NMR | nuclear magnet resonance |
| NY | non-extracted baker’s yeast (Saccharomyces cerevisiae) |
| OPLS-DA | orthogonal partial least squares-discriminant analysis |
| PCA | principal component analysis |
| PL | phospholipids |
| PUFA | polyunsaturated fatty acid(s) |
| REF | reference diet |
| SAFA | saturated fatty acid(s) |
| SEM | standard error of mean |
| SGR | specific growth rate |
| TAG | triacylglycerol |
| TOCSY | total correlation spectroscopy |
| TSP | d4-sodium-3-(trimethylsilyl)-2,2,3,3-tetradeuteriopropionate |
| VIP | variable influences in projection |
| ZYG | zygomycete fungi (Rhizopus oryzae) |
References
- FAO. The State of World Fisheries and Aquaculture 2018—Meeting the Sustainable Development Goals; Food and Agriculture Organization of the United Nations: Rome, Italy, 2018; p. 227. [Google Scholar]
- Schock, T.B.; Newton, S.; Brenkert, K.; Leffler, J.; Bearden, D.W. An NMR-based metabolomic assessment of cultured cobia health in response to dietary manipulation. Food Chem. 2012, 133, 90–101. [Google Scholar] [CrossRef]
- Wilson, R. Amino acids and proteins. In Fish Nutrition, 3rd ed.; Hardy, J.H.A.R., Ed.; Elsevier Science (Department of Biochemistry, Mississippi State University): Starkville, MS, USA, 2002; pp. 143–179. [Google Scholar]
- Kristofersson, D.; Anderson, J.L. Is there a relationship between fisheries and farming? Interdependence of fisheries, animal production and aquaculture. Mar. Policy 2006, 30, 721–725. [Google Scholar] [CrossRef]
- Berge, G.M.; Austreng, E. Blue Mussel in Feed for Rainbow-Trout. Aquaculture 1989, 81, 79–90. [Google Scholar] [CrossRef]
- Jönsson, L.; Elwinger, K. Mussel meal as a replacement for fish meal in feeds for organic poultry—A pilot short-term study. Acta Agric. Scand. Sect. A Anim. Sci. 2009, 59, 22–27. [Google Scholar] [CrossRef]
- Pan, J. Effects of Non-Fish Based Raw Materials on the Fish Muscle Quality of Salmonids; Swedish University of Agricultal Sciences: Uppsala, Sweden, 2013. [Google Scholar]
- Langeland, M.; Vidakovic, A.; Vielma, J.; Lindberg, J.E.; Kiessling, A.; Lundh, T. Digestibility of microbial and mussel meal for Arctic charr (Salvelinus alpinus) and Eurasian perch (Perca fluviatilis). Aquac. Nutr. 2016, 22, 485–495. [Google Scholar] [CrossRef]
- Kuhad, R.C.; Singh, A.; Tripathi, K.K.; Saxena, R.K.; Eriksson, K.E. Microorganisms as an alternative source of protein. Nutr. Rev. 1997, 55, 65–75. [Google Scholar] [CrossRef] [PubMed]
- Edebo, L. Zygomycetes for fish feed. Google Patents 12/338,295, 18 December 2008. [Google Scholar]
- Van Leeuwen, J.; Rasmussen, M.L.; Sankaran, S.; Koza, C.R.; Erickson, D.T.; Mitra, D.; Jin, B. Fungal Treatment of Crop Processing Wastewaters with Value-Added Co-Products. In Sustainable Bioenergy and Bioproducts; Gopalakrishnan, K., van Leeuwen, J., Brown, R.C., Eds.; Springer London: London, UK, 2012; pp. 13–44. [Google Scholar]
- Abro, R.; Moazzami, A.A.; Lindberg, J.E.; Lundh, T. Metabolic insights in Arctic charr (Salvelinus alpinus) fed with zygomycetes and fish meal diets as assessed in liver using nuclear magnetic resonance (NMR) spectroscopy. Int. Aquat. Res. 2014, 6, 1–11. [Google Scholar] [CrossRef]
- Mydland, L.T.; Landsverk, T.; Zimonja, T.; Kiessling, A.; Edebo, L.; Storebakken, T. Mycelium biomass from fungi (rhizopus oryzae) grown on spent sulphite liquor from paper pulp as a protein source in diets for rainbow trout (Oncorhynchus mykiss). In Proceedings of the Aquaculture Europe 2007, Istanbul, Turkey, 24–27 October 2007; pp. 375–376. [Google Scholar]
- Bankefors, J.; Kaszowska, M.; Schlechtriem, C.; Pickova, J.; Brannas, E.; Edebo, L.; Kiessling, A.; Sandstrom, C. A comparison of the metabolic profile on intact tissue and extracts of muscle and liver of juvenile Atlantic salmon (Salmo salar L.)—Application to a short feeding study. Food Chem. 2011, 129, 1397–1405. [Google Scholar] [CrossRef]
