Full-Length Transcriptome Reveals Heterologous Sperm Fragments in Natural Gynogenetic Grass Carp
Abstract
1. Introduction
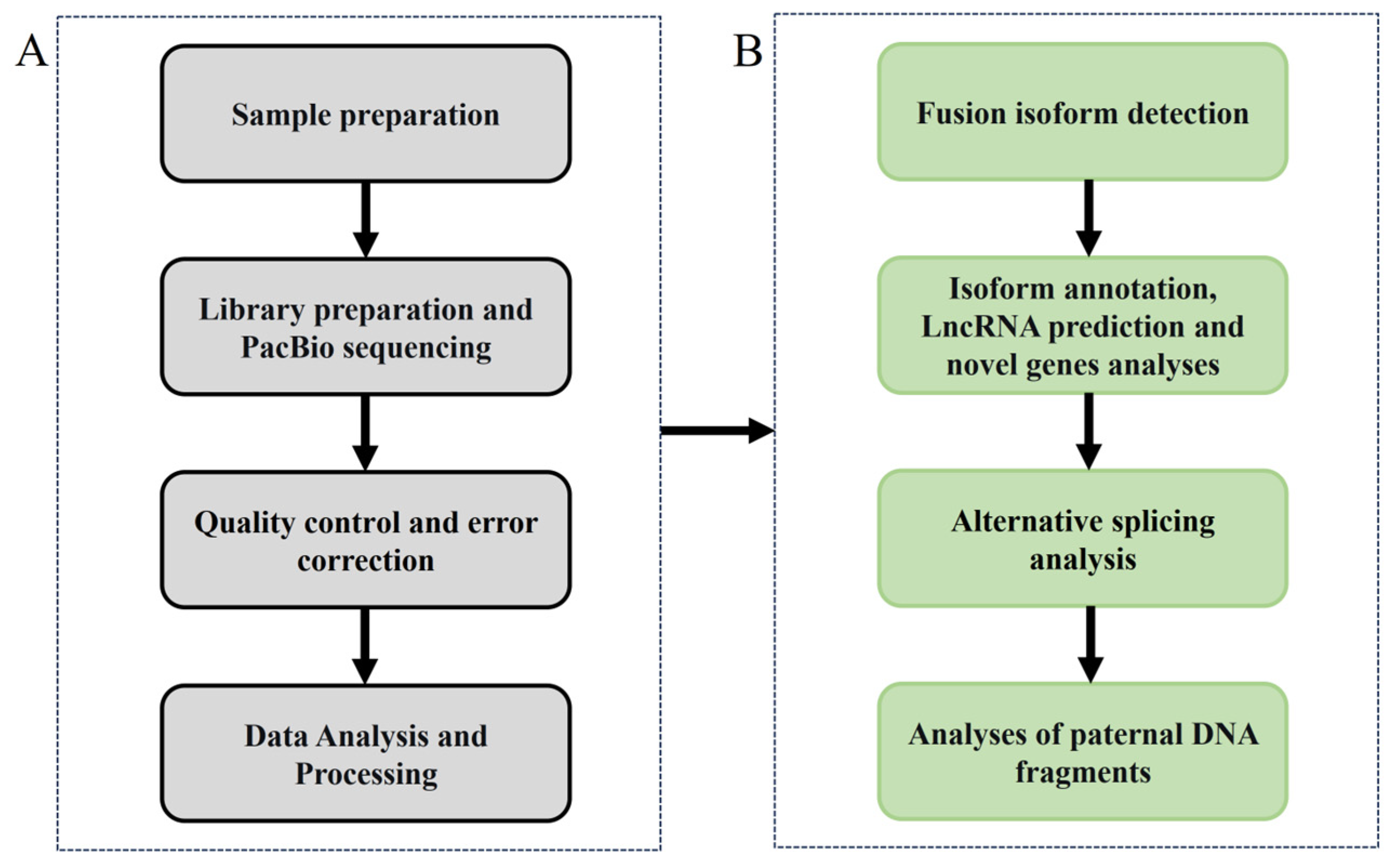
2. Materials and Methods
2.1. Materials
2.2. Library Preparation and PacBio Sequencing
2.3. Quality Control and Error Correction
2.4. Mapping to the Reference Genome and Fusion Isoforms Detection
2.5. Annotation, LncRNA Prediction and Novel Genes Analysis
2.6. Alternative Splicing Events Analysis and Validation
2.7. Analyses of Paternal DNA Fragments
3. Results
3.1. Pacbio Sequencing and Data Output
3.2. Fusion Isoform Detection
3.3. Isoform Annotation, LncRNA Prediction and Novel Genes Analyses
3.4. Alternative Splicing Analysis
3.5. Analyses of Paternal DNA Fragments
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| 2nGGC | 2n Gynogenetic Grass Carp |
| GC | Grass Carp |
| Pacbio | Pacific Biosciences |
| SNP | Single-Nucleotide Polymorphism |
| AS | Alternative Splicing |
| PCR | Polymerase Chain Reaction |
| ICE | In-Context Learning |
| GMAP | Genomic Mapping and Alignment Program |
| LncRNA | Long non-coding RNA |
| GO | Gene Ontology |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| COG | Cluster of Orthologous Groups |
| NR | Non-Redundant |
| CDS | Coding Sequence |
| HQ | Higher Quality |
| ExonS | Exon Skip |
| AltD | Alternate Donor Site |
| AltA | Alternate Acceptor Site |
| IntronR | Intron Retention |
| AltP | Alternate Promoter |
| AMPK | AMP-activated Protein Kinase |
References
- Li, L.; Liang, X.F.; He, S.; Sun, J.; Wen, Z.Y.; He, Y.H.; Cai, W.J.; Wang, Y.P.; Tao, Y.X. Transcriptome analysis of grass carp (Ctenopharyngodon idella) fed with animal and plant diets. Gene 2015, 574, 371–379. [Google Scholar] [CrossRef]
- Mao, Z.; Fu, Y.; Wang, Y.; Wang, S.; Zhang, M.; Gao, X.; Luo, K.; Qin, Q.; Zhang, C.; Tao, M.; et al. Evidence for paternal DNA transmission to gynogenetic grass carp. BMC Genet. 2019, 20, 3. [Google Scholar] [CrossRef]
