Distribution and Comparative Analysis of Genomic Microsatellites in Nine Species of Family Sillaginidae
Abstract
1. Introduction
2. Materials and Methods
2.1. Genomic Sequences
2.2. Microsatellite Identification
3. Results
3.1. The Number of SSRs
3.2. The Frequency of SSRs
3.3. The Number of Repetitions of Microsatellite Loci
3.4. Microsatellite Sequence Length and Variation Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Mckay, R.J. Silaginid fishes of the world (family Sillaginidae): An annotated and illustrated catalogue of the Sillago, smelt or Indo-Pacific whiting species known to date. In FAO Fish Synop; Food and Agriculture Organization of the United Nations: Rome, Italy, 1992; Volume 14, pp. 1–82. [Google Scholar]
- Tetsuji, N. Fish Retrieval from Japan; Tokai University Press Association: Tokyo, Japan, 2013; pp. 974–975. [Google Scholar]
- Nelson, J.S.; Grande, T.; Wilson, M.V.H. Fishes of the World; John Wiley and Sons, Inc.: Hoboken, NJ, USA, 2016; p. 707. [Google Scholar]
- Wu, H.L.; Zhong, J.S. Marine and Estuarine Fish System Retrieval in China; China Agriculture Press: Beijing, China, 2021; pp. 680–682. [Google Scholar]
- Tatsuya, K. Phylogenetic systematics of the family Sillaginidae (Percomorpha: Order Perciformes). Zootaxa 2013, 3642, 1–105. [Google Scholar] [CrossRef]
- Ye, M.; Kong, L.; Jian, Z.; Qiu, Z.; Lin, X.; Zhang, Y.; Huang, Y.; Li, G.; Tian, C. Genetic insights into hypoxia tolerance in silver sillago (Sillago sihama) through QTL mapping and SNP association analysis. Aquaculture 2024, 592, 741174. [Google Scholar] [CrossRef]
- Cheng, Q.T.; Zheng, B.S. Systematic Synopsis of Chinese Fishes; Science Press: Beijing, China, 1987. [Google Scholar]
- Konishi, H.; Nakabo, T. Color Guide to the Japanese Fishes for SportFishermen; Enterbrain, Inc.: Tokyo, Japan, 2007; p. 400. [Google Scholar]
- Yu, J.W.; Wang, Y.R.; Lin, X.H.; Shen, Y.J.; Li, G.L.; Huang, Y.; Zhu, C.H.; Tian, C.X. Effects of morphological traits on body weight in 12-month-old silver sillago (Sillago sihama). J. Guangdong Ocean Univ. 2022, 42, 137–143. (In Chinese) [Google Scholar]
- Tautz, D. Hypervariability of simple sequences as a general source for polymorphic DNA markers. Nucl. Acids. Res. 1989, 17, 6463–6471. [Google Scholar] [CrossRef] [PubMed]
