Protocol to Establish Estrogen Receptor-Negative Heterozygous BRCA1 Organoids
Abstract
1. Introduction
2. Experimental Design
2.1. Cell Lines
- BRCA1.1 MCF10A—heterozygous BRCA1 mammary cell line, which has the 185delAG mutation (Founder mutation) in one allele.
- BRCA1.2 MCF10A—heterozygous BRCA1 mammary cell line, which has the R71G mutation (Arg to Gly change at codon 71) in one allele.
2.2. Materials
- Greiner Bio-One CELLSTAR cell culture multi-well plates for suspension cultures, 6-well (Fisher Scientific, Cat #07-000-646, Waltham, MA, USA)
- Atrazine (Sigma Aldrich, Cat #49085, St Louis, MI, USA)
- Cholera toxin (Sigma Aldrich, Cat #C-8052, St Louis, MI, USA)
- DMEM/F12 (ThermoFisher Scientific Cat #11330-032, Waltham, MA, USA)
- Epidermal growth factor (EGF, Peprotech, Cat# AF-100-15, Cranberry, NJ, USA)
- Ethylene glycol tetraacetic acid (EGTA-pH 8 Alfa Aesar, Cat #J60869, Lancashire, UK)
- Estrogen (β-estradiol, Sigma Aldrich, Cat #E2758, St Louis, MI, USA)
- Human embryonic stem cells (hESC) Matrigel (Corning, Cat #354277, Charlotte, NC, USA)
- Horse serum (ThermoFisher Scientific, Cat #16050-122, Waltham, MA, USA)
- Hydrocortisone (Sigma Aldrich, Cat #H088, St Louis, MI, USA)
- Insulin (Sigma Aldrich, Cat #I-1882, St Louis, MI, USA)
- Matrigel, growth factor reduced (Corning, Cat # 356231, Charlotte, NC, USA)
- Matrigel, organoid (Corning, Cat # 356255, Charlotte, NC, USA)
- 1 M magnesium chloride (MgCl2 Sigma Aldrich, Cat #M1028, St Louis, MI, USA)
- Paraformaldehyde (Merck, Cat #1004960700, Rahway, NJ, USA)
- Phosphate-buffered saline (Sigma, Cat #D8537, St Louis, MI, USA)
- Penicillin-streptomycin (ThermoFisher Scientific, Cat #15070063, Waltham, MA, USA)
- PIPES buffer (pH 6.8) (Alfa Aesar, Cat #J60300, Lancashire, UK)
- ProLong Gold antifade mountant with DAPI (ThermoFisher Scientific, Cat #P36935, Waltham, MA, USA)
- TrypLE (ThermoFisher Scientific, Cat #12604013, Waltham, MA, USA)
- Triton X-100 (Sigma Aldrich, Cat #X100, St Louis, MI, USA)
2.3. Equipment
- Biosafety cabinet (various manufacturers)
- 37 °C incubator with 5% CO2 (various manufacturers)
- Echo brightfield microscope
- Confocal microscope (various manufacturers)
3. Procedure
3.1. Cell Culture Maintenance
- Place complete growth medium in a 37 °C water bath to warm up.
- Add 9 mL of media (DMEM/F12 supplemented with 5% horse serum, 20 ng/mL epidermal growth factor, 10 μg/mL insulin, 0.5 μg/mL hydrocortisone, 0.1 μg/mL cholera toxin, and 1% penicillin-streptomycin) to a new 15 mL conical tube.
- Remove the cell lines from liquid nitrogen storage.
- Thaw the cell lines by swirling the tubes in a 37 °C water bath for 3–5 min. The tubes were kept at 37 °C until the suspension was completely thawed. Remove the tubes immediately.
- Gently mix the cells by tapping and transfer to a 15 mL tube.
- Gently mix by pipetting.
- Centrifuge the suspension at 125 g for 10 min.
- Aspirate the supernatant gently.