- He, W.G.; Xie, L.; Li, T.; Liu, S.; Xiao, J.; Hu, J.; Wang, J.; Qin, Q.; Liu, Y. The formation of diploid and triploid hybrids of female grass carp x male blunt snout bream and their 5S rDNA analysis. BMC Genet. 2013, 14, 110. [Google Scholar] [CrossRef]
- Chilton, E.W.; Muoneke, M.I. Biology and management of grass carp (Ctenopharyngodon idella, Cyprinidae) for vegetation control: A North American perspective. Rev. Fish Biol. Fish. 1992, 2, 283–320. [Google Scholar] [CrossRef]
- Wang, Y.; Lu, Y.; Zhang, Y.; Ning, Z.; Li, Y.; Zhao, Q.; Lu, H.; Huang, R.; Xia, X.; Feng, Q.; et al. The draft genome of the grass carp (Ctenopharyngodon idellus) provides insights into its evolution and vegetarian adaptation. Nat. Genet. 2015, 47, 625–631. [Google Scholar] [CrossRef]
- Chen, Y.; Shi, M.; Zhang, W.; Cheng, Y.; Wang, Y.; Xia, X.Q. The Grass Carp Genome Database (GCGD): An online platform for genome features and annotations. Database 2017, 2017, bax051. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, W.; Li, Z.; Qiu, B.; Li, J.; Geng, G.; Hu, B.; Liao, A.; Cai, Y.; Wen, M. Improvement and application of genetic resources of grass carp (Ctenopharyngodon idella). Reprod. Breed. 2024, 4, 126–133. [Google Scholar] [CrossRef]
- Tan, H.; Wang, Y.; Hu, B.; Zhang, Y.; Liao, A.m.; Liu, W.; Gen, C.; Luo, K.; Tao, M.; Zhang, C. Analysis of the genetic characteristics and variations in disease-resistant grass carp based on whole-genome resequencing and transcriptome sequencing. Reprod. Breed. 2024, 4, 22–31. [Google Scholar] [CrossRef]
- Rhoads, A.; Au, K.F. PacBio Sequencing and Its Applications. Genom. Proteom. Bioinform. 2015, 13, 278–289. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Dai, C.; Hu, C.; Liu, Z.; Kang, C. Global identification of alternative splicing via comparative analysis of SMRT- and Illumina-based RNA-seq in strawberry. Plant J. 2017, 90, 164–176. [Google Scholar] [CrossRef] [PubMed]