- Litt, M.; Luty, J.A. A hypervariable microsatellite revealed by in vitro amplification of a dinucleotide repeat within the cardiac muscle actin gene. Am. J. Hum. Genet. 1989, 44, 397–401. [Google Scholar] [PubMed]
- Madesis, P.; Ganopoulos, I.; Tsaftaris, A. Microsatellites: Evolution and contribution. Methods Mol. Biol. 2013, 1006, 1–13. [Google Scholar]
- Fan, S.G.; Huang, H.; Liu, Y.; Wang, P.F.; Zhao, C.; Yan, L.L.; Qiao, X.T.; Qiu, L.H. Genome-wide identification of microsatellite and development of polymorphic SSR markers for spotted sea bass (Lateolabrax maculatus). Aquacult. Rep. 2021, 20, 100677. [Google Scholar]
- Serbezov, D.; Bernatchze, L.; Olsen, E.M.; Vøllestad, L.A. Mating patterns and determinants of individual reproductive success in brown trout (Salmo trutta) revealed by parentage analysis of an entire stream living population. Conserv. Genet. Resour. 2010, 19, 3193–3205. [Google Scholar] [CrossRef]
- Shen, X.Y.; Yang, G.P.; Liu, Y.J.; Liao, M.J.; Wang, X.C.; Zhu, M.Z.; Song, W.B.; Zou, G.W.; Wei, Q.W.; Wang, D.Q.; et al. Construction of genetic linkage maps of guppy (Poecilia reticulata) based on AFLP and microsatellite DNA markers. Aquaculture 2007, 271, 178–187. [Google Scholar] [CrossRef]
- Xia, J.H.; Liu, F.; Zhu, Z.Y.; Fu, J.J.; Feng, J.B.; Li, J.L.; Yue, G.H. A consensus linkage map of the grass carp (Ctenopharyngodon idella) based on microsatellites and SNPs. BMC Genom. 2010, 11, 135–151. [Google Scholar] [CrossRef]
- Kuo, J.; Chen, C.Y.; Han, C.C.; Ju, Y.M.; Tew, K.S. Analyses of diet preference of larval orange-spotted grouper (Epinephelus coioides) grown under inorganic fertilization method using next-generation sequencing. Aquaculture 2021, 531, 735916. [Google Scholar] [CrossRef]
- Wang, H.Y.; Fu, Y.; Gu, P.; Zhang, Y.Y.; Tu, W.L.; Chao, Z.; Wu, H.L.; Cao, J.G.; Zhou, X.; Liu, B.; et al. Genome-wide characterization and comparative analyses of simple sequence repeats among four miniature pig breeds. Animals 2020, 10, 1792. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.C.; Xu, Y.T.; Li, Z.K.; Fan, J.; Yang, Y. Genome-wide mining of microsatellites in king cobra (Ophiophagus hannah) and cross-species development of tetranucleotide SSR markers in Chinese cobra (Naja atra). Mol. Biol. Rep. 2019, 46, 6087–6098. [Google Scholar] [CrossRef] [PubMed]
