A Protocol for Modeling Human Bone Inflammation: Co-Culture of Osteoblasts and Osteoclasts Exposed to Different Inflammatory Microenvironments
Abstract
1. Introduction
2. Experimental Design
2.1. Materials
- Primary Human Osteoblasts (Promocell GmbH, Heidelberg, Germany, Cat. No.: C-12720)
- SupplementMix/Osteoblast Growth Medium (Promocell GmbH, Heidelberg, Germany, Cat. No.: C-39615)
- Buffy Coat (Our Blood Institute, Rantoul, IL, USA, Cat. No.: BC-8L)
- Cytica Ficoll-Paque™ Premium, 1.085 g/mL (Fisher Scientific Company LLC, Pittsburgh, PA, USA, Cat. No.: 45-001-755)
- DMEM (Dulbecco’s Modified Eagle’s Medium) (Corning, Manassas, VA, USA, Cat. No.: 10-013-CV)
- CD14 MicroBeads, human (Miltenyi Biotec, Bergisch Gladbach, Germany, Cat. No.: 130-050-201)
- Human TRANCE (RANKL) (soluble), Animal-Free Recombinant Protein (PeproTech, Secaucus, NJ, USA, Cat. No.: AF-310-01-10UG)
- Human M-CSF, Animal-Free Recombinant Protein (PeproTech, Secaucus, NJ, USA, Cat. No.: AF-300-25-10UG)
- Lipopolysaccharides from Escherichia coli O55:B5 (Sigma-Aldrich, Saint Louis, MO, USA, Cat. No.: L2880-10MG)
- Aa LPS was kindly provided by Dr. Keith Kirkwood (Department of Oral Biology, School of Dental Medicine, University at Buffalo, Buffalo, NY, USA) and characterized previously [19]
- Human IL-6 Recombinant Protein, PeproTech® (PeproTech, Secaucus, NJ, USA, Cat. No.: 200-06)
- Human TNF-alpha Recombinant Protein, PeproTech® (Peproech, Secaucus, NJ, USA, Cat. No.: 300-01A)
- Phosphate-Buffered Saline, 1× without calcium and magnesium, pH 7.4 ± 0.1 (Corning, Manassas, VA, USA, Cat. No.: 21-040-CM)
- Fetal Bovine Serum (Life Technologies Corporation, Carlsbard, CA, USA, Cat. No.: 26140-079)
- L-Glutamine 200 mM (100X) (Life Technologies Corporation, Carlsbard, CA, USA, Cat. No.: 25030-081)
- Penicillin Streptomycin Solution, 100× (Corning, Manassas, VA, USA, Cat. No.: 30-002-Cl)
- Trypsin-EDTA (0.25%), phenol red (Life Technologies Corporation, Carlsbard, CA, USA, Cat. No.: 25200056)
- UltraPure™ 0.5 M EDTA, pH 8.0 (Invitrogen, Grand Island, NY, USA, Cat. No.: 15575-038)
- ACK Lysing Buffer (Life Technologies Corporation, Carlsbard, CA, USA, Cat. No.: A10492-01)
