The Complete Mitochondrial Genome of Acrossocheilus spinifer (Osteichthyes: Cyprinidae) and Its Phylogenetic Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Mitogenome Sequencing, Assembly, and Annotation
2.3. Genome Analysis
2.4. Phylogenetic Analysis
3. Results
3.1. Genome Organization and Base Composition
3.2. Repeat Sequence Analysis
3.3. Protein-Coding Genes and Codon Usage
3.4. Transfer RNA and Ribosomal RNA Genes
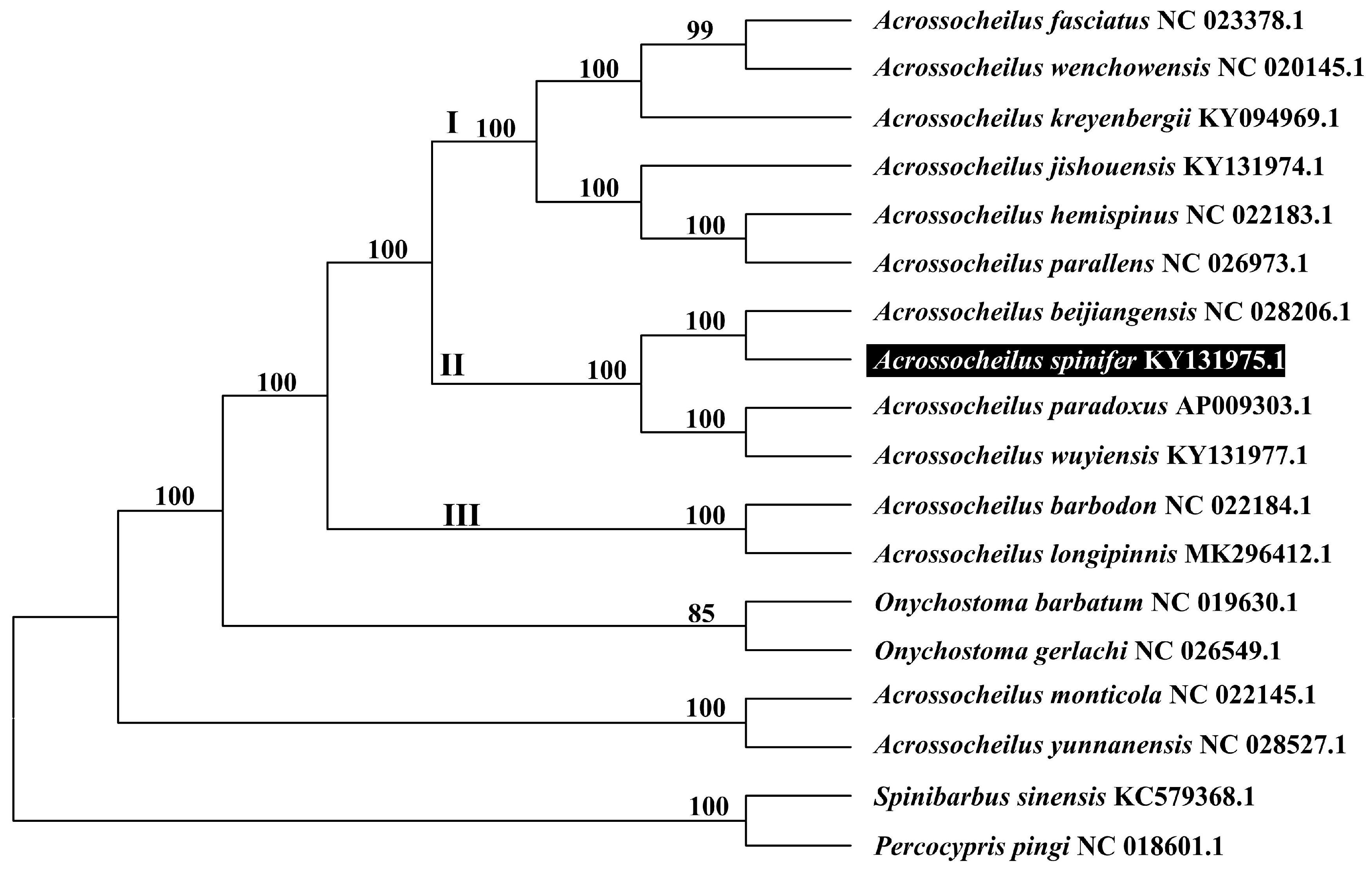
3.5. Phylogenetic Tree
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nelson, J.S.; Grande, T.C.; Wilson, M.V.H. Fishes of the World, 5th ed.; John Wiley & Sons: Hoboken, NJ, USA, 2016. [Google Scholar]
- Yue, P.Q. Fauna Sinica, Osteichthyes: Cypriniformes III; Science Press: Beijing, China, 2000. [Google Scholar]
- Kottelat, M. Fishes of the Nam Theun and Xe Bangfai basins, Laos, with diagnoses of twenty-two new species (Teleostei: Cyprinidae, Baltoridae, Cobitidae, Coiidae and Odontobutidae). Ichthyol. Explor. Freshw. 1998, 9, 1–128. [Google Scholar]
- Kottelat, M. Diagnoses of a new genus and 64 new species of fishes from Laos (Teleostei: Cyprinidae, Balitoridae, Bagridae, Syngnathidae, Chauhuriidae and Tetraodontidae). J. South Asian Nat. Hist. 2000, 5, 37–82. [Google Scholar]
- Kottelat, M. Fishes of Laos; Wildlife Heritage Trust Publications: Colombo, Sri Lanka, 2001; pp. 1–198. [Google Scholar]
- Kottelat, M. Freshwater Fishes of Northern Vietnam. A Preliminary Check-List of the Fishes Known or Expected to Occur in Northern Vietnam with Comments on Systematics and Nomenclature; The World Bank: Washington, DC, USA, 2001; pp. 1–122. [Google Scholar]