- Bastías, A.; Correa, F.; Rojas, P.; Martin, C.; Pérez-Diaz, J.; Yáñez, C.; Cuevas, M.; Verdugo, R.; Sagredo, B. Draft genome sequence data of maqui (Aristotelia chilensis) and identification of SSR markers. Data Brief. 2019, 27, 104545. [Google Scholar] [CrossRef]
- Peng, Y.; Li, J.; Wang, T.; Zhang, K.; Ning, X.H.; Ji, J.; Yin, S.W. Preliminary study on distribution characteristics and positioning of microsatellites in whole genome of Pelteobagrus vachelli. South China Fish. Sci. 2022, 18, 90–98. (In Chinese) [Google Scholar]
- Fu, J.J.; Shen, Y.B.; Xu, X.Y.; Chen, Y.; Li, D.; Li, J.L. Multiplex microsatellite PCR sets for parentage assignment of grass carp (Ctenopharyngodon idella). Aquacult. Int. 2013, 21, 1195–1207. [Google Scholar] [CrossRef]
- Zhong, L.Q.; Zhang, S.Y.; Jiang, H.C.; Wang, M.H.; Chen, X.H. Bioinformatics analysis of perfect microsatellite in the whole genome of channel catfish, Ictalurus punctatus. Genom. Appl. Biol. 2022, 41, 504–513. (In Chinese) [Google Scholar]
- Wang, X.L.; Gao, J.; Qi, X.; Wang, Y.B.; Chen, F.X.; Liu, J.Y.; Fu, S.Y. Screening and characteristics analysis of microsatellites in whole genome of five groupers. Prog. Fish. Sci. 2023, 45, 149–158. (In Chinese) [Google Scholar]
- Jian, J.B.; Yang, L.D.; Gan, X.N.; Wu, B.; Gao, L.; Zeng, H.H.; Wang, X.Z.; Liang, Z.Q.; Wang, Y.; Fang, L.H.; et al. Whole genome sequencing of silver carp (Hypophthalmichthys molitrix) and bighead carp (Hypophthalmichthys nobilis) provide novel insights into their evolution and speciation. Mol. Ecol. Resour. 2021, 21, 912–923. [Google Scholar] [CrossRef]
- Xiao, J.G.; Song, N.; Gao, T.X.; McKay, R.J. Redescription and DNA barcoding of Sillago indica (Perciformes: Sillaginidae) from the Coast of Pakistan. Pak. J. Zool. 2016, 48, 317–323. [Google Scholar]
- Bae, S.E.; Kwun, H.J.; Kim, J.K.; Kweon, S.M.; Kang, C.B. New record of Sillago sinica (Pisces: Sillaginidae) in Korean waters, and re-identification of Sillago parvisquamis previously reported from Korea as S. sinica. Anim. Syst. Evol. Divers. 2013, 29, 288–293. [Google Scholar] [CrossRef]
- Gao, T.X.; Ji, D.P.; Xiao, Y.S.; Xue, T.Q.; Yanagimoto, T.; Setoguma, T. Description and DNA barcoding of a new Sillago species, Sillago sinica (Perciformes: Sillaginidae) from coastal waters of China. Zool. Stud. 2011, 50, 254–263. [Google Scholar]
- Xiao, J.G.; Song, N.; Han, Z.Q.; Gao, T.X. Description and DNA barcoding of a new Sillago species, Sillago shaoi (Perciformes: Sillaginidae), in the Taiwan Strait. Zool. Stud. 2016, 55, e47. [Google Scholar]