- Dimethyl sulfoxide (Sigma-Aldrich, Saint Louis, MO, USA, Cat. No.: D8418)
- Trypan Blue Solution, 0.4% (Life Technologies Corporation, Carlsbard, CA, USA, Cat. No.: 15250061)
- Molecular Biology Grade Water (Corning, Manassas, VA, USA, Cat. No.: 46-000-Cl)
- Ethanol, Absolute (200 Proof), Molecular Biology Grade, Fisher BioReagents™ (ThermoFisher Scientific, Fair Lawn, NJ, USA, Cat. No.: BP2818-500)
- 2% Alizarin Red Stain (Lifeline Cell Technology, Frederick, MD, USA, Cat. No.: CM-0058)
- Leukocyte Acid Phosphatase (TRAP) Kit (Sigma-Aldrich, Saint Louis, MO, USA, Cat. No.: 387A-1KT)
- CellTiter 96 AQueous Cell Proliferation Assay Kit (Promega, Madison, WI, USA, Cat. No.: G3580)
- Invitrogen™ TRIzol™ Reagent (Invitrogen, Grand Island, NY, USA, Cat. No.: 15-596-026)
- miRNeasy Kit for miRNA Purification (Qiagen, Gaithersburg, MD, US, Cat. No.: 217084)
- High-capacity cDNA Reverse transcription kit (Life Technologies Corporation, Carlsbard, CA, USA, Cat. No.: 4374967)
- PowerUp™ SYBR™ Green Master Mix for qPCR (Life Technologies Corporation, Carlsbard, CA, USA, Cat. No.: A25743)
2.2. Equipment
- Cell Incubator (NuAire, Horsham, PA, USA, Cat. No.: NU-5810)
- Flow laminar cabinet (ThermoFisher Scientific, Fair Lawn, NJ, USA, Cat. No.: 1307)
- Cellometer Auto 1000 (Nexcelom Bioscience, Lawrence, MA, USA, Cat. No.: 9943)
- EVOS Fluorescence Microscope (EVOS, ThermoFisher Scientific, Fair Lawn, NJ, USA, Cat. No.: AMF4300)
- StepOne 7500 thermocycler (Life Technologies Corporation, Carlsbard, CA, USA, Cat. No.: 4376373)
- Eppendorf High Capacity Refrigerated (Eppendorf North America, Inc., Enfield, CT, USA, Cat. No.: 5810R)
- T75 Cell Culture Flask, Vented, Sterile, 100/CS (Thomas Scientific, Swedesboro, NJ, USA, Cat. No.: 21A00M453)
- 15 mL Centrifuge Tubes, Polypropylene, 50/Tray, 500/CS (Thomas Scientific, Swedesboro, NJ, USA, Cat. No.: 1159M36)
- 50 mL Centrifuge Tubes, Polypropylene, 25/tray, 500/CS (Thomas Scientific, Swedesboro, NJ, USA, Cat. No.: 1158R10)
- Corning® HTS Transwell® 96-well permeable supports (Sigma-Aldrich, Milwaukee, WI, USA, Cat. No.: CSL3381-1EA)
- LS Columns (Miltenyi Biotec, Bergisch Gladbach, Germany Cat. No.: 130-042-401)
- MidiMACS™ Starting Kit (LS) (Miltenyi Biotec, Bergisch Gladbach, Germany Cat. No.: 130-042-301)