- Kottelat, M. The fishes of the inland waters of Southeast Asia: A catalogue and core bibliography of the fishes known to occur in freshwaters, mangroves and estuaries. Raffles Bull. Zool. 2013, 27, 1–663. [Google Scholar]
- Froese, R.; Pauly, D. FishBase. World Wide Web Electronic Publication. 2025. Available online: https://www.fishbase.org (accessed on 6 May 2025).
- Xiao, G.B.; Lu, W.J.; Liao, L.; Mao, Q.; Wang, L.J.; Zhang, H.R.; Zhou, L.; Bao, J. A new Species of Acrossocheilus (Teleostei: Cyprinidae): A. furongjiangensis sp. nov. from Wujiang River Basin in Northern Guizhou. Sichuan J. Zool. 2024, 43, 205–214. [Google Scholar] [CrossRef]
- Chen, T.E.; Xu, J.X.; Li, P.J.; Hu, H.F.; Gao, K.; Zhao, H.P. A New Species of the Genus Acrossocheilus Oshima, 1919 (Cypriniformes: Cyprinidae) from the Dabie Mountains. Animals 2025, 15, 734. [Google Scholar] [CrossRef]
- Yuan, L.Y. Monophyly, Affinity and Taxonomic Revision of the Cyprinid Genus Acrossocheilus Oshima, 1919. Ph.D. Thesis, Graduate School of the Chinese Academy of Sciences, Beijing, China, 2009. [Google Scholar]
- Yuan, L.Y.; Wu, Z.Q.; Zhang, E. Acrossocheilus spinifer, a new species of barred cyprinid fish from south China (Pisces: Teleostei). J. Fish Biol. 2006, 8, 163–173. [Google Scholar] [CrossRef]
- Yuan, L.Y.; Liu, X.X.; Zhang, E. Mitochondrial phylogeny of Chinese barred species of the cyprinid genus Acrossocheilus Oshima, 1919 (Teleostei: Cypriniformes) and its taxonomic implications. Zootaxa 2015, 4059, 151–168. [Google Scholar] [CrossRef]
- Miya, M.; Friedman, M.; Satoh, T.P.; Takeshima, H.; Sado, T.; Iwasaki, W.; Yamanoue, Y.; Nakatani, M.; Mabuchi, K.; Inoue, J.G.; et al. Evolutionary origin of the Scombridae (tunas and mackerels): Members of a paleogene adaptive radiation with 14 other pelagic fish families. PLoS ONE 2013, 8, e73535. [Google Scholar] [CrossRef]