- Ardui, S.; Ameur, A.; Vermeesch, J.R.; Hestand, M.S. Single molecule real-time (SMRT) sequencing comes of age: Applications and utilities for medical diagnostics. Nucleic Acids Res. 2018, 46, 2159–2168. [Google Scholar] [CrossRef]
- Zhang, J.; Chen, L.L.; Sun, S.; Kudrna, D.; Copetti, D.; Li, W.; Mu, T.; Jiao, W.B.; Xing, F.; Lee, S. Building two indica rice reference genomes with PacBio long-read and Illumina paired-end sequencing data. Sci. Data 2016, 3, 160076. [Google Scholar] [CrossRef]
- Korlach, J.; Gedman, G.; Kingan, S.B.; Chin, C.S.; Howard, J.T.; Audet, J.N.; Cantin, L.; Jarvis, E.D. De novoPacBio long-read and phased avian genome assemblies correct and add to reference genes generated with intermediate and short reads. Gigascience 2017, 6, gix085. [Google Scholar] [CrossRef]
- Jung-Ting, C.; Pakala, S.B.; Geraldo, J.A.; Lapp, S.A.; Humphrey, J.C.; Barnwell, J.W.; Kissinger, J.C.; Galinski, M.R. High-Quality Genome Assembly and Annotation forPlasmodium coatneyi, Generated Using Single-Molecule Real-Time PacBio Technology. Genome Announc. 2016, 4, e00883-16. [Google Scholar]
- Giordano, F.; Aigrain, L.; Quail, M.A.; Coupland, P.; Bonfield, J.K.; Davies, R.M.; Tischler, G.; Jackson, D.K.; Keane, T.M.; Li, J. De novo yeast genome assemblies from MinION, PacBio and MiSeq platforms. Sci. Rep. 2017, 7, 3935. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Sebra, R.; Pullman, B.S.; Qiao, W.; Peter, I.; Desnick, R.J.; Geyer, C.R.; Decoteau, J.F.; Scott, S.A. Quantitative and multiplexed DNA methylation analysis using long-read single-molecule real-time bisulfite sequencing (SMRT-BS). BMC Genom. 2015, 16, 350. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Scott, S.A. DNA Methylation Profiling Using Long-Read Single Molecule Real-Time Bisulfite Sequencing (SMRT-BS); Humana Press: New York, NY, USA, 2017; pp. 125–134. [Google Scholar]
