Strain-Specific Effects of Bacillus subtilis, Enterococcus faecium, and Pediococcus pentosaceus Supplementation on Growth Performance, Immunity, and Disease Resistance in Olive Flounder (Paralichthys olivaceus)
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolation, Identification, and Characterization of Bacterial Strains
2.2. Whole-Genome Sequencing and Secondary Metabolite Gene Cluster Analysis
2.3. Experimental Fish
2.4. Experimental Diet
2.5. Growth Performance
2.6. Sampling Procedures
2.7. Biochemical Analysis
2.8. Measurement of Phagocytic Activity
2.9. Measurement of Lysozyme Activity
2.10. Measurement of Anti-Protease Activity
2.11. Gene Expression Analysis
2.12. Challenge Test
2.13. Statistical Analysis
3. Results
3.1. Antimicrobial and Enzymatic Activities of Probiotic Strains
3.2. Genome Sequencing and Secondary Metabolite Profiling of Probiotic Strains
3.3. Growth Performance of Olive Flounder Following Probiotic Supplementation
3.4. Blood Biochemical Parameters of Olive Flounder Fed Probiotic-Supplemented Diets
3.5. Non-Specific Immune Responses of Olive Flounder Fed Probiotic-Supplemented Diets
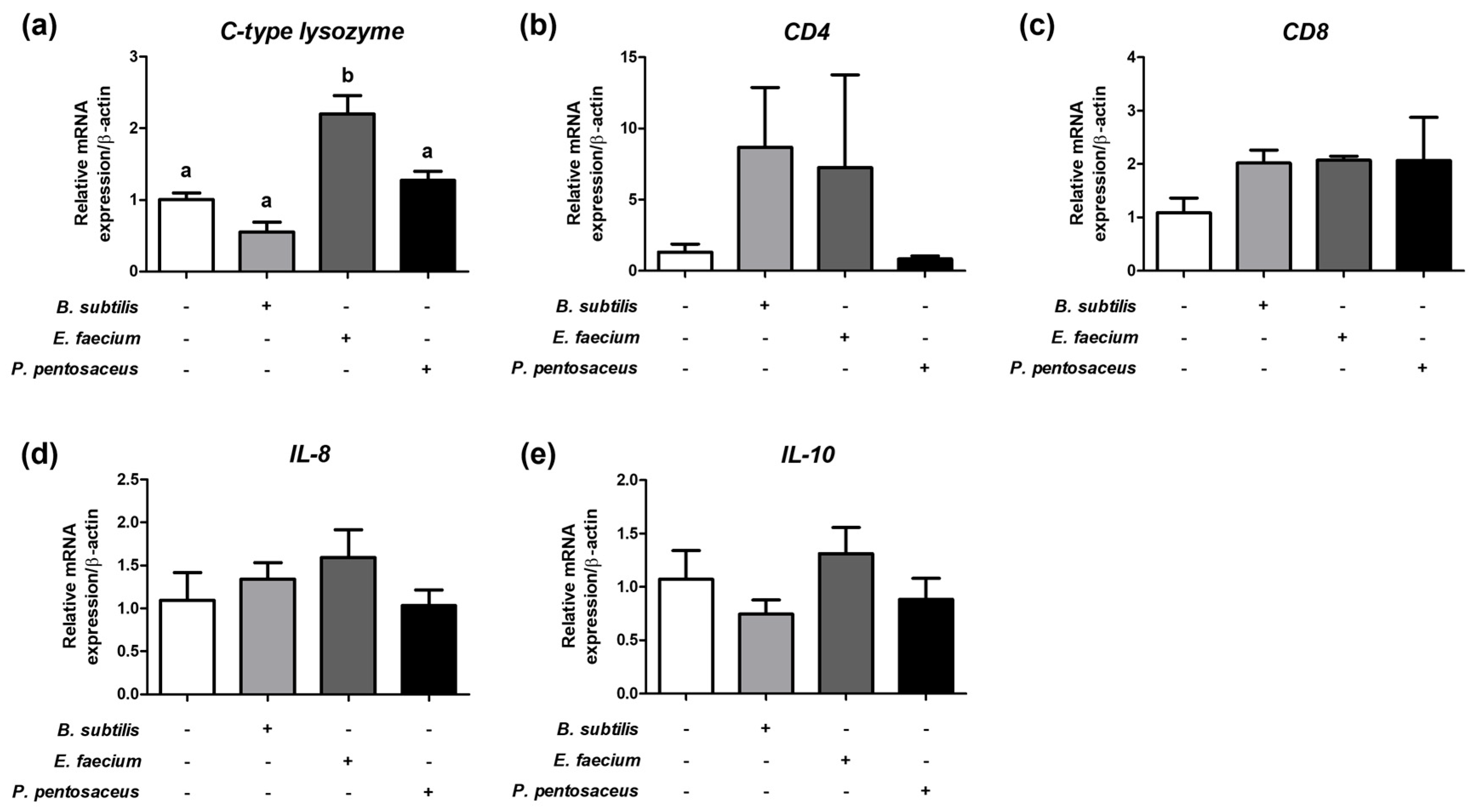
3.6. Expression of Immune-Related Genes in the Liver of Olive Flounder
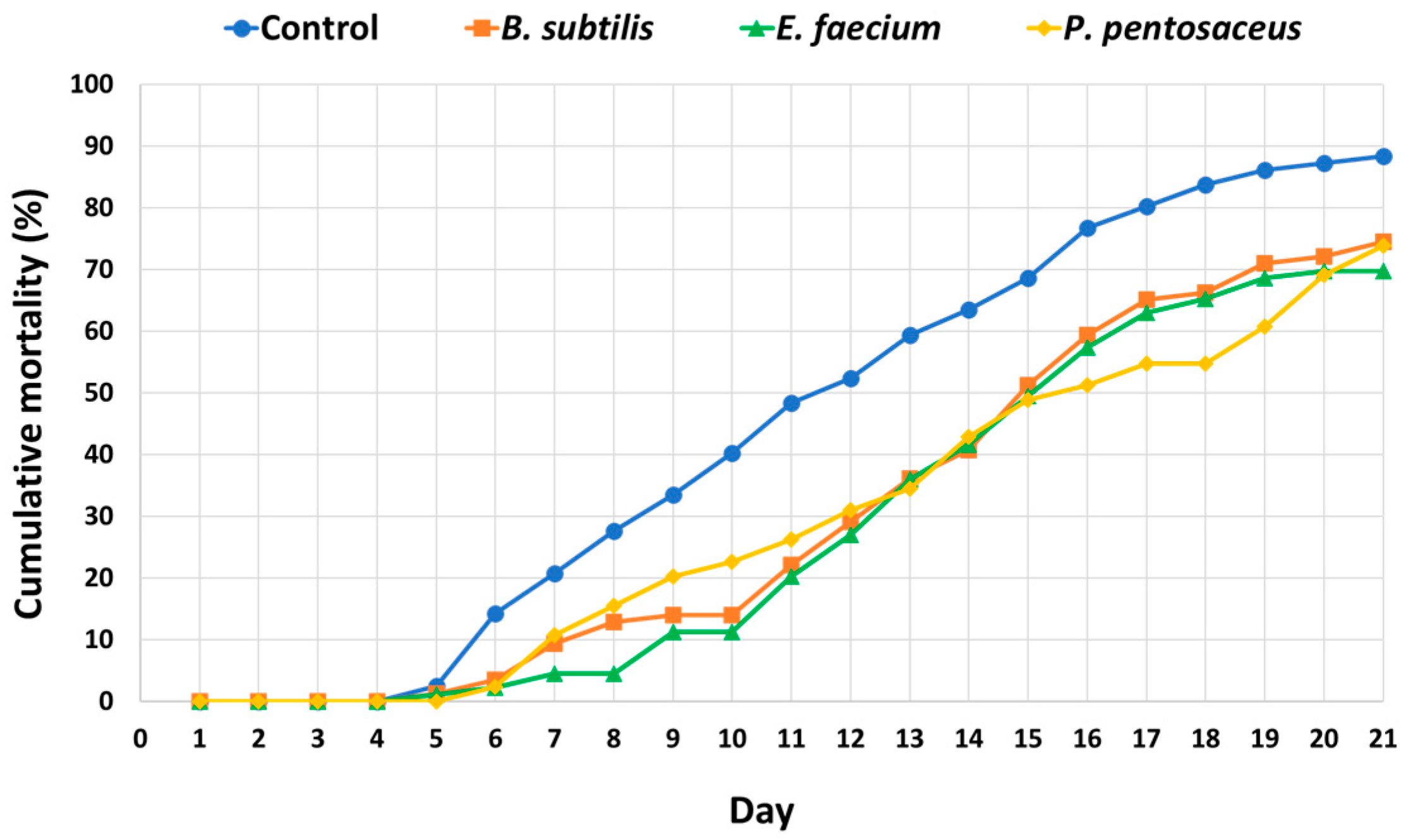
3.7. Survival Analysis Following E. tarda Challenge
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jang, W.J.; Hasan, M.T.; Choi, W.; Hwang, S.; Lee, Y.; Hur, S.W.; Lee, S.; Lee, B.-J.; Choi, Y.H.; Lee, J.M. Comparison of growth performance, non-specific immunity, and intestinal microbiota of olive flounder (Paralichthys olivaceus) fed with extruded pellet and moist pellet diets under field conditions in South Korea. Front. Microbiol. 2022, 13, 979124. [Google Scholar] [CrossRef]
- Oh, S.; Lee, S. Fish Welfare-Related Issues and Their Relevance in Land-Based Olive Flounder (Paralichthys olivaceus) Farms in Korea. Animals 2024, 14, 1693. [Google Scholar] [CrossRef] [PubMed]
- Ciji, A.; Akhtar, M.S. Stress management in aquaculture: A review of dietary interventions. Rev. Aquac. 2021, 13, 2190–2247. [Google Scholar] [CrossRef]
- Barton, B.A. Stress in fishes: A diversity of responses with particular reference to changes in circulating corticosteroids. Integr. Comp. Biol. 2002, 42, 517–525. [Google Scholar] [CrossRef]