- Near, T.J.; Eytan, R.I.; Dornburg, A.; Kuhn, K.L.; Moore, J.A.; Davis, M.P.; Wainwright, P.C.; Friedman, M.; Smith, W.L. Resolution of ray-finned fish phylogeny and timing of diversification. Proc. Natl. Acad. Sci. USA 2012, 109, 13698–13703. [Google Scholar] [CrossRef]
- Boore, J.L. Animal mitochondrial genomes. Nucleic Acids Res. 1999, 27, 1767–1780. [Google Scholar] [CrossRef]
- Saccone, C.; Pesole, G.; Sbisa, E. The main regulatory region of mammalian mitochondrial DNA: Structure-function model and evolutionary pattern. J. Mol. Evol. 1991, 33, 83–91. [Google Scholar] [CrossRef] [PubMed]
- Hou, X.J.; Lin, H.D.; Tang, W.Q.; Liu, D.; Han, C.C.; Yang, J.Q. Complete mitochondrial genome of the freshwater fish Acrossocheilus longipinnis (Teleostei: Cyprinidae): Genome characterization and phylogenetic analysis. Biologia 2020, 75, 1871–1880. [Google Scholar] [CrossRef]
- Shen, K.N.; Yen, T.C.; Chen, C.H.; Li, H.Y.; Chen, P.L.; Hsiao, C.D. Next generation sequencing yields the complete mitochondrial genome of the flathead mullet, Mugil cephalus cryptic species NWP2 (Teleostei: Mugilidae). Mitochondrial DNA Part A 2016, 27, 1758–1759. [Google Scholar] [CrossRef] [PubMed]
- Bernt, M.; Donath, A.; Juhling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Putz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef]
- Wyman, S.K.; Jansen, R.K.; Boore, J.L. Automatic annotation of organellar genomes with DOGMA. Bioinformatics 2004, 20, 3252–3255. [Google Scholar] [CrossRef]