2.3. Cell Viability Assay
2.4. Total RNA Isolation, cDNA Synthesis, and Quantitative PCR
2.5. Statistical Analysis
3. Procedure
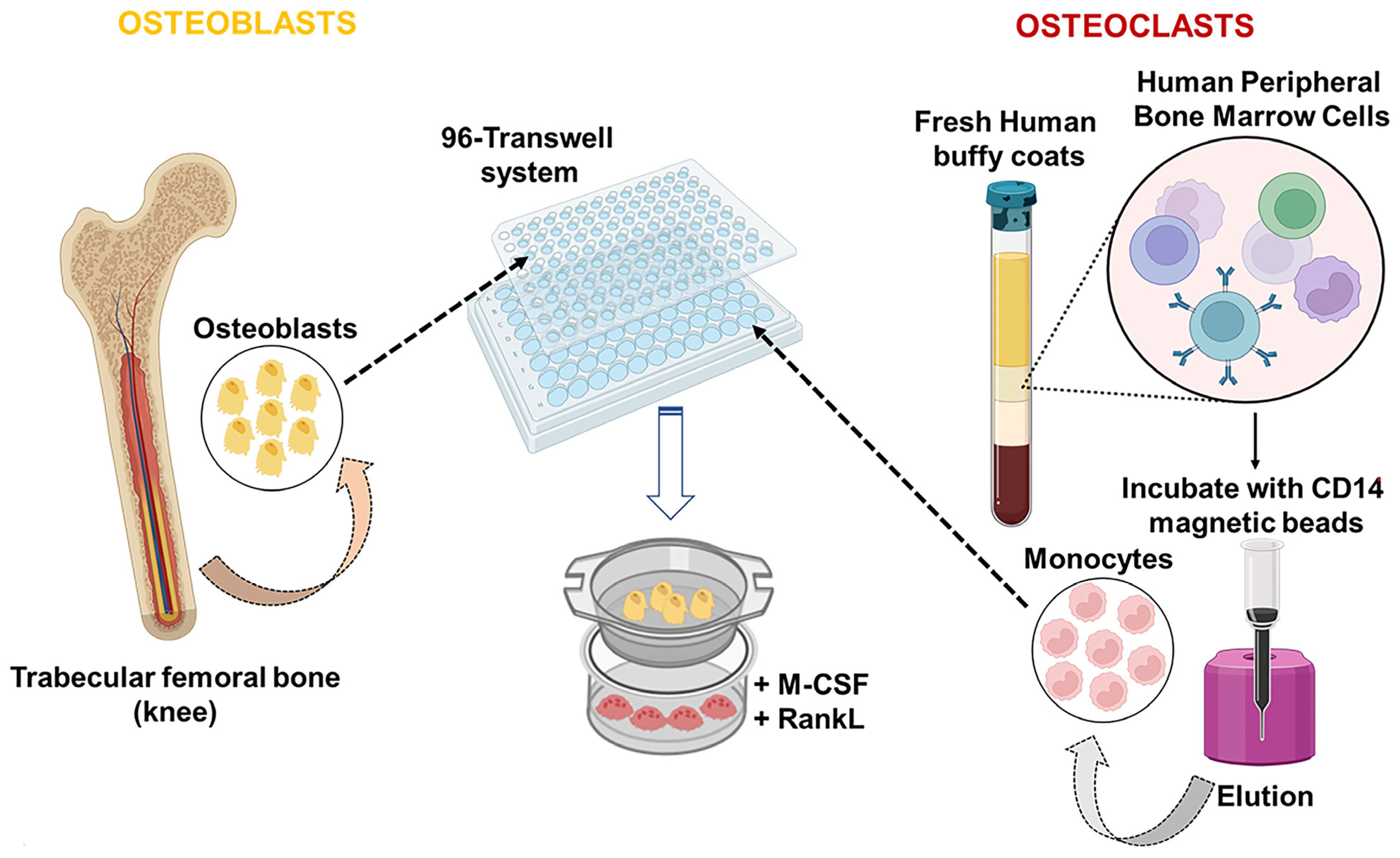
3.1. Peripheral Blood Mononuclear Cell Isolation Procedure
- Prepare 1× PBS solution containing 2 mM EDTA. Add 4 mL EDTA to 1 L of 1× PBS.
- Label 6 tubes/donor (50 mL centrifuge tubes) and add 20 mL of Ficoll-Hypaque.
- Fill a T75 cell culture flask with 150 mL of 1× PBS-EDTA solution (described above).
- 4.
- Dilute the buffy coat sample with an equal amount of 1× PBS (1:1 final dilution).
- 5.
- Gently pour 30 mL of the diluted buffy coat overlaying the Ficoll layer, being careful not to break the gradient layer.
- 6.
- Gradient centrifugation: centrifuge the tubes at 1329 rpm (RCF 300) for 30 min with breaks off to enable the PBMCs to stay on the gradient layer without being forced into the erythrocyte layer. Maintain the centrifuge temperature at 4 °C and keep the brakes off.
- 7.
- At the end of the first gradient centrifugation, PBMCs form a “ring” between Ficoll and plasma, as illustrated in Figure 1 Harvest each layer of PBMCs and transfer it into 3 new 50 mL tubes/donor. Then, add up to 50 mL of PBS + 5% FBS (500 μL/L of PBS).
- 8.
- Centrifuge the tubes at RT and 1500 rpm (RCF 382) for 10 min.
- 9.
- Discard the supernatant and harvest each PBMC pellet and transfer it into 3 new 15 mL tubes. Then, add up to 15 mL of PBS + 5% FBS.
- 10.
- Centrifuge the tubes at RT and 1500 rpm (RCF 382) for 5 min.
- 11.
- Discard the supernatant and resuspend the PBMC pellet in 1 mL ACK lysing buffer and transfer it into 2 new 15 mL tubes. Then, add up to 15 mL of ACK lysing buffer.
- 12.
- Centrifuge the tubes at RT and 1500 rpm (RCF 382) for 5 min.
- 13.
- Discard the supernatant and resuspend the PBMC pellet in 1 mL of PBS + 5% FBS and transfer it into 1 new 15 mL tube.
- 14.
- Repeat steps 12 and 13.
- 15.
- PBMCs are now ready for CD14+ sorting using the MidiMACS system.
3.2. Sorting of CD14+ from PBMCs Using the MidiMACS System and Osteoclast Culture
- Place the LS columns in the MidiMACS magnetic stand and add 4 mL of PBS + 5% FBS to hydrate the sorting columns.