- Kelleher, P.; Murphy, J.; Mahony, J.; Sinderen, D.V. Identification of DNA Base Modifications by Means of Pacific Biosciences RS Sequencing Technology; Humana Press: New York, NY, USA, 2018. [Google Scholar]
- Abdel-Ghany, S.E.; Hamilton, M.; Jacobi, J.L.; Ngam, P.; Devitt, N.; Schilkey, F.; Ben-Hur, A.; Reddy, A.S. A survey of the sorghum transcriptome using single-molecule long reads. Nat. Commun. 2016, 7, 11706. [Google Scholar] [CrossRef]
- Wang, B.; Tseng, E.; Regulski, M.; Clark, T.A.; Hon, T.; Jiao, Y.; Lu, Z.; Olson, A.; Stein, J.C.; Ware, D. Unveiling the complexity of the maize transcriptome by single-molecule long-read sequencing. Nat. Commun. 2016, 7, 11708. [Google Scholar] [CrossRef]
- Wang, M.; Wang, P.; Liang, F.; Ye, Z.; Li, J.; Shen, C.; Pei, L.; Wang, F.; Hu, J.; Tu, L.; et al. A global survey of alternative splicing in allopolyploid cotton: Landscape, complexity and regulation. New Phytol. 2017, 217, 163–178. [Google Scholar] [CrossRef]
- Filichkin, S.A.; Hamilton, M.; Dharmawardhana, P.D.; Singh, S.K.; Sullivan, C.; Ben-Hur, A.; Reddy, A.S.N.; Jaiswal, P. Abiotic Stresses Modulate Landscape of Poplar Transcriptome via Alternative Splicing, Differential Intron Retention, and Isoform Ratio Switching. Front. Plant Sci. 2018, 9, 5. [Google Scholar] [CrossRef]
- Liu, S.J.; Qin, Q.B.; Wang, Y.; Zhang, H.; Zhao, R.; Zhang, C.; Wang, J.; Li, W.; Chen, L.; Xiao, J.; et al. Evidence for the formation of the male gynogenetic fish. Mar. Biotechnol. 2010, 12, 160–172. [Google Scholar] [CrossRef] [PubMed]
- Xiao, J.; Zou, T.M.; Chen, Y.B.; Chen, L.; Liu, S.J.; Tao, M.; Zhang, C.; Zhao, R.R.; Zhou, Y.; Long, Y.; et al. Coexistence of diploid, triploid and tetraploid crucian carp (Carassius auratus) in natural waters. BMC Genet. 2011, 12, 20. [Google Scholar] [CrossRef]
- Zhou, L.; Wang, Y.; Gui, J.-F. Genetic evidence for gonochoristic reproduction in gynogenetic silver crucian carp (Carassius auratus gibelio Bloch) as revealed by RAPD assays. J. Mol. Evol. 2000, 51, 498–506. [Google Scholar] [CrossRef]