- Cabello, F.C.; Godfrey, H.P.; Tomova, A.; Ivanova, L.; Dölz, H.; Millanao, A.; Buschmann, A.H. Antimicrobial use in aquaculture re-examined: Its relevance to antimicrobial resistance and to animal and human health. Environ. Microbiol. 2013, 15, 1917–1942. [Google Scholar] [CrossRef]
- De la Casa-Resino, I.; Empl, M.T.; Villa, S.; Kolar, B.; Fabrega, J.; Lillicrap, A.D.; Karamanlis, X.N.; Carapeto-García, R. Environmental risk assessment of veterinary medicinal products intended for use in aquaculture in Europe: The need for developing a harmonised approach. Environ. Sci. Eur. 2021, 33, 84. [Google Scholar] [CrossRef]
- Balcázar, J.L.; De Blas, I.; Ruiz-Zarzuela, I.; Cunningham, D.; Vendrell, D.; Múzquiz, J.L. The role of probiotics in aquaculture. Vet. Microbiol. 2006, 114, 173–186. [Google Scholar] [CrossRef]
- Panigrahi, A.; Kiron, V.; Kobayashi, T.; Puangkaew, J.; Satoh, S.; Sugita, H. Immune responses in rainbow trout Oncorhynchus mykiss induced by a potential probiotic bacteria Lactobacillus rhamnosus JCM 1136. Vet. Immunol. Immunopathol. 2004, 102, 379–388. [Google Scholar] [CrossRef]
- Li, Y.; Yang, Y.; Song, L.; Wang, J.; Hu, Y.; Yang, Q.; Cheng, P.; Li, J. Effects of dietary supplementation of Lactobacillus plantarum and Bacillus subtilis on growth performance, survival, immune response, antioxidant capacity and digestive enzyme activity in olive flounder (Paralichthys olivaceus). Aquac. Fish. 2021, 6, 283–288. [Google Scholar] [CrossRef]
- Abbas, M.A.; Kim, H.-J.; Lee, G.-Y.; Cho, H.-Y.; Al Jawad Sayem, S.; Lee, E.-B.; Lee, S.-J.; Park, S.-C. Development and Application of Lactobacillus plantarum PSCPL13 Probiotics in Olive Flounder (Paralichthys olivaceus) Farming. Microorganisms 2025, 13, 61. [Google Scholar] [CrossRef] [PubMed]
- Ahmadifar, E.; Sadegh, T.H.; Dawood, M.A.; Dadar, M.; Sheikhzadeh, N. The effects of dietary Pediococcus pentosaceus on growth performance, hemato-immunological parameters and digestive enzyme activities of common carp (Cyprinus carpio). Aquaculture 2020, 516, 734656. [Google Scholar] [CrossRef]
- Hoseinifar, S.H.; Sun, Y.-Z.; Wang, A.; Zhou, Z. Probiotics as means of diseases control in aquaculture, a review of current knowledge and future perspectives. Front. Microbiol. 2018, 9, 2429. [Google Scholar] [CrossRef]