- Yu, Z.S.; Guo, T.; Xiao, J.G.; Song, N.; Gao, T.X. Identification and DNA barcoding of a new Sillago species in Beihai and Zhanjiang, China, with a key to related species. J. Ocean Univ. China 2022, 21, 1334–1342. [Google Scholar] [CrossRef]
- Liu, Q.; Mao, W.H.; Wang, Y.T.; Xiao, J.G.; Saha, S.; Gao, T.X.; Liu, F. Whole genome sequencing and phylogenetic analyses of the Sillaginidae family fish. Mol. Phylogenet. Evol. 2025, 207, 108340. [Google Scholar] [CrossRef]
- Liu, S.X.; Hou, W.; Sun, T.L.; Xu, Y.T.; Li, P.; Yue, B.S.; Fan, Z.X.; Li, J. Genome-wide mining and comparative analysis of microsatellites in three macaque species. Mol. Gene. Genom. 2017, 292, 537–550. [Google Scholar] [CrossRef]
- Subramanian, S.; Mishra, R.K.; Singh, L. Genome-wide analysis of microsatellite repeats in humans: Their abundance and density in specific genomic regions. Genom. Biol. 2003, 4, R13. [Google Scholar] [CrossRef]
- Xu, Y.T.; Li, W.J.; Hu, Z.X.; Zeng, T.; Shen, Y.M.; Liu, S.X.; Zhang, X.Y.; Li, J.; Yue, B.S. Genome-wide mining of perfect microsatellites and tetranucleotide orthologous microsatellites estimates in six primate species. Gene 2018, 643, 124–132. [Google Scholar] [CrossRef]
- Xu, Y.T.; Hu, Z.X.; Wang, C.; Zhang, X.Y.; Li, J.; Yue, B.S. Characterization of perfect microsatellite based on genome-wide and chromosome level in Rhesus monkey (Macaca mulatta). Gene 2016, 592, 269–275. [Google Scholar] [CrossRef]
- Huang, J.; Li, W.J.; Jian, Z.Y.; Yue, B.S.; Yan, Y.F. Genome-wide distribution and organization of microsatellites in six species of birds. Biochem. Syst. Ecol. 2016, 67, 95–102. [Google Scholar] [CrossRef]
- Karaoglu, H.; Lee, C.M.Y.; Meyer, W. Survey of simple sequence repeats in completed fungal genomes. Mol. Biol. Evol. 2005, 22, 639–649. [Google Scholar] [CrossRef]
- Li, C.Y.; Liu, L.; Yang, J.; Li, J.B.; Su, Y.; Zhang, Y.; Wang, Y.Y.; Zhu, Y.Y. Genome-wide analysis of microsatellite sequence in seven filamentous fungi. Interdiscip. Sci. Comput. Life Sci. 2009, 1, 141–150. [Google Scholar] [CrossRef]
- Qian, J.; Xu, H.B.; Song, J.Y.; Xu, J.; Zhu, Y.J.; Chen, S.L. Genome-wide analysis of simple sequence repeats in the model medicinal mushroom Ganoderma lucidum. Gene 2013, 512, 331–336. [Google Scholar] [CrossRef]
