A Six-Step Protocol for Monitoring Antimicrobial Resistance Trends Using WHONET and R: Real-World Application and R Code Integration
Abstract
1. Introduction
1.1. Antimicrobial Resistance as a Global Health Issue
1.2. The WHOnet Software
- Enhancing the local use of laboratory data for guiding therapy, assisting infection control, characterizing resistance epidemiology and identifying laboratory testing errors.
- Promoting collaboration in surveillance activities through the exchange of data.
1.3. R Software
1.4. Purpose of This Paper
2. Experimental Design
Materials and Equipment
- Any data management software (for this paper, we used Microsoft Excel® V.2504, but any other software with similar functionalities can be used effectively);
- WHOnet software (WHOnet V.25.04.25, freeware, available from WHO) [31];
- BacLink software (BacLink V.25.04.25, freeware, is usually downloaded and installed automatically with WHOnet; it can also be downloaded from WHO) [31];
- R software (R version 4.4.0 and R-Studio 2025.05.0 Build 496, freeware, available from the R Foundation and Posit® website) [32].
3. Procedure
- Data extraction from microbiology laboratory software;
- Data import with BacLink;
- Configuration and import of data in WHOnet;
- Data analysis in WHOnet;
- Preparation for analysis in R;
- Data Analysis in R.
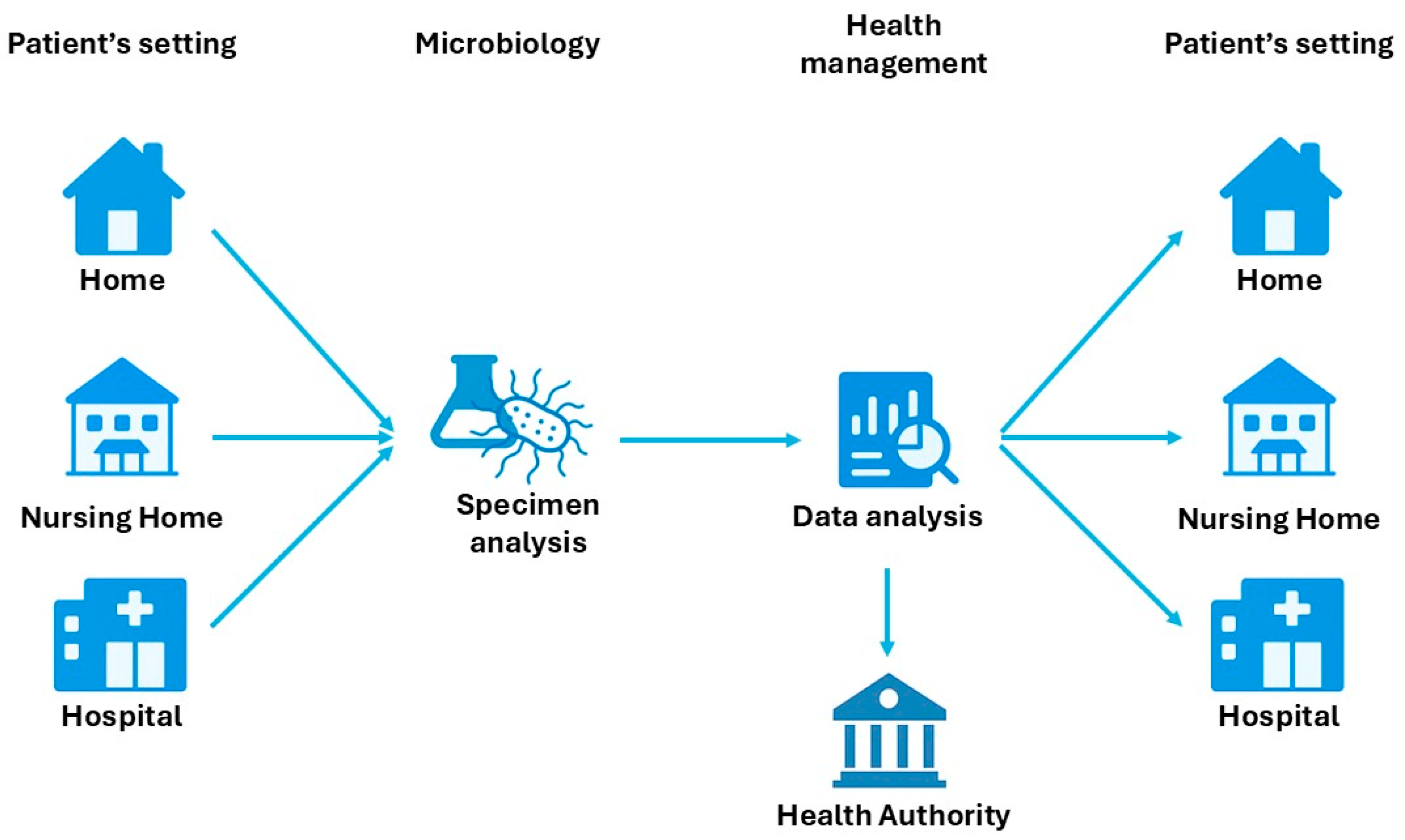
3.1. Data Extraction from Microbiology Laboratory Software
 CRITICAL STEP. Data extraction from laboratory software is a variable procedure, as it depends on the platform adopted in each laboratory. In our context, it was available Vitek 2 (bioMérieux, Inc., Hazelwood, MO, USA) and Matrix-Assisted Laser Desorption/Ionization—Time of Flight (MALDI-TOF, Bruker Daltonics, Bremen, Germany). This phase is critical because an incorrect or incomplete export may lead to data loss or compromise subsequent steps of the protocol. The investigator responsible for the export must always verify that all variables of interest—such as isolate identification number, sampling date, microorganism name, and corresponding antibiotic susceptibility results—are present and correctly formatted.