- Madhulika; Ngasotter, S.; Meitei, M.M.; Kara, T.; Meinam, M.; Sharma, S.; Rathod, S.K.; Singh, S.B.; Singh, S.K.; Bhat, R.A.H. Multifaceted Role of Probiotics in Enhancing Health and Growth of Aquatic Animals: Mechanisms, Benefits, and Applications in Sustainable Aquaculture—A Review and Bibliometric Analysis. Aquac. Nutr. 2025, 2025, 5746972. [Google Scholar]
- Choi, W.; Moniruzzaman, M.; Bae, J.; Hamidoghli, A.; Lee, S.; Choi, Y.-H.; Min, T.; Bai, S.C. Evaluation of dietary probiotic bacteria and processed yeast (GroPro-Aqua) as the alternative of antibiotics in juvenile olive flounder Paralichthys olivaceus. Antibiotics 2022, 11, 129. [Google Scholar] [CrossRef] [PubMed]
- Niu, K.-M.; Kothari, D.; Lee, W.-D.; Zhang, Z.; Lee, B.-J.; Kim, K.-W.; Wu, X.; Han, H.-S.; Khosravi, S.; Lee, S.-M. Probiotic potential of the farmed olive flounder, Paralichthys olivaceus, autochthonous gut microbiota. Probiotics Antimicrob. Proteins 2021, 13, 1106–1118. [Google Scholar] [CrossRef]
- Han, S.-R.; Munang’andu, H.M.; Yeo, I.-K.; Kim, S.-H. Bacillus subtilis inhibits viral hemorrhagic septicemia virus infection in olive flounder (Paralichthys olivaceus) intestinal epithelial cells. Viruses 2020, 13, 28. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Yoon, S.-H.; Ha, S.-M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef]
- Klančnik, A.; Piskernik, S.; Jeršek, B.; Možina, S.S. Evaluation of diffusion and dilution methods to determine the antibacterial activity of plant extracts. J. Microbiol. Methods 2010, 81, 121–126. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Kloosterman, A.M.; Charlop-Powers, Z.; Van Wezel, G.P.; Medema, M.H.; Weber, T. antiSMASH 6.0: Improving cluster detection and comparison capabilities. Nucleic Acids Res. 2021, 49, W29–W35. [Google Scholar] [CrossRef] [PubMed]
- Heo, W.-S.; Kim, Y.-R.; Kim, E.-Y.; Bai, S.C.; Kong, I.-S. Effects of dietary probiotic, Lactococcus lactis subsp. lactis I2, supplementation on the growth and immune response of olive flounder (Paralichthys olivaceus). Aquaculture 2013, 376, 20–24. [Google Scholar] [CrossRef]
- Wang, J.; Fan, D.; Zhao, R.; Lu, T.; Li, S.; Wang, D. Effects of dietary supplementation with endogenous probiotics Bacillus subtilis on growth performance, immune response and intestinal histomorphology of juvenile rainbow trout (Oncorhynchus mykiss). Fishes 2024, 9, 229. [Google Scholar] [CrossRef]