- Add 120 μL of CD14+ beads to the PBMCs (previous step 15) and incubate at 4 °C for 30 min.
- Before starting the sorting, add up to 4 mL of PBS + 5% FBS to the PBMCs.
- Load 4 mL (1 mL at a time) in a column. CD14- monocytes will pass through the column, while CD14+ monocytes remain stuck in the column.
- For elution of the CD14+ monocytes, remove the columns from the MidiMACS and add 4 mL of PBS + 5% FBS. Use a plunger provided by the manufacturer to elute the CD14+ monocytes into new 15 mL tubes.
- Centrifuge the tubes at RT and 1500 rpm (RCF 382) for 5 min, keeping the brakes on.
- Discard the supernatant and resuspend the CD14+ pellet in 10 mL of an incomplete DMEM media.
- Seed the cells at the bottom of the 96 transwell at a density of 50,000 cells/well.
- After 2 h, replace the media with complete DMEM media containing 50 ng/mL of Monocyte Colony Stimulant Factor (MSCF) and 50 ng/mL of Receptor Activator of NF-κB Ligand (RANKL).
- Refresh the complete DMEM media plus the components MSCF and RANKL at the concentrations described above every 2 days for 5 days.
3.3. Osteoblast Culture
- Fill a 50 mL tube with 50 mL of complete PromoCell Growth Medium. Place the tube in an incubator (37 °C, 5% CO2) for 30 min.
- Remove the cryovial from the liquid nitrogen container and immediately immerse the vial in a water bath (37 °C) up to the height of the screw cap for 2 min. Ensure that no water enters the thread of the screw cap.
- Thoroughly rinse the cryovial with 70% ethanol under a laminar flow bench. Open the vial and transfer the cells to a 15 mL tube containing the prewarmed medium from step 1.
- Centrifuge the tubes at RT and 1500 rpm (RCF 382) for 5 min.
- Discard supernatant and resuspend the cell pellet with 1 mL of prewarmed medium from step 1 and place the resuspended cells into the T75 cell culture flask. Add up to 10 mL of prewarmed medium from step 1.
- Place the T75 cell culture flask containing the OB in an incubator (37 °C, 5% CO2) for cell attachment. Replace the medium every two days. The cells should be subcultured once they have reached 70–90% confluency.
- Aspirate the medium from the T75 culture cell flask, wash with 5 mL of PBS, and add 2 mL of Trypsin/EDTA for a few seconds.
- Remove the Trysin/EDTA and place the T75 cell culture flask in an incubator for cell detachment for 5 min.
- Collect the OB with 10 mL of complete medium to neutralize the leftover Trypsin/EDTA and centrifuge the tubes at RT and 1500 rpm (RCF 382) for 5 min.
- Discard the supernatant and resuspend the cell pellet with 1 mL of fresh complete media and count the cells using a Cellometer.
- OB were plated on the transwell insert at a density of 2500 cells/well. Replace every two days for 5 days.