- Wu, P.; Zeng, Y.; Qin, Q.; Wu, C.; Wang, Y.; Zhao, R.; Tao, M.; Zhang, C.; Tang, C.; Liu, S. Comparative analysis of the texture, composition, antioxidant capacity and nutrients of natural gynogenesis blunt snout bream and its parent muscle. Reprod. Breed. 2022, 2, 149–155. [Google Scholar] [CrossRef]
- Fan, S.; Tang, Z.; Wang, Y.; Zhou, Z.; Wu, C.; Luo, K.; Hu, J.; Gong, D.; Li, S.; Tao, M.; et al. The bisexual natural gynogenetic blunt snout bream lineage derived from the distant hybridization of female blunt snout bream and male Bleeker’s yellow tail. Aquac. Rep. 2024, 37, 102206. [Google Scholar] [CrossRef]
- Mao, Z.; Fu, Y.; Wang, S.; Wang, Y.; Luo, K.; Zhang, C.; Tao, M.; Liu, S. Further evidence for paternal DNA transmission in gynogenetic grass carp. Sci. China Life Sci. 2020, 63, 1287–1296. [Google Scholar] [CrossRef]
- Liu, Q.; Liao, A.; Tao, M.; Qin, Q.; Luo, K.; Zhang, C.; Wang, S.; Zhou, Y.; Hu, F.; Wang, Y.; et al. Macro-Hybrid and Micro-Hybrid of Fish. Rev. Aquac. 2025, 18, e70106. [Google Scholar] [CrossRef]
- Liu, Q.; Wang, S.; Tang, C.; Tao, M.; Zhang, C.; Zhou, Y.; Qin, Q.; Luo, K.; Wu, C.; Hu, F.; et al. The research advances in distant hybridization and gynogenesis in fish. Rev. Aquac. 2025, 17, e12972. [Google Scholar] [CrossRef]
- Sahlin, K.; Medvedev, P. De novo clustering of long-read transcriptome data using a greedy, quality-value based algorithm. In Proceedings of the International Conference on Research in Computational Molecular Biology, Washington, DC, USA, 5–8 May 2019; pp. 227–242. [Google Scholar]
- Weirather, J.L.; Afshar, P.T.; Clark, T.A.; Tseng, E.; Powers, L.S.; Underwood, J.G.; Zabner, J.; Korlach, J.; Wong, W.H.; Au, K.F. Characterization of fusion genes and the significantly expressed fusion isoforms in breast cancer by hybrid sequencing. Nucleic Acids Res. 2015, 43, e116. [Google Scholar] [CrossRef]