- Add 10 mL fresh medium and mix gently. Collect an aliquot for cell count.
- Plate the cells in pre-labeled T-75 flask at about 2.5 × 104 cells/cm2.
- Incubate at 37 °C in 5% CO2 incubator.
- Observe cells daily. Change media every 2 days.
- Confluent cells are observed after 4–5 days. Use these to generate organoids.
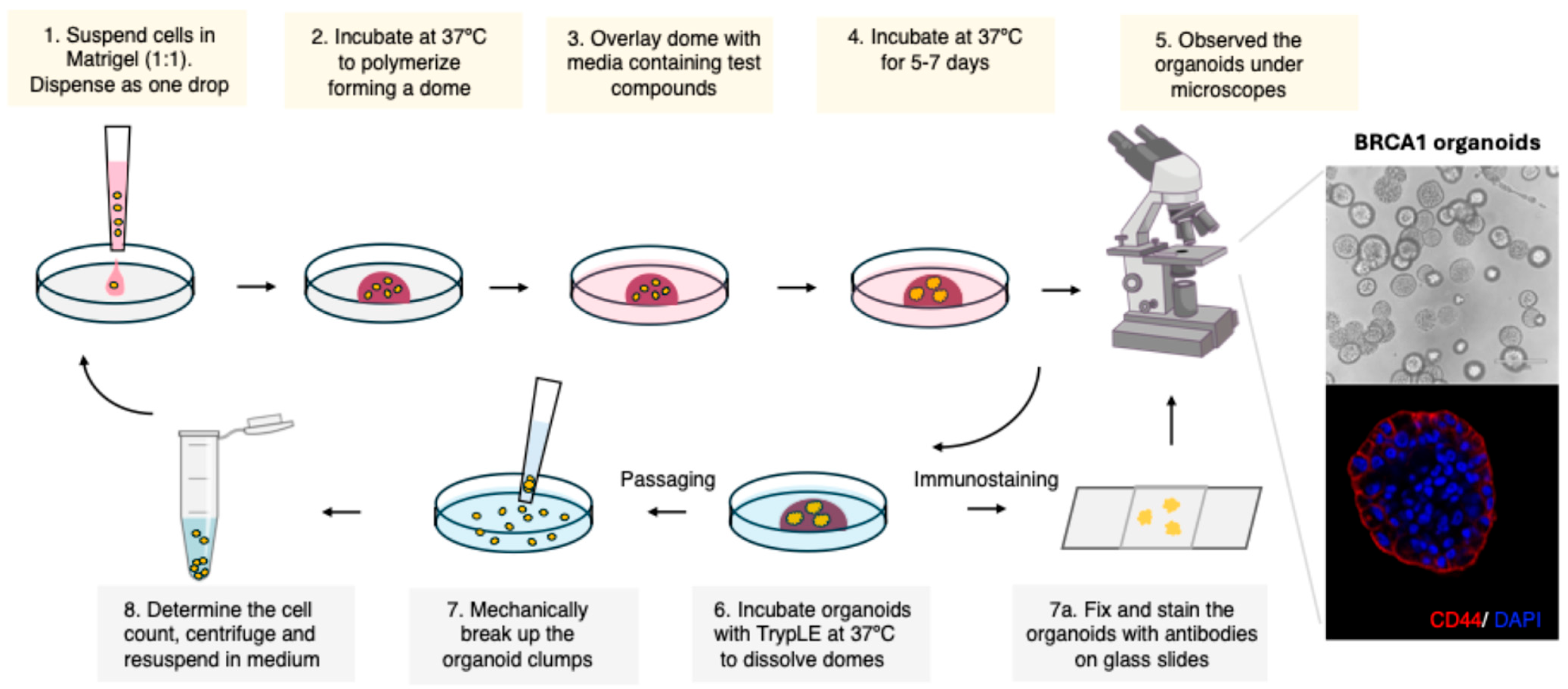
3.2. Initial Organoid Establishment (Figure 1)
- Thaw Matrigel on ice.