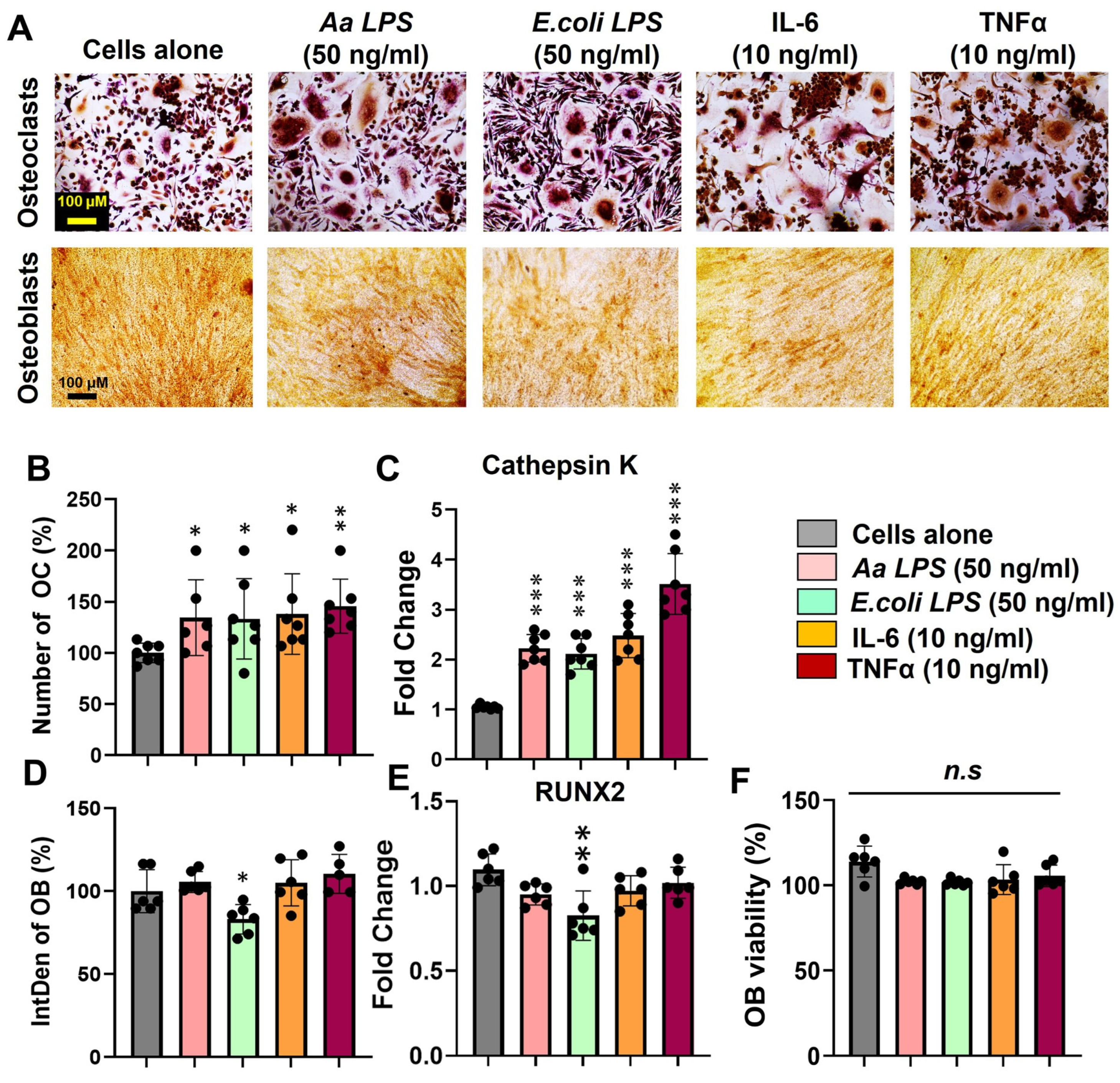
3.4. Assembly of OB/CD14+ Indirect Co-Culture and Phenotypic Evaluation of OB and OC Treated with Aa LPS, E. coli LPS, IL-6, and TNF-α
- Remove the medium and wash the cells with PBS.
- Fix the cells with absolute ethanol for 30 min at room temperature. Ethanol was removed, and wells were air-dried.
- 2% Alizarin Red S solution was added per well and incubated for 15 min. Wells were rinsed three times with distilled water and allowed to dry before imaging.
- Air dry and evaluate microscopically.
- Remove the medium and wash the cells with PBS.
- Fix the OC with Fixative Solution for 30 s. Rinse thoroughly in deionized water: Do not allow slides to dry.
- Prepare 2 test tubes and add 0.111 mL Fast Garnet GBC Base Solution and 0.111 mL Sodium Nitrite Solution. Mix by gentle inversion for 30 s.
- Prepare 2 test tubes (A and B) and add:
- Tubes A and B:
- -
- 10 mL of deionized water prewarmed to 37 °C
- -
- 0.222 mL of Diazotized Fast Garnet GCB Solution (Step 3)
- -
- 0.111 mL of Naphthol AS-Bl Phosphate Solution
- -
- 0.444 mL of Acetate Solution
- -
- 0.222 mL of Tartrate Solution (Only in tube B)
- Incubate for 1 h in 37 °C protected from light.
- Rinse the wells in deionized water, then counterstain for 2 min in Hematoxylin Solution, Gill No. 3.
- Rinse for several minutes in alkaline tap water to blue nuclei.
- Air dry and evaluate microscopically.