- Laslett, D.; Canback, B. ARWEN: A program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics 2008, 24, 172–175. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Greiner, S.; Lehwark, P.; Bock, R. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: Expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 2019, 47, W59–W64. [Google Scholar] [CrossRef]
- Benson, G. Tandem Repeats Finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar] [CrossRef]
- Beier, S.; Thiel, T.; Munch, T.; Scholz, U.; Mascher, M. MISA-web: A web server for microsatellite prediction. Bioinformatics 2017, 33, 2583–2585. [Google Scholar] [CrossRef]
- Kurtz, S.; Choudhuri, J.V.; Ohlebusch, E.; Schleiermacher, C.; Stoye, J.; Giegerich, R. REPuter: The manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001, 29, 4633–4642. [Google Scholar] [CrossRef] [PubMed]
- Ponce, M.; Infante, C.; Jimenez-Cantizano, R.M.; Perez, L.; Manchado, M. Complete mitochondrial genome of the blackspot seabream, Pagellus bogaraveo (Perciformes: Sparidae), with high levels of length heteroplasmy in the WANCY region. Gene 2008, 409, 44–52. [Google Scholar] [CrossRef]
- Janke, A.; Paabo, S. Editing of a tRNA anticodon in marsupial mitochondria changes its codon recognition. Nucleic Acids Res. 1993, 21, 1523–1525. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Mayden, R.L. Phylogenetic relationships, subdivision, and biogeography of the cyprinid tribe Labeonini (sensu) (Teleostei: Cypriniformes), with comments on the implications of lips and associated structures in the labeonin classification. Mol. Phylogenet. Evol. 2010, 54, 254–265. [Google Scholar] [CrossRef]