- Pre-warm low attachment 6-well plates at 37 °C.
- Harvest confluent cells and determine cell count.
- Adjust the cell count to 1 × 104 cells/drop (50 µL), i.e., 2 × 105 cells/mL. Keep the cell suspension on ice.
- Add an equal volume of cold Matrigel to the cell suspension (Media:Matrigel ratio 1:1)
- Mix well gently but avoid introducing any air bubbles.
- Gently drop 50 µL of the cell suspension (containing 0.5–1 × 104 cells for each well) to pre-warmed low attachment 6-well plates.
- Incubate the plates at 37 °C for 30 min to allow the Matrigel dome to solidify.
- Add 2 mL of pre-warmed media to the well. Ensure that the domes are solidified.
- Additions to the media can be made to check the effect of the compounds on organoid formation. In our study, we exposed the organoids to estradiol or atrazine and replaced the media every 2–3 days.
- Incubate the plate at 37 °C in 5% CO2 incubator.
- Observe the organoids and record the morphology.
- The images of the organoids were taken using an Echo brightfield microscope on days 0, 2, 4, 6, 8, and 10. The media was changed every 2–3 days.
- After 10–15 days, the organoids were harvested for passaging as well as for staining.
3.3. Passaging Organoids (This Step Is Performed Every 10–15 Days, Depending on the Growth Rate and Concentration of the Organoids per Well)
- Thaw Matrigel on ice.
- Pre-warm low attachment 6-well plates at 37 °C.
- Remove media from the organoid plate. Wash with cold PBS.
- Add cold TrypLE and incubate for 1-3 min at RT.
- Add 1 mL of cold media and use large-bore tips (You can cut a 1 mL tip) to break the clumps
- Do not over pipett. Ensure that you see cell fragments as well under the microscope.
- Add 5 mL of cold media and centrifuge.
- Remove the supernatant and add cold media; then keep the mixture on ice.
- Determine live cell count.
- Adjust the cell count to obtain 1 × 104 single cells or small fragments/drop (50 µL).
- Add the cells to cold Matrigel-Media mix (1:1 ratio).
- Mix well but avoid introducing any air bubbles.
- Gently drop 50 µL of the cell suspension (containing 0.5–1 × 104 cells for each well) to pre-warmed low attachment 6-well plates.
- Incubate the plates at 37 °C for 30 min to allow the Matrigel dome to solidify.
- Add 2 mL of pre-warmed media to the well. Ensure that the domes are solidified.
- Incubate the plate at 37 °C in 5% CO2 incubator.
- Observe the organoids and record the morphology.
- The images of the organoids were taken using an Echo brightfield microscope on days 0, 2, 4, 6, 8, and 10. The media was changed every 4 days.
- After 10–15 days, the organoids were harvested for passaging as well as for staining.
3.4. Immunostaining of Organoids
- Thaw Matrigel on ice.
- Pre-warm low attachment 6-well plates at 37 °C.
- Remove media from the organoid plate. Wash with cold PBS.
- With a large tip, pipette gently to dislodge the dome. Try to extract the organoids detached from Matrigel.
- Add 5 mL cold PBS and transfer to a 50 mL tube. Centrifuge.
- Remove supernatant, add cold PBS, and repeat the wash.
- Depending upon the experimental set-up, you can aliquot the organoids into individual tubes and proceed.
- Transfer the organoids to a microfuge tube (for ease of handling) and centrifuge to remove PBS.
- Fix the organoids with the PMTEF buffer for 30–40 min at RT (4% paraformaldehyde, 200 mM PIPES-pH 6.8, 200 mM MgCl2, 10 mM EGTA, 0.2% Triton X).
- Wash gently with PBS and block with 1% BSA and 0.3% Triton X in PBS (PBST) for 1 hr.