- Müller, J.; Alföldy, P.; Lemmel, E.-M. Nitroblue-tetrazolium test for the functional evaluation of phagocytic cells: A critical analysis of the methodology. Agents Actions 1981, 11, 384–390. [Google Scholar] [CrossRef]
- Hikima, J.-i.; Minagawa, S.; Hirono, I.; Aoki, T. Molecular cloning, expression and evolution of the Japanese flounder goose-type lysozyme gene, and the lytic activity of its recombinant protein. Biochim. Et Biophys. Acta (BBA)-Gene Struct. Expr. 2001, 1520, 35–44. [Google Scholar] [CrossRef]
- Coêlho, D.F.; Saturnino, T.P.; Fernandes, F.F.; Mazzola, P.G.; Silveira, E.; Tambourgi, E.B. Azocasein substrate for determination of proteolytic activity: Reexamining a traditional method using bromelain samples. BioMed Res. Int. 2016, 2016, 8409183. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.-G.; Gunathilaka, B.E.; Lee, S.; Kim, Y.; Lee, K.-J. Effects of Bacillus SW1-1 coated diets on innate immunity and disease resistance of olive flounder Paralichthys olivaceus against Edwardsiella tarda infection. Fish. Aquat. Sci. 2022, 25, 243–249. [Google Scholar] [CrossRef]
- Yi, C.-C.; Liu, C.-H.; Chuang, K.-P.; Chang, Y.-T.; Hu, S.-Y. A potential probiotic Chromobacterium aquaticum with bacteriocin-like activity enhances the expression of indicator genes associated with nutrient metabolism, growth performance and innate immunity against pathogen infections in zebrafish (Danio rerio). Fish Shellfish Immunol. 2019, 93, 124–134. [Google Scholar] [CrossRef]
- Ding, F.; Zhou, N.; Luo, Y.; Wang, T.; Li, W.; Qiao, F.; Du, Z.; Zhang, M. Probiotic Pediococcus pentosaceus restored gossypol-induced intestinal barrier injury by increasing propionate content in Nile tilapia. J. Anim. Sci. Biotechnol. 2024, 15, 54. [Google Scholar] [CrossRef]
- Dawood, M.; Koshio, S.; Ishikawa, M.; Yokoyama, S. Effects of dietary inactivated Pediococcus pentosaceus on growth performance, feed utilization and blood characteristics of red sea bream, Pagrus major juvenile. Aquac. Nutr. 2016, 22, 923–932. [Google Scholar] [CrossRef]
- Daboor, M.; Esmael, N.A.; Lall, S.P. Effect of Different Dietary Probiotics on Growth, Feed Utilization and Digestive Enzymes Activities of Nile Tilapia, Oreochromis niloticus Mohamed A Essa1, Sabry S EL-Serafy2, Magda M El-Ezabi2, Said. Aquaculture 2010, 454, 243–251. [Google Scholar]