- Cai, G.; Leadbetter, C.W.; Muehlbauer, M.F.; Molnar, T.J.; Hillman, B.I. Genome-wide microsatellite identification in the fungus Anisogramma anomala using Illumina sequencing and genome assembly. PLoS ONE 2013, 8, e82408. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, M.J.; Wang, H.; Wang, J.F.; Bao, D.P. Microsatellites in the genome of the edible mushroom, Volvariella volvacea. BioMed Res. Int. 2014, 2014, 281912. [Google Scholar] [CrossRef]
- Ellegren, H. Microsatellites: Simple sequences with complex evolution. Nat. Rev. Genet. 2004, 5, 435–445. [Google Scholar] [CrossRef]
- Katti, M.V.; Ranjekar, P.K.; Gupta, V.S. Differential distribution of simple sequence repeats in eukaryotic genome sequences. Mol. Biol. Evol. 2001, 18, 1161–1167. [Google Scholar] [CrossRef]
- Tóth, G.; Gáspári, Z.; Jurka, J. Microsatellites in different eukaryotic genomes: Survey and analysis. Genome Res. 2000, 10, 967–981. [Google Scholar] [CrossRef] [PubMed]
- Hancock, J.M. Simple sequences in a ‘minimal’ genome. Nat. Genet. 1996, 14, 14–15. [Google Scholar] [CrossRef] [PubMed]
- Burranboina, K.; Abraham, S.; Murugan, K. Genome wide identification and analysis of microsatellite repeats in the largest DNA viruses (Poxviridae Family): An insilico approach. Annu. Res. Rev. Biol. 2018, 22, 1–11. [Google Scholar] [CrossRef]
- Qi, W.H.; Jiang, X.M.; Du, L.M.; Xiao, G.S.; Hu, T.Z.; Yue, B.S.; Quan, Q.M. Genome-wide survey and analysis of microsatellite sequences in bovid species. PLoS ONE 2015, 10, e0133667. [Google Scholar] [CrossRef]
- Perinchery, G.; Nojima, D.; Goharderakhshan, R.; Tanaka, Y.; Alonzo, J.; Dahiya, R. Microsatellite instability of dinucleotide tandem repeat sequences is higher than trinucleotide, tetranucleotide and pentanucleotide repeat sequences in prostate cancer. Int. J. Oncol. 2000, 16, 1203–1212. [Google Scholar] [CrossRef] [PubMed]
- Kumari, R.; Wankhede, D.P.; Bajpai, A.; Maurya, A.; Prasad, K.; Gautam, D.; Rangan, P.; Latha, M.; Joseph, J.K.; Suma, A.; et al. Genome wide identification and characterization of microsatellite markers in black pepper (Piper nigrum): A valuable resource for boosting genomics applications. PLoS ONE 2019, 14, e0226002. [Google Scholar] [CrossRef] [PubMed]