CRITICAL STEP. Data extraction from laboratory software is a variable procedure, as it depends on the platform adopted in each laboratory. In our context, it was available Vitek 2 (bioMérieux, Inc., Hazelwood, MO, USA) and Matrix-Assisted Laser Desorption/Ionization—Time of Flight (MALDI-TOF, Bruker Daltonics, Bremen, Germany). This phase is critical because an incorrect or incomplete export may lead to data loss or compromise subsequent steps of the protocol. The investigator responsible for the export must always verify that all variables of interest—such as isolate identification number, sampling date, microorganism name, and corresponding antibiotic susceptibility results—are present and correctly formatted.3.2. Data Import with BacLink
 CRITICAL STEP. When importing data for the first time, it is necessary to create a new format using the BacLink Configuration wizard. This tool enables the configuration of laboratory data for import into WHONET. Through the “File structure” option, the user specifies the file type, the file path, and the antimicrobial information, and links the relevant variables from the source file to the corresponding “Data fields” of the new file to be imported into WHONET. The “New data file” option allows the selection of the path and format for the newly generated file.
CRITICAL STEP. When importing data for the first time, it is necessary to create a new format using the BacLink Configuration wizard. This tool enables the configuration of laboratory data for import into WHONET. Through the “File structure” option, the user specifies the file type, the file path, and the antimicrobial information, and links the relevant variables from the source file to the corresponding “Data fields” of the new file to be imported into WHONET. The “New data file” option allows the selection of the path and format for the newly generated file.3.3. Configuration and Import of Data in WHOnet
 CRITICAL STEP. Before importing the file generated with BacLink into WHONET, the software must first be configured to receive data from the converted .dbf file. WHONET provides the option to configure a virtual laboratory through the “New laboratory” function. This configuration tool enables researchers to define the molecules of interest (e.g., antibiotics or antifungals) and to select the interpretive guidelines to be applied, such as CLSI, EUCAST, or other standards. The selection of “Antibiotics” is mandatory, as it defines the antimicrobial panel for analysis. Other options, such as “Location”, “Data fields”, and “Alerts”, are optional and allow customization or stratification of the dataset if these variables have been previously set.