- Paritova, A.; Nurgaliyev, A.; Nurgaliyeva, G.; Abekeshev, N.; Abuova, A.; Zakirova, F.; Zwierzchowski, G.; Kuanchaleyev, Z.; Issabekova, S.; Kizatova, M. The dietary effects of two strain probiotics (Leuconostoc mesenteroides, Lactococcus lactis) on growth performance, immune response and gut microbiota in Nile tilapia (Oreochromis niloticus). PLoS ONE 2024, 19, e0312580. [Google Scholar] [CrossRef]
- Kunst, F.; Ogasawara, N.; Moszer, I.; Albertini, A.; Alloni, G.; Azevedo, V.; Bertero, M.; Bessières, P.; Bolotin, A.; Borchert, S. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 1997, 390, 249–256. [Google Scholar] [CrossRef] [PubMed]
- Kompramool, S.; Singkhamanan, K.; Pomwised, R.; Chaichana, N.; Suwannasin, S.; Wonglapsuwan, M.; Jitpakdee, J.; Kantachote, D.; Yaikhan, T.; Surachat, K. Genomic Insights into Pediococcus pentosaceus ENM104: A Probiotic with Potential Antimicrobial and Cholesterol-Reducing Properties. Antibiotics 2024, 13, 813. [Google Scholar] [CrossRef]
- Qin, X.; Galloway-Peña, J.R.; Sillanpaa, J.; Roh, J.H.; Nallapareddy, S.R.; Chowdhury, S.; Bourgogne, A.; Choudhury, T.; Muzny, D.M.; Buhay, C.J. Complete genome sequence of Enterococcus faecium strain TX16 and comparative genomic analysis of Enterococcus faecium genomes. BMC Microbiol. 2012, 12, 135. [Google Scholar] [CrossRef]
- Mi, J.; He, T.; Hu, X.; Wang, Z.; Wang, T.; Qi, X.; Li, K.; Gao, L.; Liu, C.; Zhang, Y. Enterococcus faecium C171: Modulating the immune response to acute lethal viral challenge. Int. J. Antimicrob. Agents 2023, 62, 106969. [Google Scholar] [CrossRef]
- Mansour, N.M.; Heine, H.; Abdou, S.M.; Shenana, M.E.; Zakaria, M.K.; El-Diwany, A. Isolation of Enterococcus faecium NM113, Enterococcus faecium NM213 and Lactobacillus casei NM512 as novel probiotics with immunomodulatory properties. Microbiol. Immunol. 2014, 58, 559–569. [Google Scholar] [CrossRef]
- Aerts, J.; Schaeck, M.; De Swaef, E.; Ampe, B.; Decostere, A. Vibrio lentus as a probiotic candidate lowers glucocorticoid levels in gnotobiotic sea bass larvae. Aquaculture 2018, 492, 40–45. [Google Scholar] [CrossRef]
- Wendelaar Bonga, S.E. The stress response in fish. Physiol. Rev. 1997, 77, 591–625. [Google Scholar] [CrossRef] [PubMed]