- Nasseri, A.T.; Rasoul-Amini, S.; Morowvat, M.H.; Ghasemi, Y. Single cell protein: Production and process. Am. J. Food Technol. 2011, 6, 472–485. [Google Scholar] [CrossRef]
- Rumsey, G.L.; Hughes, S.G.; Kinsella, J.L. Use of Dietary Yeast Saccharomyces cerevisiae Nitrogen by Lake Trout. J. World Aquac. Soc. 1990, 21, 205–209. [Google Scholar] [CrossRef]
- Oliva-Teles, A.; Goncalves, P. Partial replacement of fishmeal by brewers yeast (Saccaromyces cerevisae) in diets for sea bass (Dicentrarchus labrax) juveniles. Aquaculture 2001, 202, 269–278. [Google Scholar] [CrossRef]
- Øverland, M.; Karlsson, A.; Mydland, L.T.; Romarheim, O.H.; Skrede, A. Evaluation of Candida utilis, Kluyveromyces marxianus and Saccharomyces cerevisiae yeasts as protein sources in diets for Atlantic salmon (Salmo salar). Aquaculture 2013, 402–403, 1–7. [Google Scholar] [CrossRef]
- Goodacre, R.; Vaidyanathan, S.; Dunn, W.B.; Harrigan, G.G.; Kell, D.B. Metabolomics by numbers: Acquiring and understanding global metabolite data. Trends Biotechnol. 2004, 22, 245–252. [Google Scholar] [CrossRef] [PubMed]
- Wagner, L.; Trattner, S.; Pickova, J.; Gomez-Requeni, P.; Moazzami, A.A. (1)H NMR-based metabolomics studies on the effect of sesamin in Atlantic salmon (Salmo salar). Food Chem. 2014, 147, 98–105. [Google Scholar] [CrossRef] [PubMed]
- Cheng, K.; Wagner, L.; Pickova, J.; Moazzami, A.A. NMR-based metabolomics reveals compartmental metabolic heterogeneity in liver of Arctic char (Salvelinus alpinus). Can. J. Zool. 2016, 94, 665–669. [Google Scholar] [CrossRef]
- Jasour, M.S.; Wagner, L.; Sundekilde, U.K.; Larsen, B.K.; Greco, I.; Orlien, V.; Olsen, K.; Rasmussen, H.T.; Hjermitslev, N.H.; Hammershoj, M.; et al. A Comprehensive Approach to Assess Feathermeal as an Alternative Protein Source in Aquafeed. J. Agric. Food Chem. 2017, 65, 10673–10684. [Google Scholar] [CrossRef] [PubMed]
- Gribbestad, I.S.; Aursand, M.; Martinez, I. High-resolution 1H magnetic resonance spectroscopy of whole fish, fillets and extracts of farmed Atlantic salmon (Salmo salar) for quality assessment and compositional analyses. Aquaculture 2005, 250, 445–457. [Google Scholar] [CrossRef]
- Vidakovic, A.; Langeland, M.; Sundh, H.; Sundell, K.; Olstorpe, M.; Vielma, J.; Kiessling, A.; Lundh, T. Evaluation of growth performance and intestinal barrier function in Arctic Charr (Salvelinus alpinus) fed yeast (Saccharomyces cerevisiae), fungi (Rhizopus oryzae) and blue mussel (Mytilus edulis). Aquac. Nutr. 2016, 22, 1348–1360. [Google Scholar] [CrossRef]
- Linner, J.; Brannas, E. Growth in Arctic charr and rainbow trout fed temporally concentrated or spaced daily meals. Aquac. Int. 2001, 9, 35–44. [Google Scholar] [CrossRef]
- Moazzami, A.A.; Andersson, R.; Kamal-Eldin, A. Changes in the metabolic profile of rat liver after alpha-tocopherol deficiency as revealed by metabolomics analysis. NMR Biomed. 2011, 24, 499–505. [Google Scholar] [CrossRef] [PubMed]
- Ekman, D.R.; Teng, Q.; Villeneuve, D.L.; Kahl, M.D.; Jensen, K.M.; Durhan, E.J.; Ankley, G.T.; Collette, T.W. Profiling lipid metabolites yields unique information on sex- and time-dependent responses of fathead minnows (Pimephales promelas) exposed to 17α-ethynylestradiol. Metabolomics 2008, 5, 22–32. [Google Scholar] [CrossRef]
- Mraz, J.; Pickova, J. Differences between lipid content and composition of different parts of fillets from crossbred farmed carp (Cyprinus carpio). Fish Physiol. Biochem. 2009, 35, 615–623. [Google Scholar] [CrossRef] [PubMed]
- Appelqvist, L.-Å. Rapid methods of lipid extraction and fatty acid methyl ester preparation for seed and leaf tissue with special remarks on preventing the accumulation of lipid contaminants. Arkiv För Kemi 1968, 28, 551–570. [Google Scholar]