- Wu, T.D.; Watanabe, C.K. GMAP: A genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics 2005, 21, 1859. [Google Scholar] [CrossRef]
- Qin, Q.; Popic, V.; Wienand, K.; Yu, H.; White, E.; Khorgade, A.; Shin, A.; Georgescu, C.; Campbell, C.D.; Dondi, A. Accurate fusion transcript identification from long-and short-read isoform sequencing at bulk or single-cell resolution. Genome Res. 2025, 35, 967–986. [Google Scholar] [CrossRef]
- Chen, X.; Sun, Y.-Z.; Guan, N.-N.; Qu, J.; Huang, Z.-A.; Zhu, Z.-X.; Li, J.-Q. Computational models for lncRNA function prediction and functional similarity calculation. Brief. Funct. Genom. 2019, 18, 58–82. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Chen, Q.; Huang, X.; Hu, F.; Zhu, S.; Luo, L.; Gong, D.; Gong, K.; Zhao, R.; Zhang, C.; et al. Genomic and epigenetic alterations in diploid gynogenetic hybrid fish. Aquaculture 2019, 512, 734383. [Google Scholar] [CrossRef]
- Huang, X.; Huang, J.; Qin, L.; Long, Y.; Wu, C.; Gong, K.; Xiao, Q.; Qin, Q. Full-length transcriptome reveals rapid genetic changes in triploid hybrid grass carp derived from female grass carp × male topmouth culter. Reprod. Breed. 2023, 3, 82–87. [Google Scholar] [CrossRef]
- Liu, S. Distant hybridization leads to different ploidy fishes. Sci. China Life Sci. 2010, 53, 416–425. [Google Scholar] [CrossRef]
- Liu, S.; Duan, W.; Tao, M.; Zhang, C.; Sun, Y.; Shen, J.; Wang, J.; Luo, K.; Liu, Y. Establishment of the diploid gynogenetic hybrid clonal line of red crucian carp × common carp. Sci. China Ser. C Life Sci. 2007, 50, 186–193. [Google Scholar] [CrossRef]
- Hubbs, C.L.; Hubbs, L.C. Apparent parthenogenesis in nature, in a form of fish of hybrid origin. Science 1932, 76, 628–630. [Google Scholar] [CrossRef]