4. Expected Results
Enhanced Osteoclast Differentiation and Activity Driven by Periodontal Pathogens and Proinflammatory Cytokines
5. Strengths and Limitations
6. Discussion and Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
Abbreviations
| 2D | Two-dimensional |
| 3D | Three-dimensional |
| Aa LPS | Aggregatibacter actinomycetemcomitans lipopolysaccharide |
| ACK | Ammonium-Chloride-Potassium |
| E. coli LPS | Escherichia coli lipopolysaccharide |
| EDTA | Ethylenediaminetetraacetic acid |
| IL-6 | Interleukin-6 (IL-6) |
| IntDen | Integrated Density |
| MSCF | Monocyte Colony Stimulant Factor |
| OB | Osteoblast |
| OC | Osteoclast |
| PBMC | Peripheral blood mononuclear cells |
| PBS | Phosphate-Buffered Saline |
| RANKL | Receptor Activator of NF-κB Ligand |
| TNF-α | Tumor necrosis factor-alpha |
| TLR4 | Toll-Like Receptor 4 |
| TRAP | Tartrate-resistant acid phosphatase |
References
- Chen, X.; Wang, Z.; Duan, N.; Zhu, G.; Schwarz, E.M.; Xie, C. Osteoblast-Osteoclast Interactions. Connect. Tissue Res. 2018, 59, 99–107. [Google Scholar] [CrossRef]
- Tamma, R.; Zallone, A. Osteoblast and osteoclast crosstalks: From OAF to Ephrin. Inflamm. Allergy Drug Argets 2012, 11, 196–200. [Google Scholar] [CrossRef]
- Kim, J.M.; Lin, C.; Stavre, Z.; Greenblatt, M.B.; Shim, J.H. Osteoblast-Osteoclast Communication and Bone Homeostasis. Cells 2020, 9, 2073. [Google Scholar] [CrossRef] [PubMed]
- Matsuo, K.; Irie, N. Osteoclast-osteoblast communication. Arch. Biochem. Biophys. 2008, 473, 201–209. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, Z.; Goultschin, J.; Dean, D.D.; Boyan, B.D. Mechanisms of alveolar bone destruction in periodontitis. Periodontology 2000 1997, 14, 158–172. [Google Scholar] [CrossRef] [PubMed]
- Hienz, S.A.; Paliwal, S.; Ivanovski, S. Mechanisms of Bone Resorption in Periodontitis. J. Immunol. Res. 2015, 2015, 615486. [Google Scholar] [CrossRef]
- Könönen, E.; Gursoy, M.; Gursoy, U.K. Periodontitis: A Multifaceted Disease of Tooth-Supporting Tissues. J. Clin. Med. 2019, 8, 1135. [Google Scholar] [CrossRef]
- Valverde, A.; George, A.; Nares, S.; Naqvi, A.R. Emerging Therapeutic Strategies Targeting Bone Signaling Pathways in Periodontitis. J. Periodontal Res. 2024, 60, 101–120. [Google Scholar] [CrossRef]
- Mukherjee, P.; Roy, S.; Ghosh, D.; Nandi, S.K. Role of animal models in biomedical research: A review. Lab. Anim. Res. 2022, 1, 18. [Google Scholar] [CrossRef]
- Marshall, L.J.; Bailey, J.; Cassotta, M.; Herrmann, K.; Pistollato, F. Poor Translatability of Biomedical Research Using Animals—A Narrative Review. Altern. Lab. Anim. 2023, 51, 102–135. [Google Scholar] [CrossRef]
- Sieberath, A.; Della Bella, E.; Ferreira, A.M.; Gentile, P.; Eglin, D.; Dalgarno, K. A Comparison of Osteoblast and Osteoclast In Vitro Co-Culture Models and Their Translation for Preclinical Drug Testing Applications. Int. J. Mol. Sci. 2020, 21, 912. [Google Scholar] [CrossRef]
- Jolly, J.J.; Chin, K.Y.; Farhana, M.F.N.; Alias, E.; Chua, K.H.; Hasan, W.N.W.; Ima-Nirwana, S. Optimization of the Static Human Osteoblast/Osteoclast Co-culture System. Iran. J. Med. Sci. 2018, 43, 208–213. [Google Scholar] [PubMed]