- Fredriksson Eriksson, S.; Pickova, J. Fatty acids and tocopherol levels in M. Longissimus dorsi of beef cattle in Sweden—A comparison between seasonal diets. Meat Sci. 2007, 76, 746–754. [Google Scholar] [CrossRef] [PubMed]
- Eriksson, L.; Trygg, J.; Wold, S. CV-ANOVA for significance testing of PLS and OPLS (R) models. J. Chemom. 2008, 22, 594–600. [Google Scholar] [CrossRef]
- Bell, J.G.; Henderson, R.J.; Tocher, D.R.; Sargent, J.R. Replacement of dietary fish oil with increasing levels of linseed oil: Modification of flesh fatty acid compositions in Atlantic salmon (Salmo salar) using a fish oil finishing diet. Lipids 2004, 39, 223–232. [Google Scholar] [CrossRef] [PubMed]
- Henderson, R.J.; Tocher, D.R. The Lipid-Composition and Biochemistry of Fresh-Water Fish. Prog. Lipid Res. 1987, 26, 281–347. [Google Scholar] [CrossRef]
- Gunstone, F.D.; Wijesundera, R.C. The component acids of the lipids in four commercial fish meals. J. Sci. Food Agric. 1978, 29, 28–32. [Google Scholar] [CrossRef]
- Pettersson, A.; Johnsson, L.; Brannas, E.; Pickova, J. Effects of rapeseed oil replacement in fish feed on lipid composition and self-selection by rainbow trout (Oncorhynchus mykiss). Aquac. Nutr. 2009, 15, 577–586. [Google Scholar] [CrossRef]
- Lever, M.; Slow, S. The clinical significance of betaine, an osmolyte with a key role in methyl group metabolism. Clin. Biochem. 2010, 43, 732–744. [Google Scholar] [CrossRef]
- Muona, M.; Virtanen, E. Effect of Dimethylglycine and Trimethylglycine (Betaine) on the Response of Atlantic Salmon (Salmo-salar L.) Smolts to Experimental Vibrio-Anguillarum Infection. Fish Shellfish Immunol. 1993, 3, 439–449. [Google Scholar] [CrossRef]
- Tocher, D.R.; Bendiksen, E.A.; Campbell, P.J.; Bell, J.G. The role of phospholipids in nutrition and metabolism of teleost fish. Aquaculture 2008, 280, 21–34. [Google Scholar] [CrossRef]
- Savorani, F.; Picone, G.; Badiani, A.; Fagioli, P.; Capozzi, F.; Engelsen, S.B. Metabolic profiling and aquaculture differentiation of gilthead sea bream by 1H NMR metabonomics. Food Chem. 2010, 120, 907–914. [Google Scholar] [CrossRef]
- Liu, Y.Z.; He, G.; Wang, Q.C.; Mai, K.S.; Xu, W.; Zhou, H.H. Hydroxyproline supplementation on the performances of high plant protein source based diets in turbot (Scophthalmus maximus L.). Aquaculture 2014, 433, 476–480. [Google Scholar] [CrossRef]
- Sunde, J.; Taranger, G.L.; Rungruangsak-Torrissen, K. Digestive protease activities and free amino acids in white muscle as indicators for feed conversion efficiency and growth rate in Atlantic salmon (Salmo salar L.). Fish Physiol. Biochem. 2001, 25, 335–345. [Google Scholar] [CrossRef]
- Pandhare, J.; Donald, S.P.; Cooper, S.K.; Phang, J.M. Regulation and function of proline oxidase under nutrient stress. J. Cell. Biochem. 2009, 107, 759–768. [Google Scholar] [CrossRef] [PubMed]
- Ogata, H.; Murai, T. White muscle of masu salmon, Oncorhynchus masou masou, smolts possesses a strong buffering capacity due to a high level of anserine. Fish Physiol. Biochem. 1994, 13, 285–293. [Google Scholar] [CrossRef] [PubMed]
- Ueno, S.; Morino, H.; Sano, A.; Kakimoto, Y. Purification and characterization of D-3-aminoisobutyrate-pyruvate aminotransferase from rat liver. Biochim. Biophys. Acta 1990, 1033, 169–175. [Google Scholar] [CrossRef]
- Awapara, J.; Allen, K. Occurrence of beta-aminoisobutyric acid in Mytilus edulis. Science 1959, 130, 1250. [Google Scholar] [CrossRef]
- DuBois, V.R.P.K.P. Studies on the mechanism of hydrogen transport in animal tissues. J. Gen. Physiol. 1943, 26, 391–404. [Google Scholar]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).