- Wang, S.; Ye, X.; Wang, Y.; Chen, Y.; Lin, B.; Yi, Z.; Mao, Z.; Hu, F.; Zhao, R.; Wang, J.; et al. A new type of homodiploid fish derived from the interspecific hybridization of female common carp × male blunt snout bream. Sci. Rep. 2017, 7, 4189. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.; Ji, W.; Zeng, Y.; Tang, J.; Wu, C.; Qin, Q.; Yi, T.; Zhou, Y.; Zhao, R.; Tao, M. The analysis of growth performance and expression of growth-related genes in natural gynogenic blunt snout bream muscle derived from the blunt snout bream (Megalobrama amblycephala, ♀) × Chinese perch (Siniperca chuatsi, ♂). Aquaculture 2024, 590, 741052. [Google Scholar] [CrossRef]
- Wu, C.; Huang, X.; Hu, F.; Ouyang, Y.; Zhao, L.; Wang, S.; Li, W.; Fan, J.; Zhang, C.; Ren, L.; et al. Production of diploid gynogenetic grass carp and triploid hybrids derived from the distant hybridization of female grass carp and male topmouth culter. Aquaculture 2019, 504, 462–470. [Google Scholar] [CrossRef]
- Li, X.-Y.; Liu, X.-L.; Zhu, Y.-J.; Zhang, J.; Ding, M.; Wang, M.-T.; Wang, Z.-W.; Li, Z.; Zhang, X.-J.; Zhou, L.; et al. Origin and transition of sex determination mechanisms in a gynogenetic hexaploid fish. Heredity 2018, 121, 64–74. [Google Scholar] [CrossRef] [PubMed]
- Yi, M.S.; Li, Y.Q.; Liu, J.D.; Zhou, L.; Yu, Q.X.; Gui, J.F. Molecular cytogenetic detection of paternal chromosome fragments in allogynogenetic gibel carp, Carassius auratus gibelio Bloch. Chromosome Res. 2003, 11, 665–671. [Google Scholar] [CrossRef] [PubMed]
- Ren, L.; Li, W.; Qin, Q.; Dai, H.; Han, F.; Xiao, J.; Gao, X.; Cui, J.; Wu, C.; Yan, X.; et al. The subgenomes show asymmetric expression of alleles in hybrid lineages of Megalobrama amblycephala × Culter alburnus. Genome Res. 2019, 29, 1805–1815. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Chen, J.; Li, L.; Tao, M.; Zhang, C.; Qin, Q.; Xiao, J.; Liu, Y.; Liu, S. Research advances in animal distant hybridization. Sci. China Life Sci. 2014, 57, 889–902. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Yang, S.-T.; Wei, X.-H.; Gui, J.-F. Genetic diversity among different clones of the gynogenetic silver crucian carp, Carassius auratus gibelio, revealed by transferrin and isozyme markers. Biochem. Genet. 2001, 39, 213–225. [Google Scholar] [CrossRef]