- Remove the blocking buffer by centrifugation and incubate overnight with primary antibody at 4 °C.
- The next day, remove the primary antibody solution and wash thrice with PBS.
- Following PBS wash, incubate the organoids with secondary antibody (Alexa 594; 1:1000 in PBST) at room temperature for 1 hr.
- Wash thrice with PBS and gently drop the organoids on charged slides.
- Mount with ProLong Gold antifade mountant with DAPI.
- The images can be taken using an immunofluorescence or confocal microscope. In the present study, an NL5 confocal microscope was used for imaging.
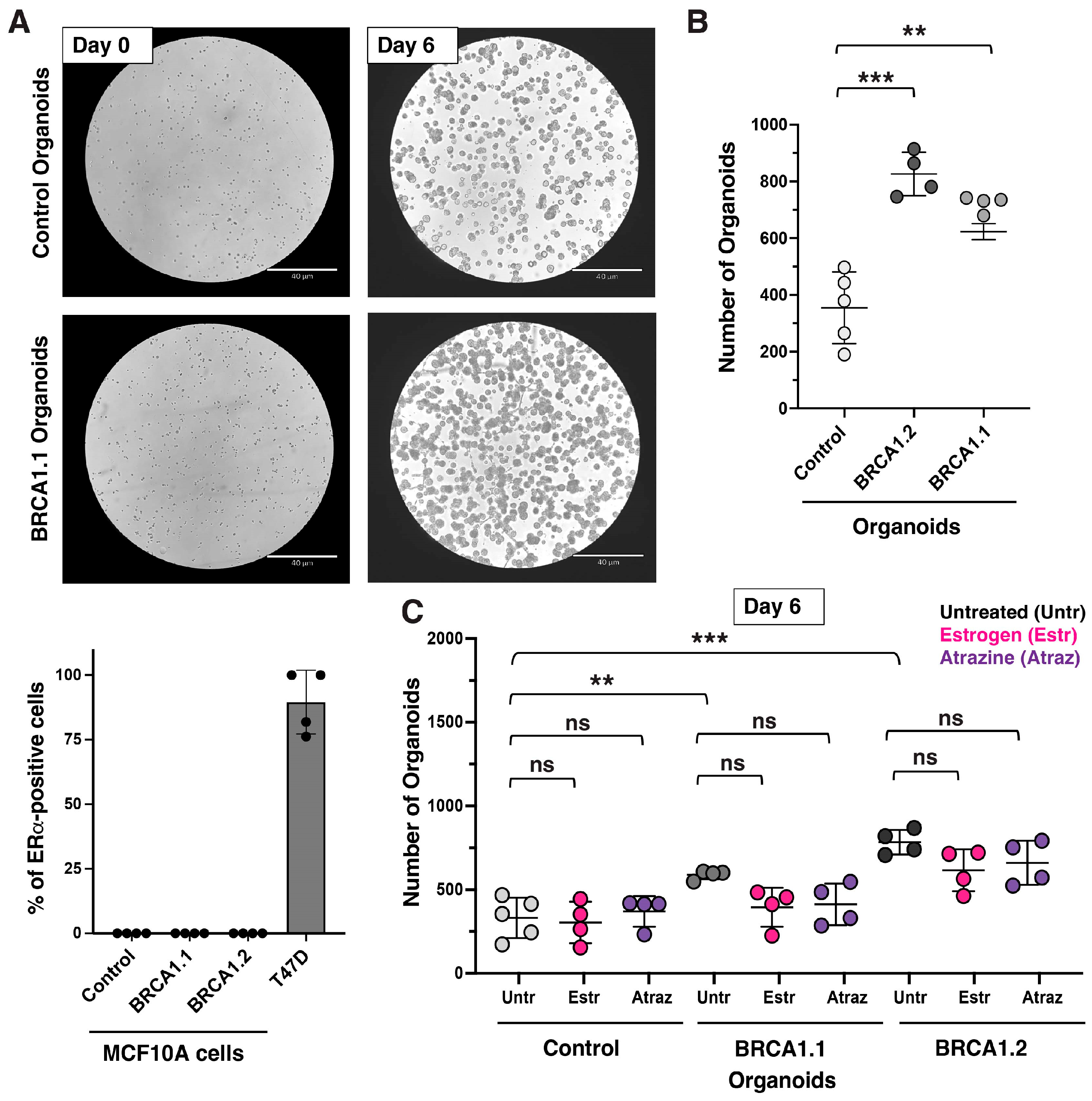
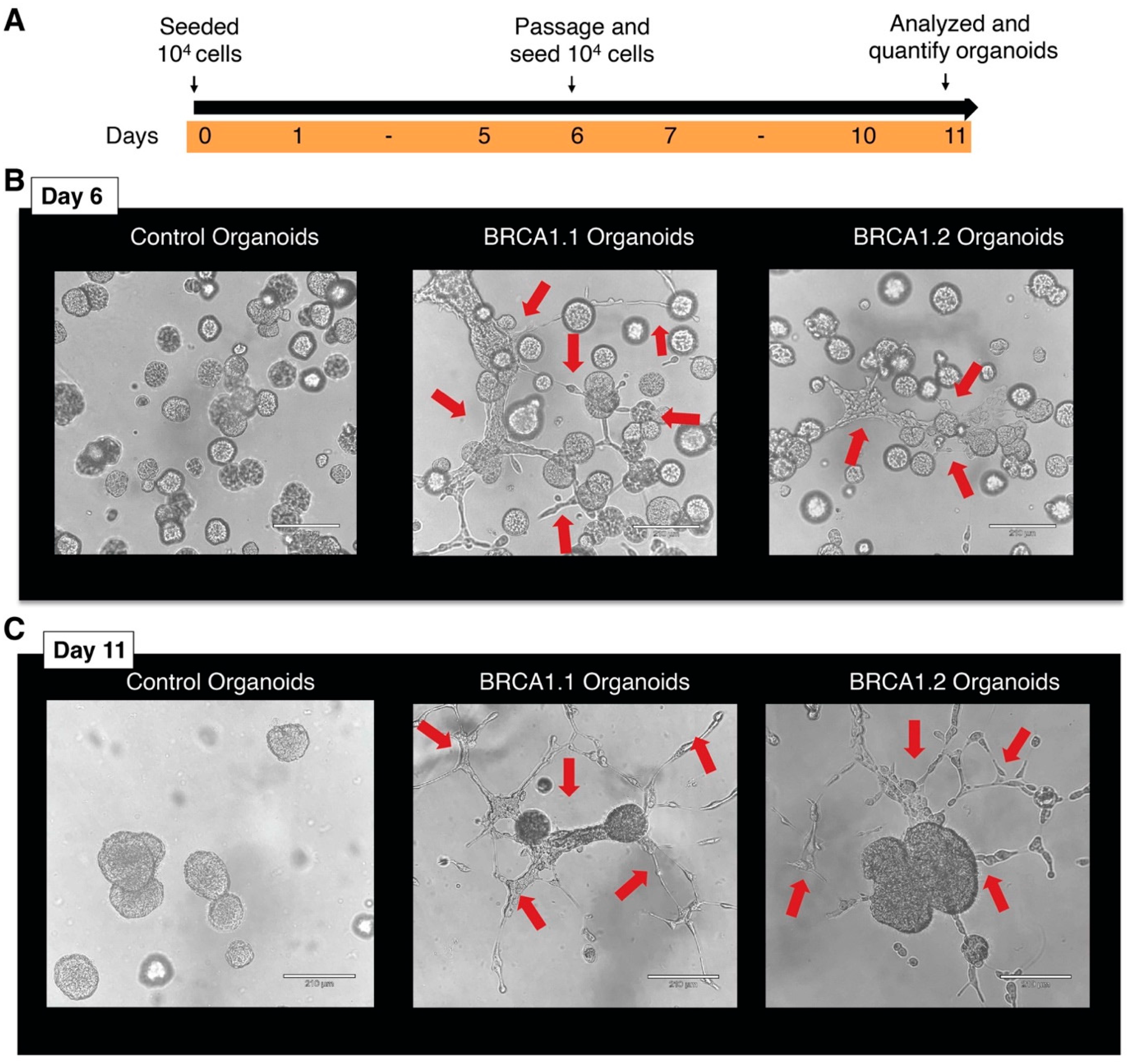
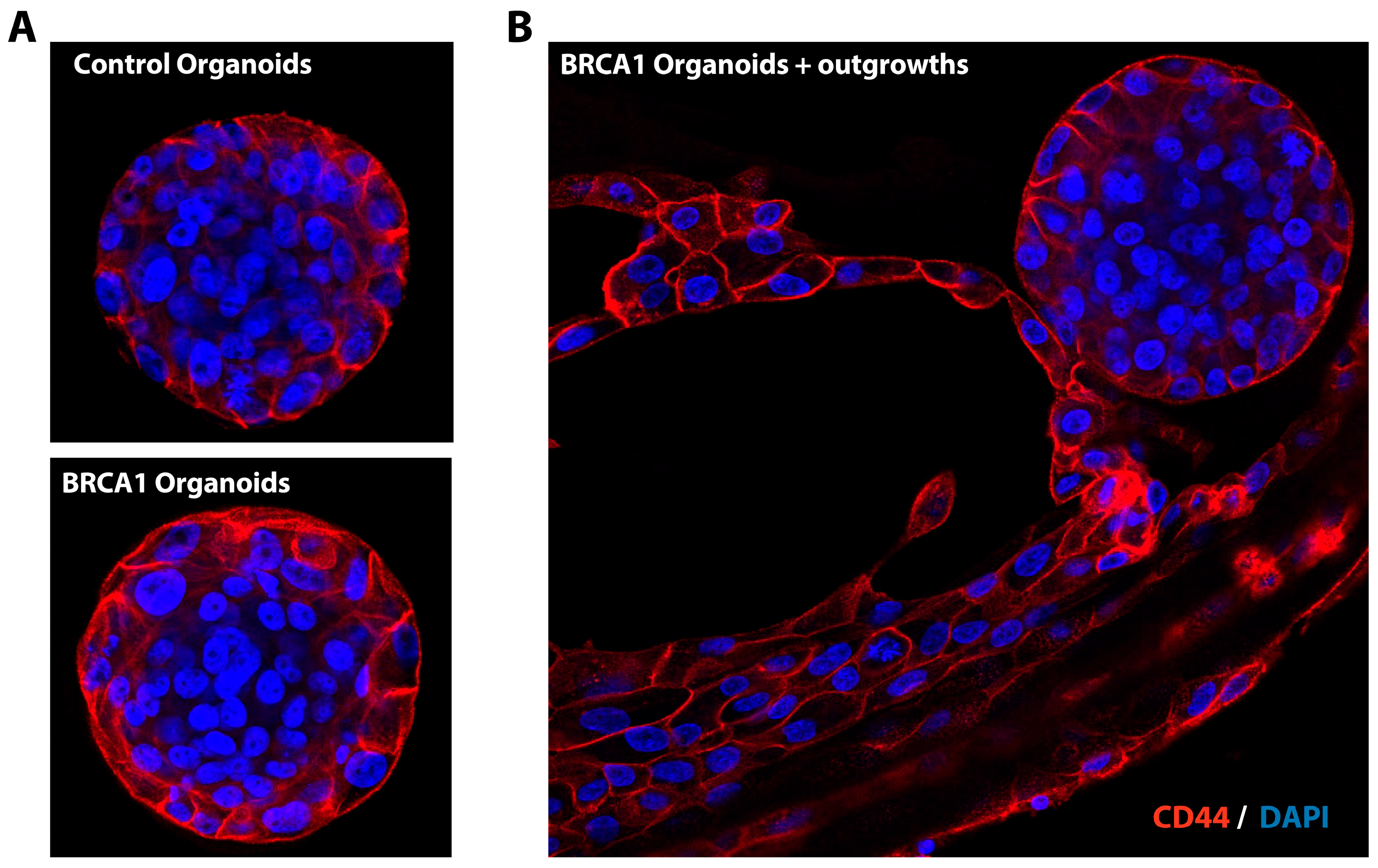
4. Expected Results and Discussion
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Welcsh, P.L.; King, M.C. BRCA1 and BRCA2 and the genetics of breast and ovarian cancer. Hum. Mol. Genet. 2001, 10, 705–713. [Google Scholar] [CrossRef] [PubMed]
- Osorio, A.; de la Hoya, M.; Rodriguez-Lopez, R.; Martinez-Ramirez, A.; Cazorla, A.; Granizo, J.J.; Esteller, M.; Rivas, C.; Caldes, T.; Benitez, J. Loss of heterozygosity analysis at the BRCA loci in tumor samples from patients with familial breast cancer. Int. J. Cancer 2002, 99, 305–309. [Google Scholar] [CrossRef] [PubMed]