- Wang, X.Z.; Li, J.B.; He, S.P. Molecular evidence for the monophyly of East Asian groups of Cyprinidae (Teleostei: Cypriniformes) derived from the nuclear recombination activating gene 2 sequences. Mol. Phylogenet. Evol. 2007, 42, 157–170. [Google Scholar] [CrossRef] [PubMed]
- Li, J.B.; Wang, X.Z.; Kong, X.H.; Zhao, K.; He, S.P.; Mayden, R.L. Variation patterns of the mitochondrial 16S rRNA gene with secondary structure constraints and their application to phylogeny of cyprinine fishes (Teleostei: Cypriniformes). Mol. Phylogenet. Evol. 2008, 47, 472–487. [Google Scholar] [CrossRef]
- Yang, L.; Sado, T.; Vincent Hirt, M.; Pasco-Viel, E.; Arunachalam, M.; Li, J.; Wang, X.; Freyhof, J.; Saitoh, K.; Simons, A.M.; et al. Phylogeny and polyploidy: Resolving the classification of cyprinine fishes (Teleostei: Cypriniformes). Mol. Phylogenet. Evol. 2015, 85, 97–116. [Google Scholar] [CrossRef]
- Tang, D.S. Fishes of Kweiyang, with descriptions of two new genera and five new species. Lingnan Sci. J. (Canton) 1942, 20, 147–166. [Google Scholar]
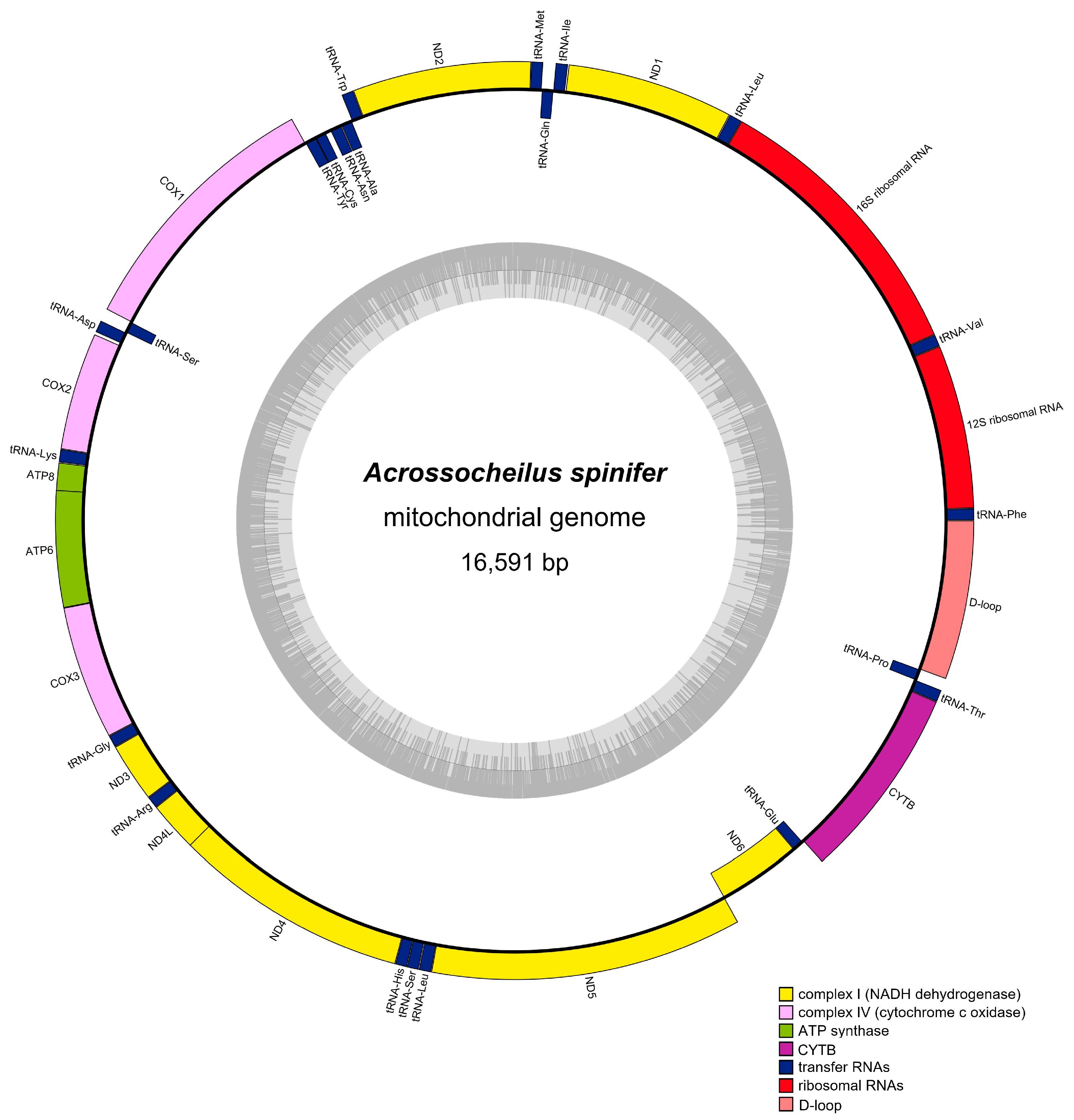
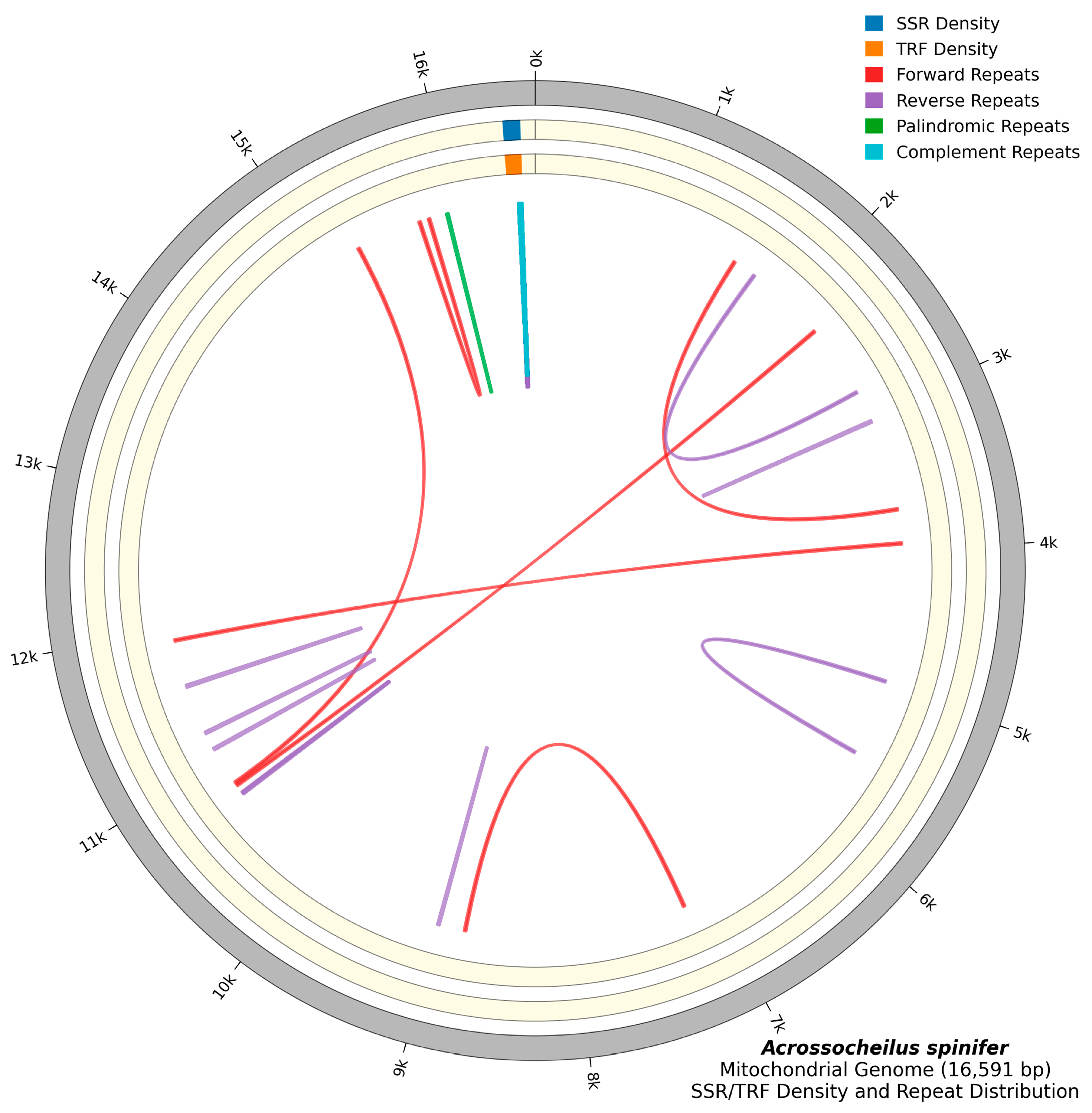
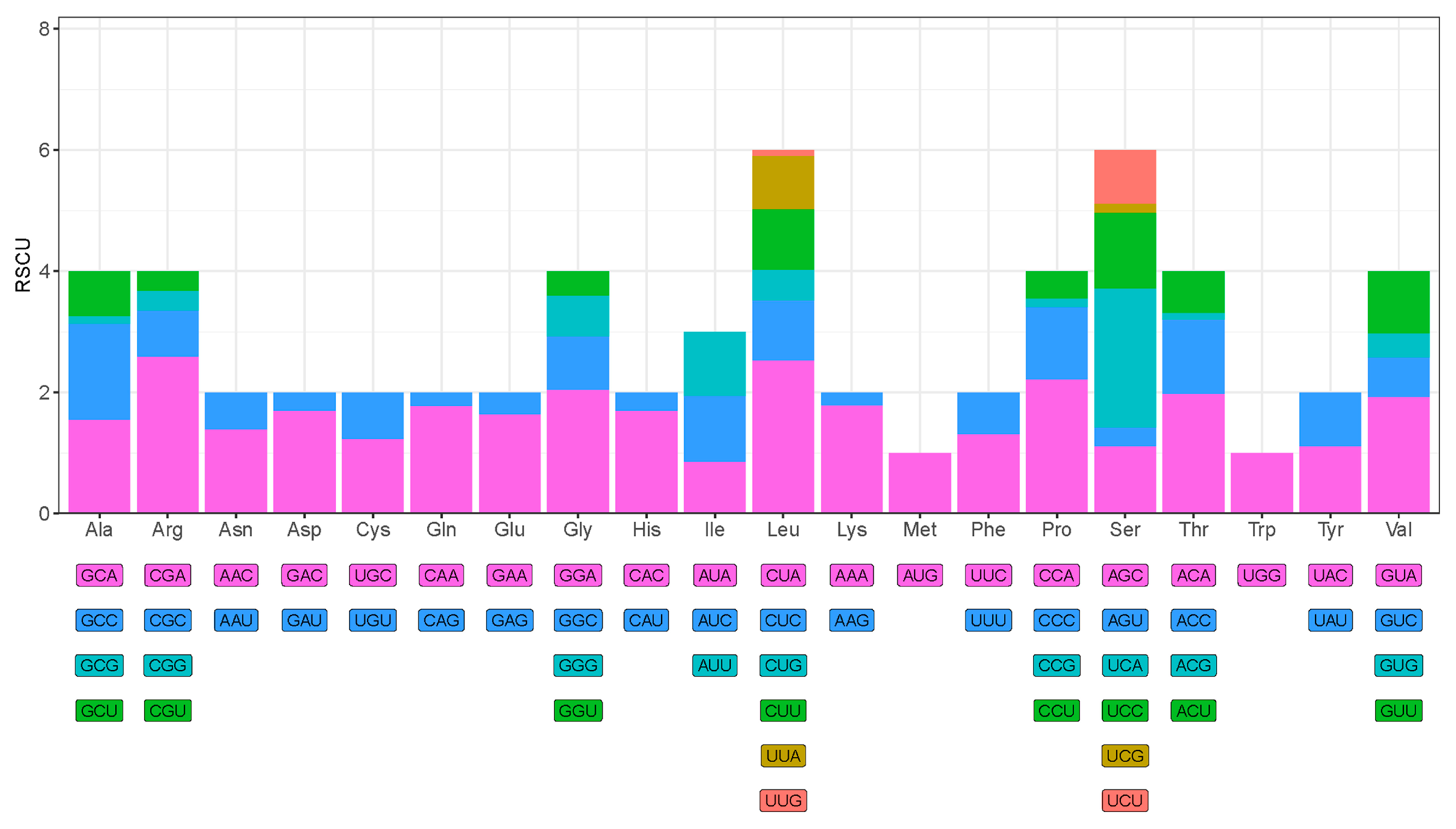
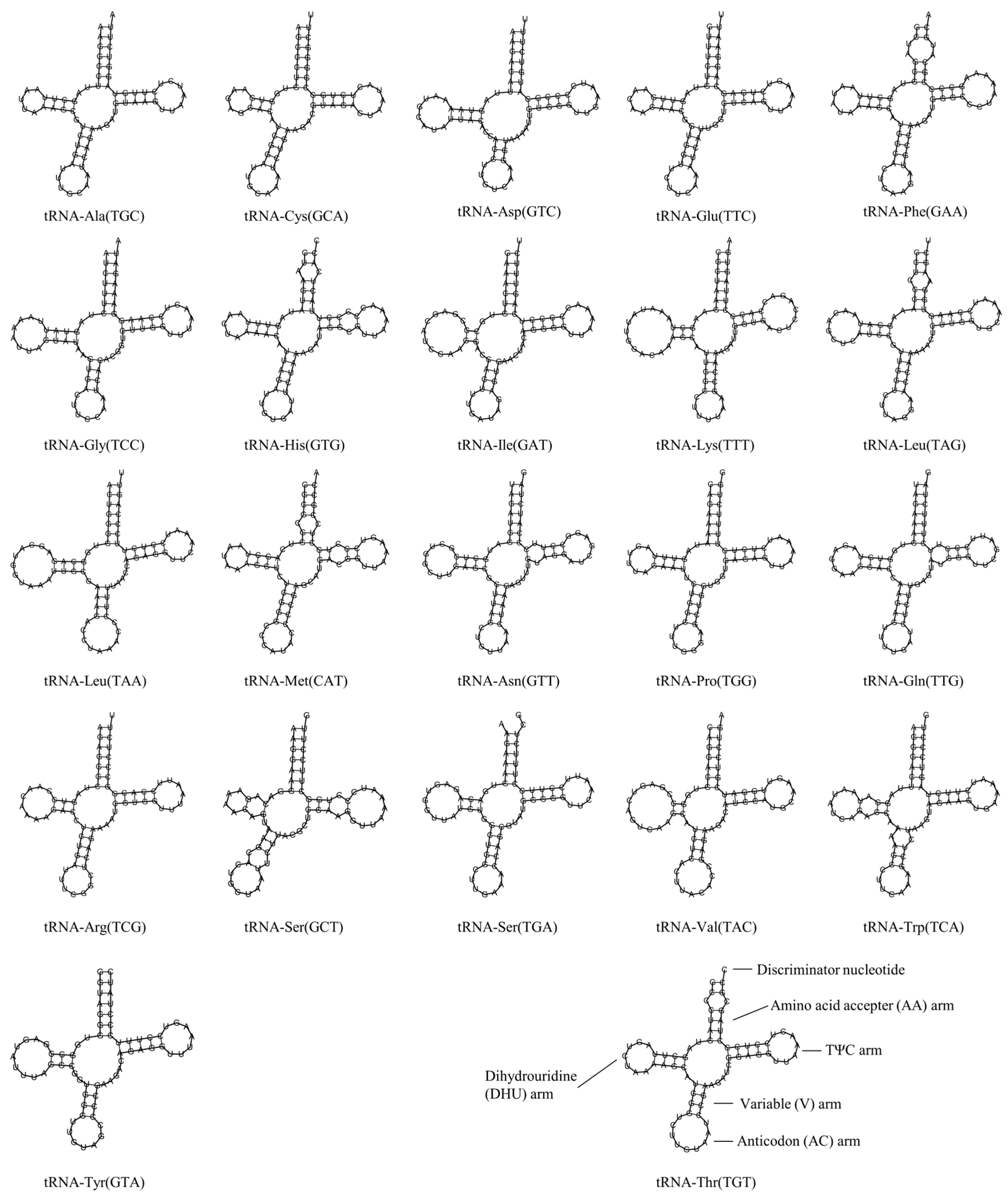
| Group | Group of Genes | Gene | Three Letter Code | Sequence | Size (bp) | Strand | No. of Amino Acids | Start Codon | Stop Codon | Anti-Codon | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Start | End | ||||||||||
| NADH dehydrogenase subunit | ND1 | - | 2870 | 3829 | 960 | H | 319 | ATG | TAA | - | |
| ND2 | - | 4052 | 5089 | 1038 | H | 345 | ATC | CCT | - | ||
| ND3 | - | 9659 | 10,006 | 348 | H | 115 | GTG | AAT | - | ||
| ND4 | - | 10,368 | 11,741 | 1374 | H | 457 | ATG | ACT | - | ||