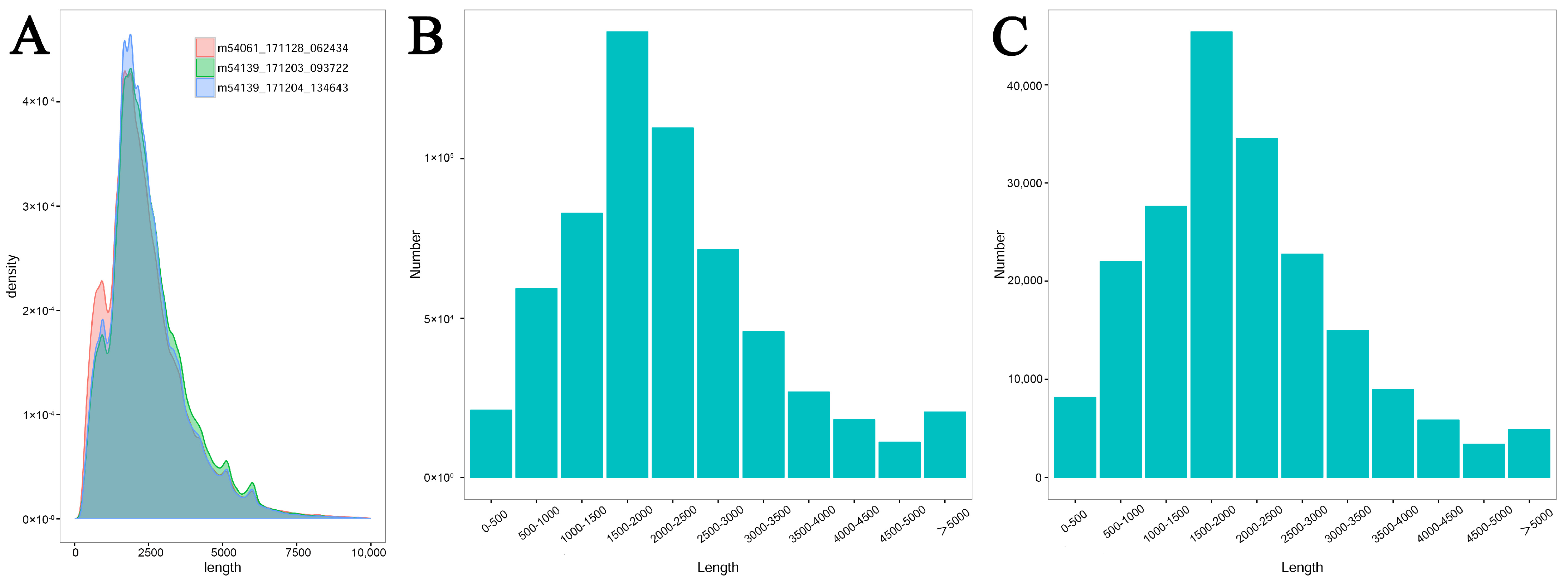
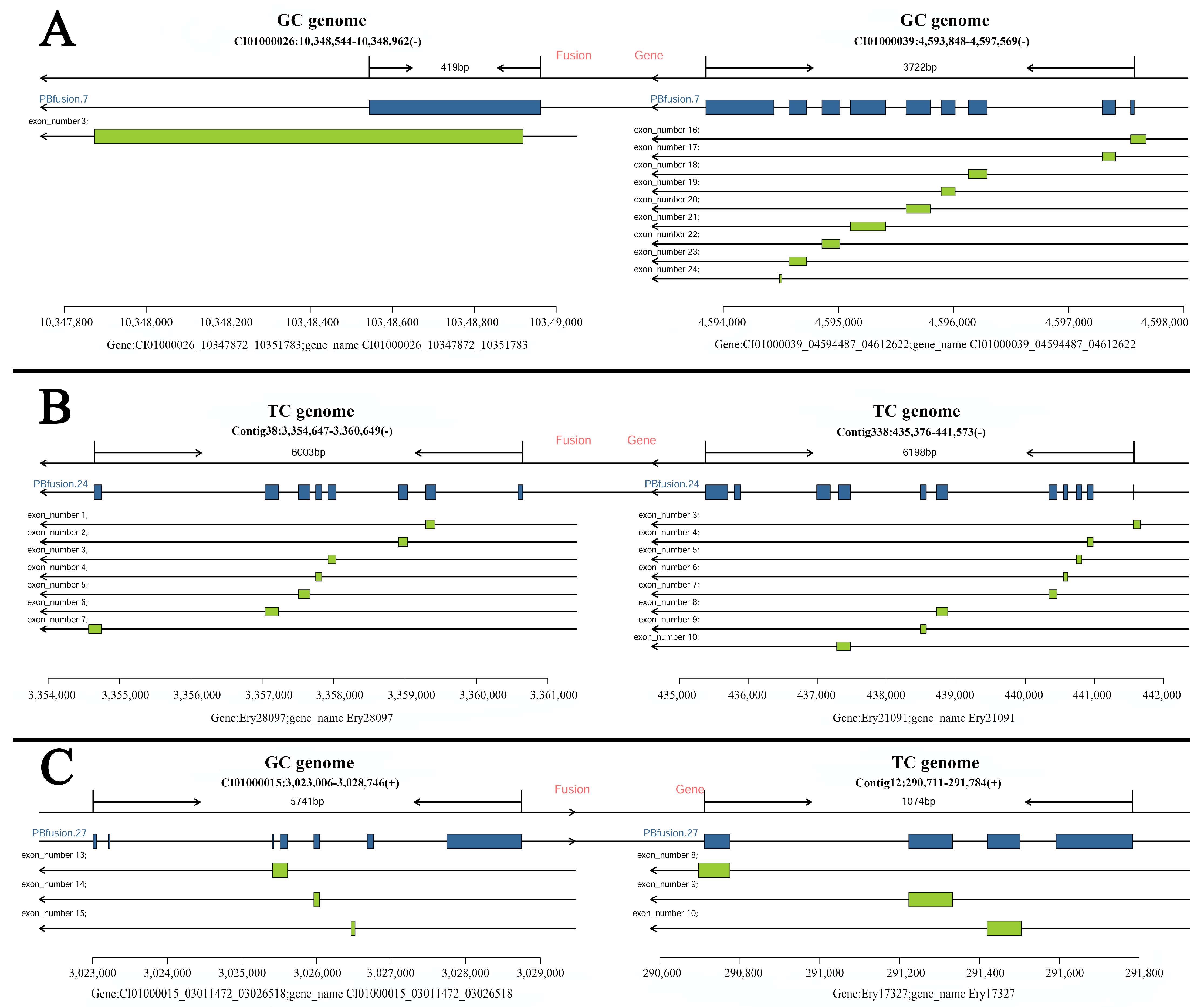

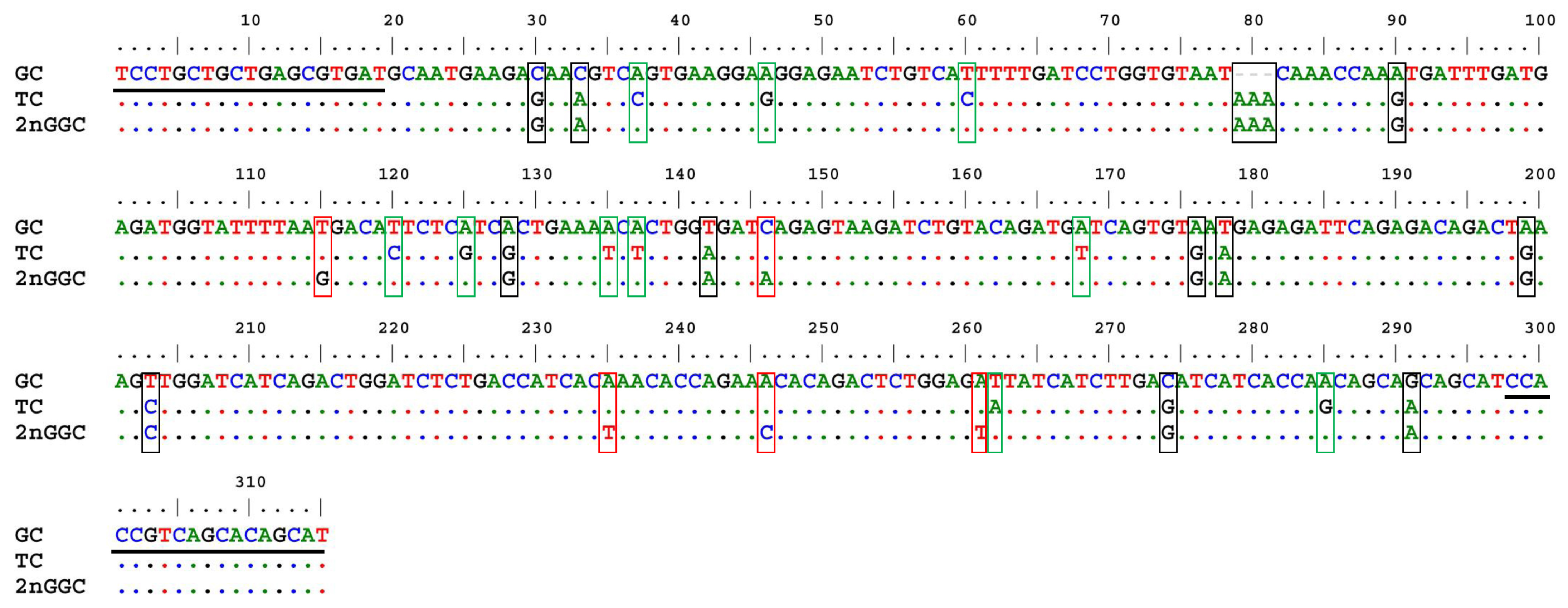
| Summary of Mapping Result | Total Mapped Number | Total Mapped Rate | Number of Collapse Isoforms | Isoforms Mapped to Known Gene | Total Mapped Gene | Isoforms Mapped to Unknown Gene | Number of LncRNAs | Number of Novel Isoforms |
|---|---|---|---|---|---|---|---|---|
| 376,948 | 374,939 | 99.47% | 107,721 | 92,628 | 21,702 | 15,093 | 5994 | 9099 |
| Type | AS_Number | Gene_Number |
|---|---|---|
| Exons | 10,934 | 5773 |
| AltD | 6651 | 3654 |
| AltA | 7582 | 4242 |
| IntronR | 32,952 | 8678 |
| Other | 59,998 | 6832 |
| AltP | 22,291 | 5382 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qin, L.; Wang, Y.; Wen, M.; Huang, J.; Huang, X.; Chen, Q.; Peng, D.; Wu, Y.; Wei, Q.; Hu, F.; et al. Full-Length Transcriptome Reveals Heterologous Sperm Fragments in Natural Gynogenetic Grass Carp. Fishes 2025, 10, 570. https://doi.org/10.3390/fishes10110570
Qin L, Wang Y, Wen M, Huang J, Huang X, Chen Q, Peng D, Wu Y, Wei Q, Hu F, et al. Full-Length Transcriptome Reveals Heterologous Sperm Fragments in Natural Gynogenetic Grass Carp. Fishes. 2025; 10(11):570. https://doi.org/10.3390/fishes10110570
Chicago/Turabian StyleQin, Lang, Yuxiang Wang, Ming Wen, Jinhui Huang, Xu Huang, Qian Chen, Dan Peng, Yang Wu, Qianye Wei, Fangzhou Hu, and et al. 2025. "Full-Length Transcriptome Reveals Heterologous Sperm Fragments in Natural Gynogenetic Grass Carp" Fishes 10, no. 11: 570. https://doi.org/10.3390/fishes10110570
APA StyleQin, L., Wang, Y., Wen, M., Huang, J., Huang, X., Chen, Q., Peng, D., Wu, Y., Wei, Q., Hu, F., Gong, K., Zhang, C., Qin, Q., Wu, C., & Liu, S. (2025). Full-Length Transcriptome Reveals Heterologous Sperm Fragments in Natural Gynogenetic Grass Carp. Fishes, 10(11), 570. https://doi.org/10.3390/fishes10110570