- Maxwell, K.N.; Wubbenhorst, B.; Wenz, B.M.; De Sloover, D.; Pluta, J.; Emery, L.; Barrett, A.; Kraya, A.A.; Anastopoulos, I.N.; Yu, S.; et al. BRCA locus-specific loss of heterozygosity in germline BRCA1 and BRCA2 carriers. Nat. Commun. 2017, 8, 319. [Google Scholar] [CrossRef] [PubMed]
- Nik-Zainal, S.; Davies, H.; Staaf, J.; Ramakrishna, M.; Glodzik, D.; Zou, X.; Martincorena, I.; Alexandrov, L.B.; Martin, S.; Wedge, D.C.; et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature 2016, 534, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Pathania, S.; Bade, S.; Le Guillou, M.; Burke, K.; Reed, R.; Bowman-Colin, C.; Su, Y.; Ting, D.T.; Polyak, K.; Richardson, A.L.; et al. BRCA1 haploinsufficiency for replication stress suppression in primary cells. Nat. Commun. 2014, 5, 5496. [Google Scholar] [CrossRef]
- Deshpande, M.; Paniza, T.; Jalloul, N.; Nanjangud, G.; Twarowski, J.; Koren, A.; Zaninovic, N.; Zhan, Q.; Chadalavada, K.; Malkova, A.; et al. Error-prone repair of stalled replication forks drives mutagenesis and loss of heterozygosity in haploinsufficient BRCA1 cells. Mol. Cell. 2022, 82, 3781–3793. [Google Scholar] [CrossRef] [PubMed]