- Chuwen, Q.; Yonghai, S.; Jianming, Z. Probiotics and their impact on mitigating high-temperature stress responses in farmed fish. J. Fish. Res. 2025, 47, 248–260. [Google Scholar]
- Fachri, M.; Amoah, K.; Huang, Y.; Cai, J.; Alfatat, A.; Ndandala, C.B.; Shija, V.M.; Jin, X.; Bissih, F.; Chen, H. Probiotics and paraprobiotics in aquaculture: A sustainable strategy for enhancing fish growth, health and disease prevention-a review. Front. Mar. Sci. 2024, 11, 1499228. [Google Scholar] [CrossRef]
- Taoka, Y.; Maeda, H.; Jo, J.-Y.; Jeon, M.-J.; Bai, S.C.; Lee, W.-J.; Yuge, K.; Koshio, S. Growth, stress tolerance and non-specific immune response of Japanese flounder Paralichthys olivaceus to probiotics in a closed recirculating system. Fish. Sci. 2006, 72, 310–321. [Google Scholar] [CrossRef]
- Nguyen, T.L.; Park, C.-I.; Kim, D.-H. Improved growth rate and disease resistance in olive flounder, Paralichthys olivaceus, by probiotic Lactococcus lactis WFLU12 isolated from wild marine fish. Aquaculture 2017, 471, 113–120. [Google Scholar] [CrossRef]
- Aly, S.M.; Mohamed, M.F.; John, G. Effect of probiotics on the survival, growth and challenge infection in Tilapia nilotica (Oreochromis niloticus). Aquac. Res. 2008, 39, 647–656. [Google Scholar] [CrossRef]
- Won, S.; Hamidoghli, A.; Choi, W.; Bae, J.; Jang, W.J.; Lee, S.; Bai, S.C. Evaluation of potential probiotics Bacillus subtilis WB60, Pediococcus pentosaceus, and Lactococcus lactis on growth performance, immune response, gut histology and immune-related genes in whiteleg shrimp, Litopenaeus vannamei. Microorganisms 2020, 8, 281. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Qiao, L.; Li, A.; Zhao, J.; Qian, R.; Luo, Q.; Xu, Q. Effects of coated Enterococcus faecium on the growth performance, immunity, digestive enzyme activity, intestinal morphometry, and microbiota of crucian carp (Carassius auratus gibelio). Aquac. Rep. 2024, 37, 102264. [Google Scholar] [CrossRef]
- Hossain, M.K.; Islam, S.M.; Rafiquzzaman, S.; Nuruzzaman, M.; Hossain, M.T.; Shahjahan, M. Multi-species probiotics enhance growth of Nile tilapia (Oreochromis niloticus) through upgrading gut, liver and muscle health. Aquac. Res. 2022, 53, 5710–5719. [Google Scholar] [CrossRef]