| ND4L | - | 10,078 | 10,371 | 294 | H | 97 | ATG | TAA | - | ||
| ND5 | - | 11,970 | 13,775 | 1806 | H | 601 | ATG | TAA | - | ||
| ND6 | - | 13,784 | 14,302 | 519 | L | 172 | ATG | TAA | - | ||
| PCGs | Cytochrome c oxidase subunit | CO1 | - | 5490 | 7022 | 1533 | H | 510 | GTG | TAA | - |
| CO2 | - | 7193 | 7873 | 681 | H | 226 | ATG | CCT | - | ||
| CO3 | - | 8802 | 9584 | 783 | H | 260 | ATG | ATA | - | ||
| ATP synthase subunit | ATP6 | - | 8119 | 8796 | 678 | H | 225 | ATG | TAA | - | |
| ATP8 | - | 7961 | 8122 | 162 | H | 53 | ATG | TAG | - | ||
| Cytochrome b | Cyt b | - | 14,378 | 15,511 | 1134 | H | 377 | ATG | CTT | - | |
| Transfer RNA genes | trnA | Ala | 5170 | 5238 | 69 | L | - | - | - | TGC | |
| trnC | Cys | 5346 | 5412 | 67 | L | - | - | - | GCA | ||
| trnD | Asp | 7107 | 7178 | 72 | H | - | - | - | GTC | ||
| RNAs | trnE | Glu | 14,303 | 14,371 | 69 | L | - | - | - | TTC | |
| trnF | Phe | 1 | 69 | 69 | H | - | - | - | GAA | ||
| trnG | Gly | 9587 | 9658 | 72 | H | - | - | - | TCC | ||
| trnH | His | 11,749 | 11,817 | 69 | H | - | - | - | GTG | ||
| trnI | Ile | 3840 | 3911 | 72 | H | - | - | - | GAT | ||
| trnK | Lys | 7884 | 7959 | 76 | H | - | - | - | TTT | ||
| trnL | Leu | 11,888 | 11,960 | 73 | H | - | - | - | TAG | ||
| trnL | Leu | 2784 | 2859 | 76 | H | - | - | - | TAA | ||
| trnM | Met | 3983 | 4051 | 69 | H | - | - | - | CAT | ||
| trnN | Asn | 5240 | 5312 | 73 | L | - | - | - | GTT | ||
| trnP | Pro | 15,590 | 15,659 | 70 | L | - | - | - | TGG | ||
| trnQ | Gly | 3910 | 3980 | 71 | L | - | - | - | TTG | ||
| trnR | Arg | 10,008 | 10,077 | 70 | H | - | - | - | TCG | ||
| trnS | Ser | 11,818 | 11,886 | 69 | H | - | - | - | GCT | ||