- Borciani, G.; Montalbano, G.; Baldini, N.; Cerqueni, G.; Vitale-Brovarone, C.; Ciapetti, G. Co-culture systems of osteoblasts and osteoclasts: Simulating in vitro bone remodeling in regenerative approaches. Acta Biomater. 2020, 108, 22–45. [Google Scholar] [CrossRef] [PubMed]
- Steller, D.; Scheibert, A.; Sturmheit, T.; Hakim, S.G. Establishment and validation of an in vitro co-culture model for oral cell lines using human PBMC-derived osteoclasts, osteoblasts, fibroblasts and keratinocytes. Sci. Rep. 2020, 10, 16861. [Google Scholar] [CrossRef] [PubMed]
- Borciani, G.; Montalbano, G.; Baldini, N.; Vitale-Brovarone, C.; Ciapetti, G. Protocol of Co-Culture of Human Osteoblasts and Osteoclasts to Test Biomaterials for Bone Tissue Engineering. Methods Protoc. 2022, 5, 8. [Google Scholar] [CrossRef]
- Kinane, D.F.; Stathopoulou, P.G.; Papapanou, P.N. Periodontal diseases. Nat. Rev. Dis. Primers 2017, 3, 17038. [Google Scholar] [CrossRef]
- Nanci, A.; Bosshardt, D.D. Structure of periodontal tissues in health and disease. Periodontology 2000 2006, 40, 11–28. [Google Scholar] [CrossRef]
- Slots, J. Periodontology: Past, present, perspectives. Periodontology 2000 2013, 62, 7–19. [Google Scholar] [CrossRef]
- Naqvi, A.R.; Fordham, J.B.; Khan, A.; Nares, S. MicroRNAs responsive to Aggregatibacter actinomycetemcomitans and Porphyromonas gingivalis LPS modulate expression of genes regulating innate immunity in human macrophages. Innate Immun. 2014, 20, 540–551. [Google Scholar] [CrossRef]
- Makkar, H.; Sriram, G. Advances in modeling periodontal host-microbe interactions: Insights from organotypic and organ-on-chip systems. Lab Chip 2025, 25, 1342–1371. [Google Scholar] [CrossRef]
- Mountcastle, S.E.; Cox, S.C.; Sammons, R.L.; Jabbari, S.; Shelton, R.M.; Kuehne, S.A. A review of co-culture models to study the oral microenvironment and disease. J. Oral Microbiol. 2020, 12, 1773122. [Google Scholar] [CrossRef]
- Wang, C.; Xu, T.; Seneviratne, C.J.; Ong, L.J.Y.; Zhou, Y. Modelling periodontitis in vitro: Engineering strategies and biofilm model development. Front. Biomater. Sci. 2024, 3, 1380153. [Google Scholar] [CrossRef]
- Florencio-Silva, R.; Sasso, G.R.; Sasso-Cerri, E.; Simões, M.J.; Cerri, P.S. Biology of Bone Tissue: Structure, Function, and Factors That Influence Bone Cells. Biomed Res. Int. 2015, 2015, 421746. [Google Scholar] [CrossRef]
- Tsourdi, E.; Zillikens, M.C.; Meier, C.; Body, J.J.; Gonzalez Rodriguez, E.; Anastasilakis, A.D.; Abrahamsen, B.; McCloskey, E.; Hofbauer, L.C.; Guañabens, N.; et al. Fracture risk and management of discontinuation of denosumab therapy: A systematic review and position statement by ECTS. J. Clin. Endocrinol. Metab. 2020, 106, 264–281. [Google Scholar] [CrossRef] [PubMed]
- Galán-Díez, M.; Cuesta-Domínguez, Á.; Kousteni, S. The Bone Marrow Microenvironment in Health and Myeloid Malignancy. Cold Spring Harb. Perspect. Med. 2018, 8, a031328. [Google Scholar] [CrossRef] [PubMed]
- Verdugo-Avello, F.; Wychowaniec, J.K.; Villacis-Aguirre, C.A.; D’Este, M.; Toledo, J.R. Bone microphysiological models for biomedical research. Lab Chip 2025, 25, 806–836. [Google Scholar] [CrossRef] [PubMed]
- Boyce, B.F.; Xing, L. Functions of RANKL/RANK/OPG in bone modeling and remodeling. Arch. Biochem. Biophys. 2008, 473, 139–146. [Google Scholar] [CrossRef]