- Lakhani, S.R.; Van De Vijver, M.J.; Jacquemier, J.; Anderson, T.J.; Osin, P.P.; McGuffog, L.; Easton, D.F. The pathology of familial breast cancer: Predictive value of immunohistochemical markers β-Estradiol receptor, progesterone receptor, HER-2, and p53 in patients with mutations in BRCA1 and BRCA2. J. Clin. Oncol. 2002, 20, 2310–2318. [Google Scholar] [CrossRef] [PubMed]
- Fu, N.Y.; Nolan, E.; Lindeman, G.J.; Visvader, J.E. Stem Cells and the Differentiation Hierarchy in Mammary Gland Development. Physiol. Rev. 2020, 100, 489–523. [Google Scholar] [CrossRef] [PubMed]
- Nolan, E.; Vaillant, F.; Branstetter, D.; Pal, B.; Giner, G.; Whitehead, L.; Lok, S.W.; Mann, G.B.; Kathleen Cuningham Foundation Consortium for Research into Familial Breast Cancer (kConFab); Rohrbach, K.; et al. RANK ligand as a potential target for breast cancer prevention in BRCA1-mutation carriers. Nat. Med. 2016, 22, 933–939. [Google Scholar] [CrossRef]
- Subik, K.; Lee, J.F.; Baxter, L.; Strzepek, T.; Costello, D.; Crowley, P.; Xing, L.; Hung, M.C.; Bonfiglio, T.; Hicks, D.G.; et al. The Expression Patterns of ER, PR, HER2, CK5/6, EGFR, Ki-67 and AR by Immunohistochemical Analysis in Breast Cancer Cell Lines. Breast Cancer (Auckl). 