| trnS | Ser | 7035 | 7105 | 71 | L | - | - | - | TGA | ||
| trnT | Thr | 15,519 | 15,590 | 72 | H | - | - | - | TGT | ||
| trnV | Val | 1029 | 1100 | 72 | H | - | - | - | TAC | ||
| trnW | Trp | 5097 | 5167 | 71 | H | - | - | - | TCA | ||
| trnY | Tyr | 5413 | 5482 | 70 | L | - | - | - | GTA | ||
| 12S rRNA | rrnS | - | 70 | 1026 | 957 | H | - | - | - | - | |
| 16S rRNA | rrnL | - | 1101 | 2783 | 1683 | H | - | - | - | - | |
| D-loop | Control region | D-loop | - | 15,660 | 16,591 | 932 | H | - | - | - | - |
| Amino_Acid | Codon | Count | RSCU | Amino_Acid | Codon | Count | RSCU |
|---|---|---|---|---|---|---|---|
| Lys | AAA | 67 | 1.787 | Asp | GAC | 62 | 2.548 |
| Asn | AAC | 84 | 2.083 | Glu | GAG | 19 | 0.362 |
| Lys | AAG | 8 | 0.213 | Asp | GAT | 11 | 0.452 |
| Asn | AAT | 37 | 0.917 | Ala | GCA | 132 | 1.935 |
| Thr | ACA | 157 | 2.476 | Ala | GCC | 135 | 1.979 |
| Thr | ACC | 97 | 1.530 | Ala | GCG | 11 | 0.161 |
| Thr | ACG | 9 | 0.142 | Ala | GCT | 63 | 0.924 |
| Thr | ACT | 54 | 0.852 | Gly | GGA | 124 | 2.562 |
| Ser | AGC | 43 | 2.039 | Gly | GGC | 53 | 1.095 |
| Ser | AGT | 12 | 0.569 | Gly | GGG | 41 | 0.847 |
| Ile | ATA | 120 | 1.718 | Gly | GGT | 24 | 0.496 |
| Ile | ATC | 152 | 2.177 | Val | GTA | 106 | 3.855 |
| Met | ATG | 40 | 2.000 | Val | GTC | 36 | 1.309 |
| Ile | ATT | 147 | 2.105 | Val | GTG | 22 | 0.800 |
| Gln | CAA | 91 | 1.784 | Val | GTT | 56 | 2.036 |
| His | CAC | 90 | 2.547 | Tyr | TAC | 64 | 2.226 |
| Gln | CAG | 11 | 0.216 | Tyr | TAT | 51 | 1.774 |
| His | CAT | 16 | 0.453 | Ser | TCA | 89 | 4.220 |
| Pro | CCA | 118 | 2.770 | Ser | TCC | 48 | 2.276 |
| Pro | CCC | 64 | 1.502 | Ser | TCG | 6 | 0.284 |