- Ji, P.F.; Zhang, Y.; Li, C.; Zhao, Z.; Wang, J.; Li, J.T.; Xu, P.; Sun, X.W. High throughput mining and characterization of microsatellites from common carp genome. Int. J. Mol. Sci. 2012, 13, 9798–9807. [Google Scholar] [CrossRef]
- Iranawati, F.; Jung, H.; Chand, V.; Hurwood, D.A.; Mather, P.B. Analysis of genome survey sequences and SSR marker development for Siamese mud carp, Henicorhynchus siamensis, using 454 pyrosequencing. Int. J. Mol. Sci. 2012, 13, 10807–10827. [Google Scholar] [CrossRef]
- Mathew, D.; Anju, P.S.; Tom, A.; Johnson, N.; George, M.L.; Davis, S.P.; Ravisankar, V.; Asha, K.N. Genome-wide microsatellites and species specific markers in genus Phytophthora revealed through whole genome analysis. 3 Biotech 2020, 10, 442. [Google Scholar] [CrossRef]
- Zhang, X.J.; Yuan, J.B.; Sun, Y.M.; Li, S.H.; Gao, Y.; Yu, Y.; Liu, C.Z.; Wang, Q.C.; Lv, X.J.; Zhang, X.X.; et al. Penaeid shrimp genome provides insights into benthic adaptation and frequent molting. Nat. Commun. 2019, 10, 356. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.T.; Zhang, Y.J.; Qiao, L.; Chen, B. Comparative analyses of simple sequence repeats (SSRs) in 23 mosquito species genomes: Identification, characterization and distribution (Diptera: Culicidae). Insect Sci. 2019, 26, 607–619. [Google Scholar] [CrossRef]
- Jiang, Q.; Li, Q.; Yu, H.; Kong, L.F. Genome-wide analysis of simple sequence repeats in marine animals—A comparative approach. Mar. Biotechnol. 2014, 16, 604–619. [Google Scholar] [CrossRef]
- Lei, Y.; Zhou, Y.; Price, M.; Song, Z.B. Genome-wide characterization of microsatellite DNA in fishes: Survey and analysis of their abundance and frequency in genome-specific regions. BMC Genom. 2021, 22, 421. [Google Scholar] [CrossRef]
- Zhao, X.Y.; Tan, Z.Y.; Feng, H.P.; Yang, R.H.; Li, M.F.; Jiang, J.H.; Shen, G.L.; Yu, R.Q. Microsatellites in different Potyvirus genomes: Survey and analysis. Gene 2011, 488, 52–56. [Google Scholar] [CrossRef]
- Shan, Y.J.; Lu, C.Y.; Li, C.; Cheng, L.; Sun, X.W. Isolation and characterization of 57 novel polynucleotide microsatellites from yellow catfish (Pelteobagrus fulvidraco) genome for genetic analysis. Conser. Genet. Resour. 2014, 6, 73–77. [Google Scholar] [CrossRef]
- Wang, J.L.; Li, H.L.; Yin, S.; Hu, Z.H.; Ye, M.; Zhang, P.P. Distribution characteristics of whole genome microsatellite of Thammaconus modestus. J. Yantai Univ. 2022, 35, 285–293. (In Chinese) [Google Scholar]