2010, 4, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Qu, Y.; Han, B.; Yu, Y.; Yao, W.; Bose, S.; Karlan, B.Y.; Giuliano, A.E.; Cui, X. Evaluation of MCF10A as a Reliable Model for Normal Human Mammary Epithelial Cells. PLoS ONE 2015, 10, e0131285. [Google Scholar] [CrossRef]
- Konishi, H.; Mohseni, M.; Tamaki, A.; Garay, J.P.; Croessmann, S.; Karnan, S.; Ota, A.; Wong, H.Y.; Konishi, Y.; Karakas, B.; et al. Mutation of a single allele of the cancer susceptibility gene BRCA1 leads to genomic instability in human breast epithelial cells. Proc. Natl. Acad. Sci. USA 2011, 108, 17773–17778. [Google Scholar] [CrossRef] [PubMed]
- Rosenbluth, J.M.; Schackmann, R.C.J.; Gray, G.K.; Selfors, L.M.; Li, C.M.; Boedicker, M.; Kuiken, H.J.; Richardson, A.; Brock, J.; Garber, J.; et al. Organoid cultures from normal and cancer-prone human breast tissues preserve complex epithelial lineages. Nat. Commun. 2020, 11, 1711. [Google Scholar] [CrossRef] [PubMed]
- Ganz, H.M.; Buchmann, B.; Engelbrecht, L.K.; Jesinghaus, M.; Eichelberger, L.; Gabka, C.J.; Schmidt, G.P.; Muckenhuber, A.; Weichert, W.; Bausch, A.R.; et al. Generation of ductal organoids from normal mammary luminal cells reveals invasive potential. J. Pathol. 2021, 255, 451–463. [Google Scholar] [CrossRef] [PubMed]