- Wang, Y.-C.; Hu, S.-Y.; Chiu, C.-S.; Liu, C.-H. Multiple-strain probiotics appear to be more effective in improving the growth performance and health status of white shrimp, Litopenaeus vannamei, than single probiotic strains. Fish Shellfish Immunol. 2019, 84, 1050–1058. [Google Scholar] [CrossRef]
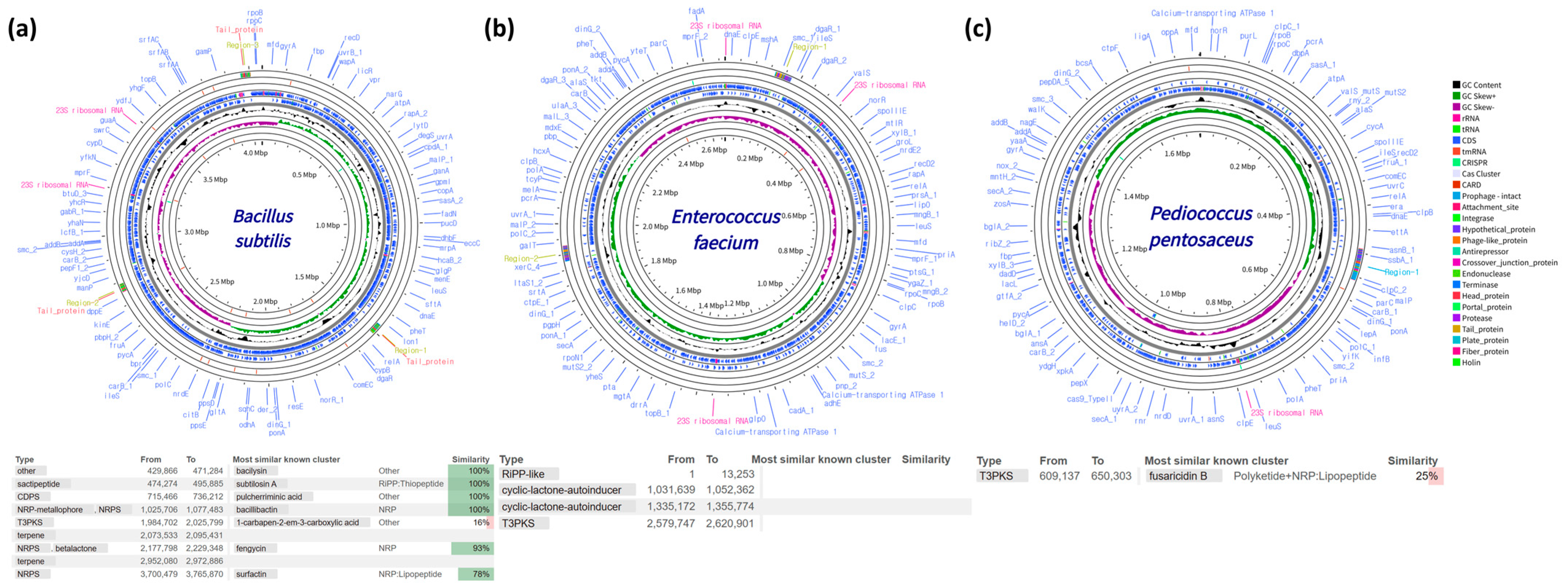
| Proximate Composition | Content (%) |
|---|---|
| Crude protein | ≥54.0% |
| Crude lipid | ≥8.0% |
| Crude ash | ≥18.0% |
| Crude fiber | ≤5.0% |
| Calcium | ≤1.0% |
| Phosphorus | ≤2.7% |
| Gene | Primer (5′-3′) | Accession Number |
|---|---|---|
| C-type lysozyme | F: CTG TGG GCA GGA AAG ACT TC R: GGA AGT GTT GGT GGA GAG G | AB050589.1 |
| CD4 | F: AGG TGC CAG TGA GGT GGT TTA T R: GCC GTC CTG TTT ACC AAA ACT C | XM_069537653.1 |
| CD8 | F: CGC TGC GCT GCA ATG AT | AB082958.1 |
| R: CAC GCC CCA CCT GTA ACC | ||
| IL-8 | F: CAT CGT TGT TGC TGT GAT GGT | AF216646.1 |
| R: AGG CTC ACC GCT TCA CTG AT | ||
| IL-10 | F: AGC GAA CGA TGA CCT AGA CAC G | AB685381.1 |
| R: ACC GTG CTC AGG TAG AAG TCC A | ||
| β-actin | F: CAT CAG GGA GTG ATG GTG GGT A R: ATA CCG TGC TCG ATG GGG TAC T | HQ386788.1 |
| Strain | Species | Antimicrobial Activity (mm) | Enzyme Activity (mm) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| E. coli | B. cereus | C. albicans | E. tarda | TCN | TBN | Skim Milk | CMC | Soluble Starch | Gelatin | ||
| E-TSA-3 | B. subtilis | - | - | - | - | - |  13.9 ± 0.10 *** |  19.6 ± 0.20 *** |  17.0 ± 0.59 *** |  14.7 ± 1.37 *** |  22.1 ± 0.15 *** |
| Jeju1 YPD-N2 | E. faecium | - |  11.5 ± 0.25 *** | - | - | - | - |  15.3 ± 0.51 *** | - | - | - |
| MRS-A4-1 | P. pentosaceus |  20.6 ± 0.60 *** |  24.2 ± 0.75 *** | - |  9.7 ± 0.57 *** | - | - | - | - | - | - |
| IBW (g) | FBW (g) | BWG (g) | WGR (%) | SGR (%) | FCR | |
|---|---|---|---|---|---|---|
| Control | 16.96 ± 3.0 | 46.57 ± 9.8 a | 29.59 ± 9.8 a | 174.26 ± 57.9 a | 2.81 ± 0.6 a | 1.55 ± 0.8 |
| B. subtilis | 16.97 ± 3.2 | 48.61 ± 10.2 ab | 32.01 ± 9.6 ab | 188.51 ± 56.5 ab | 2.97 ± 0.6 ab | 1.41 ± 0.9 |
| E. faecium | 16.98 ± 1.9 | 49.51 ± 8.8 ab | 32.53 ± 8.8 ab | 191.58 ± 51.6 ab | 3.01 ± 0.5 ab | 1.31 ± 0.4 |
| P. pentosaceus | 16.97 ± 2.7 | 50.87 ± 10.0 b | 33.89 ± 10.0 b | 199.59 ± 59.0 b | 3.07 ± 0.6 b | 1.34 ± 0.9 |
| GLU (mg/dL) | ALP (U/L) | GOT (U/L) | GPT (U/L) | TP (g/dL) | |