| Pro | CCG | 7 | 0.164 | Ser | TCT | 34 | 1.612 |
| Pro | CCT | 24 | 0.563 | Cys | TGC | 16 | 2.462 |
| Arg | CGA | 48 | 4.541 | Trp | TGG | 10 | 2.000 |
| Arg | CGC | 14 | 1.324 | Cys | TGT | 10 | 1.538 |
| Arg | CGG | 6 | 0.568 | Leu | TTA | 93 | 1.763 |
| Arg | CGT | 6 | 0.568 | Phe | TTC | 144 | 2.618 |
| Leu | CTA | 267 | 5.062 | Leu | TTG | 10 | 0.190 |
| Leu | CTC | 105 | 1.991 | Phe | TTT | 76 | 1.382 |
| Leu | CTG | 53 | 1.005 | Stop | TAA | 6 | 0.308 |
| Leu | CTT | 105 | 1.991 | Stop | TAG | 1 | 0.051 |
| Glu | GAA | 86 | 1.638 | Stop | TGA | 110 | 5.641 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gong, J.; She, S.-Q.; Liu, G.-F.; Zhao, X.-X.; Yuan, L.-Y.; Zhang, E. The Complete Mitochondrial Genome of Acrossocheilus spinifer (Osteichthyes: Cyprinidae) and Its Phylogenetic Analysis. Fishes 2025, 10, 296. https://doi.org/10.3390/fishes10060296
Gong J, She S-Q, Liu G-F, Zhao X-X, Yuan L-Y, Zhang E. The Complete Mitochondrial Genome of Acrossocheilus spinifer (Osteichthyes: Cyprinidae) and Its Phylogenetic Analysis. Fishes. 2025; 10(6):296. https://doi.org/10.3390/fishes10060296
Chicago/Turabian StyleGong, Jian, Shi-Qi She, Guang-Fu Liu, Xing-Xing Zhao, Le-Yang Yuan, and E Zhang. 2025. "The Complete Mitochondrial Genome of Acrossocheilus spinifer (Osteichthyes: Cyprinidae) and Its Phylogenetic Analysis" Fishes 10, no. 6: 296. https://doi.org/10.3390/fishes10060296
APA StyleGong, J., She, S.-Q., Liu, G.-F., Zhao, X.-X., Yuan, L.-Y., & Zhang, E. (2025). The Complete Mitochondrial Genome of Acrossocheilus spinifer (Osteichthyes: Cyprinidae) and Its Phylogenetic Analysis. Fishes, 10(6), 296. https://doi.org/10.3390/fishes10060296