- Takagi, M.; Sato, J.; Monbayashi, C.; Aoki, K.; Tsuji, T.; Hatanaka, H.; Takahashi, H.; Sakai, H. Evaluation of microsatellites identified in the tiger puffer Takifugu rubripes DNA database. Fish. Sci. 2003, 69, 1085–1095. [Google Scholar] [CrossRef]
- Wierdl, M.; Dominska, M.; Petes, T.D. Microsatellite instability in yeast: Dependence on the length of the microsatellite. Genetics 1997, 146, 769–779. [Google Scholar] [CrossRef] [PubMed]

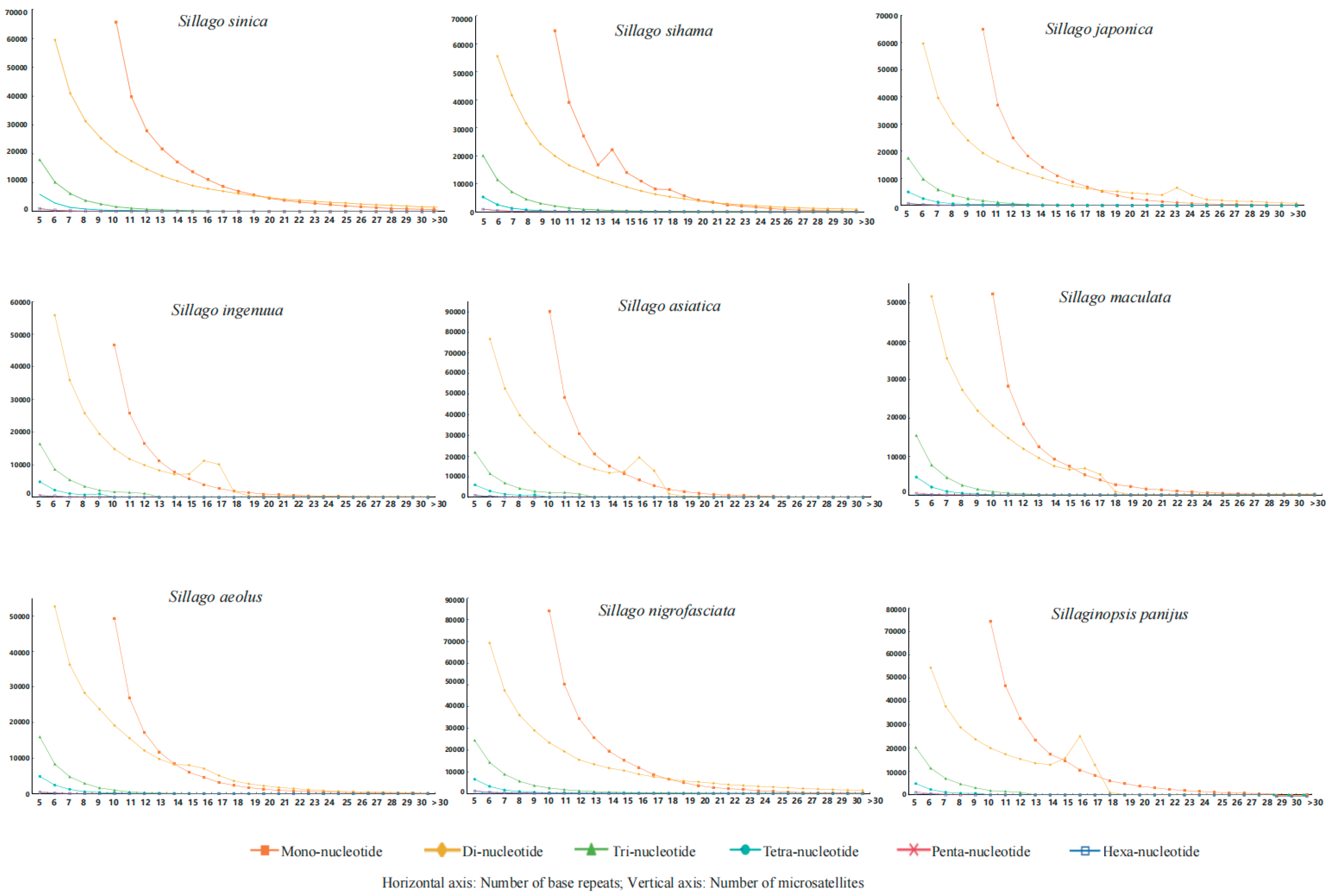
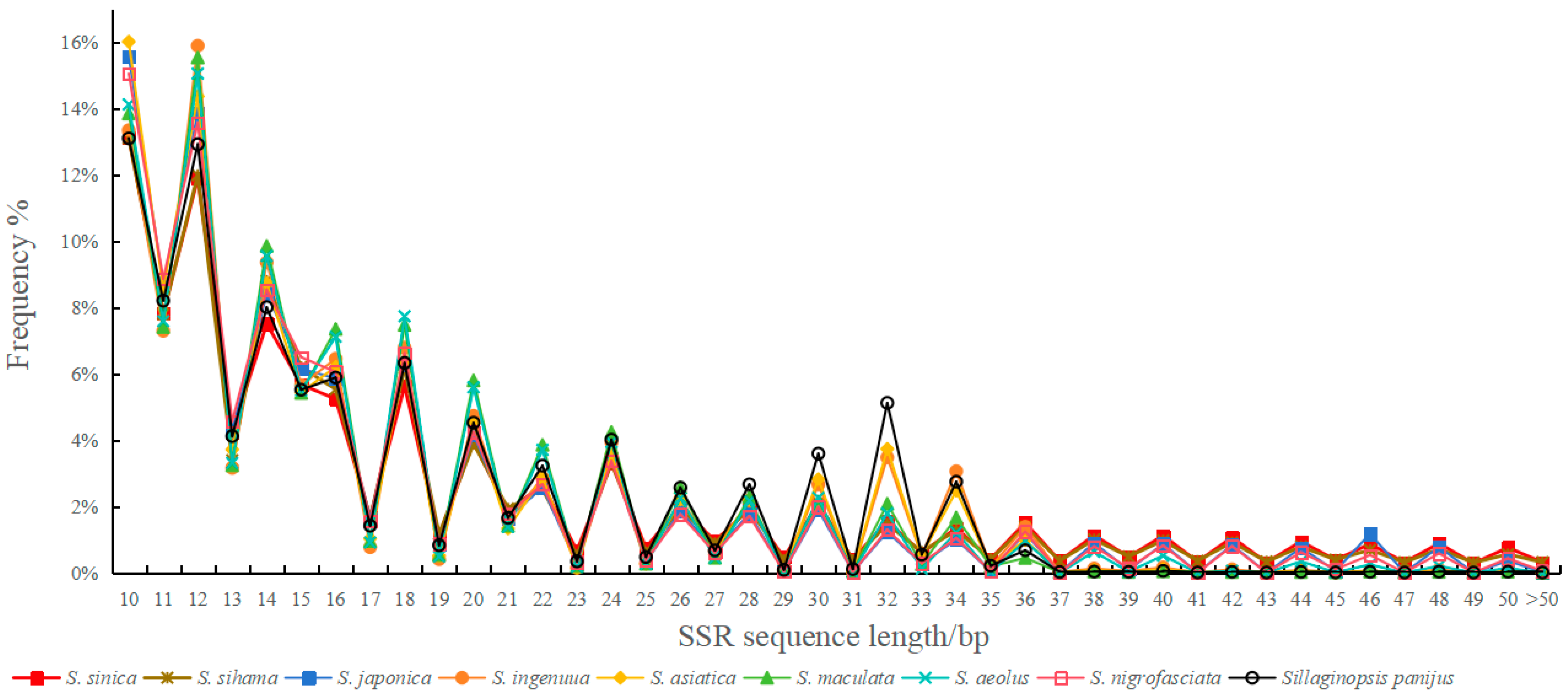
| Items | Genome Size/Mb | MC | NSPCF | TLM/bp | PGS | ADS/bp | RA/(loci/Mb) |
|---|---|---|---|---|---|---|---|
| S. sinica | 534 | 627,460 | 166,294 | 17,156,757 | 3.21% | 852 | 1174.36 |
| S. sihama | 522 | 599,804 | 153,808 | 16,005,572 | 3.07% | 869 | 1149.84 |
| S.japonica | 562 | 560,704 | 137,896 | 14,600,875 | 2.58% | 1009 | 990.78 |
| S. ingenuua | 554 | 409,623 | 63,238 | 9,139,678 | 1.65% | 1353 | 739.14 |
| S. asiatica | 701 | 657,299 | 113,405 | 14,552,136 | 2.07% | 1067 | 937.11 |
| S. maculata | 630 | 424,023 | 55,836 | 9,293,656 | 1.47% | 1485 | 673.52 |
| S. aeolus | 524 | 426,085 | 77,709 | 9,867,944 | 1.88% | 1230 | 812.57 |
| S. nigrofasciata | 633 | 723,688 | 172,203 | 19,649,687 | 3.11% | 874 | 1160.56 |
| S. panijus | 652 | 619,958 | 92,131 | 14,491,374 | 2.22% | 1051 | 951.57 |
| Items | NPM | NPD | NPTR | NPTE | NPP | NPH |
|---|---|---|---|---|---|---|
| S. sinica | 247,032 (39.37) | 316,071 (50.37) | 46,514 (7.41) | 13,640 (2.17) | 1805 (0.29) | 2398 (0.38) |
| S. sihama | 235,100 (39.20) | 294,271 (49.06) | 53,701 (8.95) | 12,318 (2.05) | 2205 (0.37) | 2209 (0.37) |
| S.japonica | 205,275 (36.61) | 294,781 (52.57) | 44,627 (7.96) | 12,711 (2.27) | 1673 (0.30) | 1637 (0.29) |
| S. ingenuua | 129,886 (31.71) | 226,368 (55.26) | 40,872 (9.98) | 10,171 (2.48) | 1320 (0.32) | 1006 (0.25) |
| S. asiatica | 247,906 (37.72) | 338,625 (51.52) | 54,205 (8.25) | 13,027 (1.98) | 2027 (0.31) | 1509 (0.23) |
| S. maculata | 152,822 (36.04) | 227,791 (53.72) | 33,905 (8.00) | 8462 (2.00) | 771 (0.18) | 272 (0.06) |
| S. aeolus | 136,660 (32.07) | 242,913 (57.01) | 35,527 (8.34) | 9701 (2.28) | 823 (0.19) | 461 (0.11) |
| S. nigrofasciata | 278,656 (38.50) | 355,441 (49.12) | 67,478 (9.23) | 16,280 (2.25) | 2947 (0.41) | 2886 (0.40) |
| S. panijus | 262,679 (42.37) | 291,764 (47.06) | 51,903 (8.37) | 9954 (1.61) | 2405 (0.39) | 1253 (0.20) |