|---|---|---|---|---|---|
| Control | 74.50 ± 32.9 a | 255.0 ± 101.7 | 27.3 ± 9.4 | 13.3 ± 1.9 | 3.4 ± 0.26 |
| B. subtilis | 54.53 ± 26.3 a | 218.3 ± 61.6 | 21.5 ± 7.3 | 12.1 ± 1.5 | 3.5 ± 0.27 |
| E. faecium | 23.43 ± 10.5 b | 220.5 ± 49.5 | 26.3 ± 9.5 | 11.1 ± 2.7 | 3.5 ± 0.20 |
| P. pentosaceus | 27.07 ± 12.6 b | 194.7 ± 40.5 | 28.2 ± 18.0 | 13.3 ± 4.7 | 3.5 ± 0.27 |
| Phagocytic Activity (O.D. 550 nm) | Lysozyme Activity (U/mL) | Anti-Protease Activity (% Inhibition) | |
|---|---|---|---|
| Control | 0.054 ± 0.007 a | 4.55 ± 9.53 a | 52.00 ± 19.7 |
| B. subtilis | 0.066 ± 0.009 ab | 7.48 ± 4.77 a | 50.22 ± 18.3 |
| E. faecium | 0.065 ± 0.007 ab | 21.52 ± 11.98 b | 42.25 ± 16.8 |
| P. pentosaceus | 0.070 ± 0.021 b | 19.13 ± 8.30 b | 56.69 ± 18.8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, M.-K.; Jung, H.-K.; Kim, D.-G.; Park, I.-S.; Heo, Y.L.; Kang, J.; Kim, Y.S. Strain-Specific Effects of Bacillus subtilis, Enterococcus faecium, and Pediococcus pentosaceus Supplementation on Growth Performance, Immunity, and Disease Resistance in Olive Flounder (Paralichthys olivaceus). Fishes 2025, 10, 465. https://doi.org/10.3390/fishes10090465
Lee M-K, Jung H-K, Kim D-G, Park I-S, Heo YL, Kang J, Kim YS. Strain-Specific Effects of Bacillus subtilis, Enterococcus faecium, and Pediococcus pentosaceus Supplementation on Growth Performance, Immunity, and Disease Resistance in Olive Flounder (Paralichthys olivaceus). Fishes. 2025; 10(9):465. https://doi.org/10.3390/fishes10090465
Chicago/Turabian StyleLee, Min-Kyeong, Hyun-Kyoung Jung, Dong-Gyun Kim, In-Suk Park, Yu Li Heo, Jungha Kang, and Young Sam Kim. 2025. "Strain-Specific Effects of Bacillus subtilis, Enterococcus faecium, and Pediococcus pentosaceus Supplementation on Growth Performance, Immunity, and Disease Resistance in Olive Flounder (Paralichthys olivaceus)" Fishes 10, no. 9: 465. https://doi.org/10.3390/fishes10090465
APA StyleLee, M.-K., Jung, H.-K., Kim, D.-G., Park, I.-S., Heo, Y. L., Kang, J., & Kim, Y. S. (2025). Strain-Specific Effects of Bacillus subtilis, Enterococcus faecium, and Pediococcus pentosaceus Supplementation on Growth Performance, Immunity, and Disease Resistance in Olive Flounder (Paralichthys olivaceus). Fishes, 10(9), 465. https://doi.org/10.3390/fishes10090465