- Fillmore, C.; Kuperwasser, C. Human breast cancer stem cell markers CD44 and CD24: Enriching for cells with functional properties in mice or in man? Breast Cancer Res. 2007, 9, 303. [Google Scholar] [CrossRef] [PubMed]
- Morath, I.; Hartmann, T.N.; Orian-Rousseau, V. CD44: More than a mere stem cell marker. Int. J. Biochem. Cell. Biol. 2016, 81, 166–173. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Deshpande, M.; Gerhardt, J. Protocol to Establish Estrogen Receptor-Negative Heterozygous BRCA1 Organoids. Methods Protoc. 2025, 8, 127. https://doi.org/10.3390/mps8060127
Deshpande M, Gerhardt J. Protocol to Establish Estrogen Receptor-Negative Heterozygous BRCA1 Organoids. Methods and Protocols. 2025; 8(6):127. https://doi.org/10.3390/mps8060127
Chicago/Turabian StyleDeshpande, Madhura, and Jeannine Gerhardt. 2025. "Protocol to Establish Estrogen Receptor-Negative Heterozygous BRCA1 Organoids" Methods and Protocols 8, no. 6: 127. https://doi.org/10.3390/mps8060127
APA StyleDeshpande, M., & Gerhardt, J. (2025). Protocol to Establish Estrogen Receptor-Negative Heterozygous BRCA1 Organoids. Methods and Protocols, 8(6), 127. https://doi.org/10.3390/mps8060127