| Items | Mono- Nucleotide | Di- Nucleotide | Tri- Nucleotide | Tetra- Nucleotide | Penta- Nucleotide | Hexa- Nucleotide |
|---|---|---|---|---|---|---|
| S. sinica | A/T (63.01) | AC/GT (79.62) | AGG/CCT (39.28) | AGGG/CCCT (13.50) | AAGCT /AGCTT (12.80) | ACACGC /CGTGTG (27.27) |
| S. sihama | A/T (72.32) | AC/GT (79.77) | AGG/CCT (39.37) | AGGG/CCCT (18.87) | AGCCG /CGGCT (11.88) | AACCCT /AGGGTT (37.89) |
| S.japonica | A/T (76.42) | AC/GT (82.19) | AGG/CCT (36.39) | AGGG/CCCT (14.48) | AGAGG /CCTCT (11.36) | ACCAGG /CCTGGT(20.04) |
| S. ingenuua | A/T (79.97) | AC/GT (77.21) | AGG/CCT (41.03) | ACAG/CTGT (19.88) | AAGCT/AGCTT (15.76) | ACCAGG /CCTGGT (26.54) |
| S. asiatica | A/T (75.82) | AC/GT (81.99) | AGG/CCT (43.09) | AGGG/CCCT (14.98) | AGCCG /CGGCT (11.30) | ACCAGG /CCTGGT (18.75) |
| S. maculata | A/T (76.53) | AC/GT (81.05) | AGG/CCT (41.65) | ACAG/CTGT (18.19) | AGAGG /CCTCT (12.58) | ACCAGG /CCTGGT (17.65) |
| S. aeolus | A/T (82.47) | AC/GT (81.80) | AGG/CCT (43.02) | ACAG/CTGT (22.67) | AGAGG /CCTCT (12.52) | ACACGC /CGTGTG (16.70) |
| S. nigrofasciata | A/T (74.30) | AC/GT (80.56) | AGG/CCT (38.74) | ACAG/CTGT (16.63) | AGCCG /CGGCT (13.27) | AACCCT /AGGGTT (25.95) |
| S. panijus | A/T (75.30) | AC/GT (81.04) | AGG/CCT (41.94) | ACAG/CTGT (15.16) | AATCT /AGATT (25.32) | ACCAGG /CCTGGT (12.21) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qu, Y.; Qu, C.; Tian, C.; Gao, T.; Zhang, Y. Distribution and Comparative Analysis of Genomic Microsatellites in Nine Species of Family Sillaginidae. Fishes 2025, 10, 536. https://doi.org/10.3390/fishes10110536
Qu Y, Qu C, Tian C, Gao T, Zhang Y. Distribution and Comparative Analysis of Genomic Microsatellites in Nine Species of Family Sillaginidae. Fishes. 2025; 10(11):536. https://doi.org/10.3390/fishes10110536
Chicago/Turabian StyleQu, Yinquan, Caihui Qu, Changxu Tian, Tianxiang Gao, and Yuan Zhang. 2025. "Distribution and Comparative Analysis of Genomic Microsatellites in Nine Species of Family Sillaginidae" Fishes 10, no. 11: 536. https://doi.org/10.3390/fishes10110536
APA StyleQu, Y., Qu, C., Tian, C., Gao, T., & Zhang, Y. (2025). Distribution and Comparative Analysis of Genomic Microsatellites in Nine Species of Family Sillaginidae. Fishes, 10(11), 536. https://doi.org/10.3390/fishes10110536