- Tatsumi, S.; Ishii, K.; Amizuka, N.; Li, M.; Kobayashi, T.; Kohno, K.; Ito, M.; Takeshita, S.; Ikeda, K. Targeted ablation of osteocytes induces osteoporosis with defective mechanotransduction. Cell Metab. 2007, 5, 464–475. [Google Scholar] [CrossRef]
- Lorenzo, J.; Horowitz, M.; Choi, Y. Osteoimmunology: Interactions of the bone and immune system. Endocr. Rev. 2008, 29, 403–440. [Google Scholar] [CrossRef]
- Langer, R.; Tirrell, D.A. Designing materials for biology and medicine. Nature 2004, 428, 487–492. [Google Scholar] [CrossRef]
- Lutolf, M.P.; Hubbell, J.A. Synthetic biomaterials as instructive extracellular microenvironments for morphogenesis in tissue engineering. Nat. Biotechnol. 2005, 23, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Ham, J.; Lever, L.; Fox, M.; Reagan, M.R. In Vitro 3D Cultures to Reproduce the Bone Marrow Niche. JBMR Plus 2019, 3, e10228. [Google Scholar] [CrossRef] [PubMed]
- Remmers, S.J.A.; de Wildt, B.W.M.; Vis, M.A.M.; Spaander, E.S.R.; de Vries, R.B.M.; Ito, K.; Hofmann, S. Osteoblast-osteoclast co-cultures: A systematic review and map of available literature. PLoS ONE 2021, 16, e0257724. [Google Scholar] [CrossRef] [PubMed]
- Owen, R.; Reilly, G.C. In vitro Models of Bone Remodeling and Associated Disorders. Front. Bioeng. Biotechnol. 2018, 6, 134. [Google Scholar] [CrossRef]
- Alonso-Pérez, A.; Franco-Trepat, E.; Guillán-Fresco, M.; Jorge-Mora, A.; López, V.; Pino, J.; Gualillo, O.; Gómez, R. Role of Toll-Like Receptor 4 on Osteoblast Metabolism and Function. Front. Physiol. 2018, 9, 504. [Google Scholar] [CrossRef]
- Komori, T. Regulation of Proliferation, Differentiation and Functions of Osteoblasts by Runx2. Int. J. Mol. Sci. 2019, 20, 1694. [Google Scholar] [CrossRef]
- Chan, W.C.W.; Tan, Z.; To, M.K.T.; Chan, D. Regulation and Role of Transcription Factors in Osteogenesis. Int. J. Mol. Sci. 2021, 22, 5445. [Google Scholar] [CrossRef]
- Valverde, A.; Naqvi, R.A.; Chen, Y.; Moshaverinia, A.; George, A.; Shukla, D.; Martinez, G.; Chapa, G.; Nares, S.; Naqvi, A.R. Herpesvirus Simplex Virus-1 Exploits Inflammation to Infect Periodontal Stem Cells and Disrupt Lineage Commitment. J. Periodontal Res. 2025. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Valverde, A.; Naqvi, A.R. A Protocol for Modeling Human Bone Inflammation: Co-Culture of Osteoblasts and Osteoclasts Exposed to Different Inflammatory Microenvironments. Methods Protoc. 2025, 8, 97. https://doi.org/10.3390/mps8050097
Valverde A, Naqvi AR. A Protocol for Modeling Human Bone Inflammation: Co-Culture of Osteoblasts and Osteoclasts Exposed to Different Inflammatory Microenvironments. Methods and Protocols. 2025; 8(5):97. https://doi.org/10.3390/mps8050097
Chicago/Turabian StyleValverde, Araceli, and Afsar Raza Naqvi. 2025. "A Protocol for Modeling Human Bone Inflammation: Co-Culture of Osteoblasts and Osteoclasts Exposed to Different Inflammatory Microenvironments" Methods and Protocols 8, no. 5: 97. https://doi.org/10.3390/mps8050097
APA StyleValverde, A., & Naqvi, A. R. (2025). A Protocol for Modeling Human Bone Inflammation: Co-Culture of Osteoblasts and Osteoclasts Exposed to Different Inflammatory Microenvironments. Methods and Protocols, 8(5), 97. https://doi.org/10.3390/mps8050097