CRITICAL STEP. Before importing the file generated with BacLink into WHONET, the software must first be configured to receive data from the converted .dbf file. WHONET provides the option to configure a virtual laboratory through the “New laboratory” function. This configuration tool enables researchers to define the molecules of interest (e.g., antibiotics or antifungals) and to select the interpretive guidelines to be applied, such as CLSI, EUCAST, or other standards. The selection of “Antibiotics” is mandatory, as it defines the antimicrobial panel for analysis. Other options, such as “Location”, “Data fields”, and “Alerts”, are optional and allow customization or stratification of the dataset if these variables have been previously set.3.4. Data Analysis in WHOnet
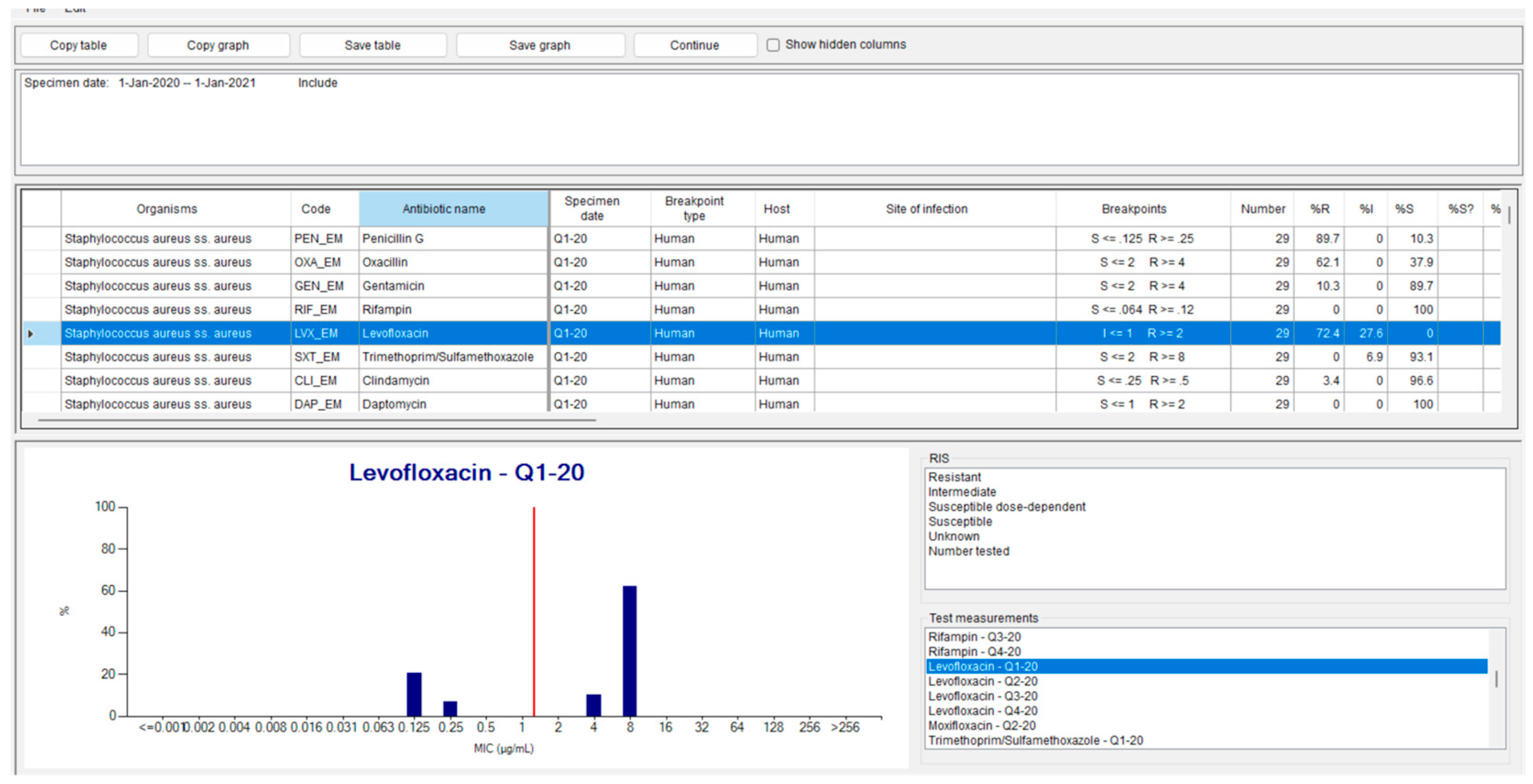
 CRITICAL STEP. Once the configuration described in the previous step has been completed, data analysis can be performed by selecting the “Data analysis” menu. This menu allows the user to choose among the different types of analyses available in WHONET (for a comprehensive list, please refer to the official WHONET technical documentation). Within this menu, the “Analysis type” option enables the evaluation of resistance for each bacterium or fungus against the previously configured molecules.
CRITICAL STEP. Once the configuration described in the previous step has been completed, data analysis can be performed by selecting the “Data analysis” menu. This menu allows the user to choose among the different types of analyses available in WHONET (for a comprehensive list, please refer to the official WHONET technical documentation). Within this menu, the “Analysis type” option enables the evaluation of resistance for each bacterium or fungus against the previously configured molecules.3.5. Preparation of Data for Analysis in R
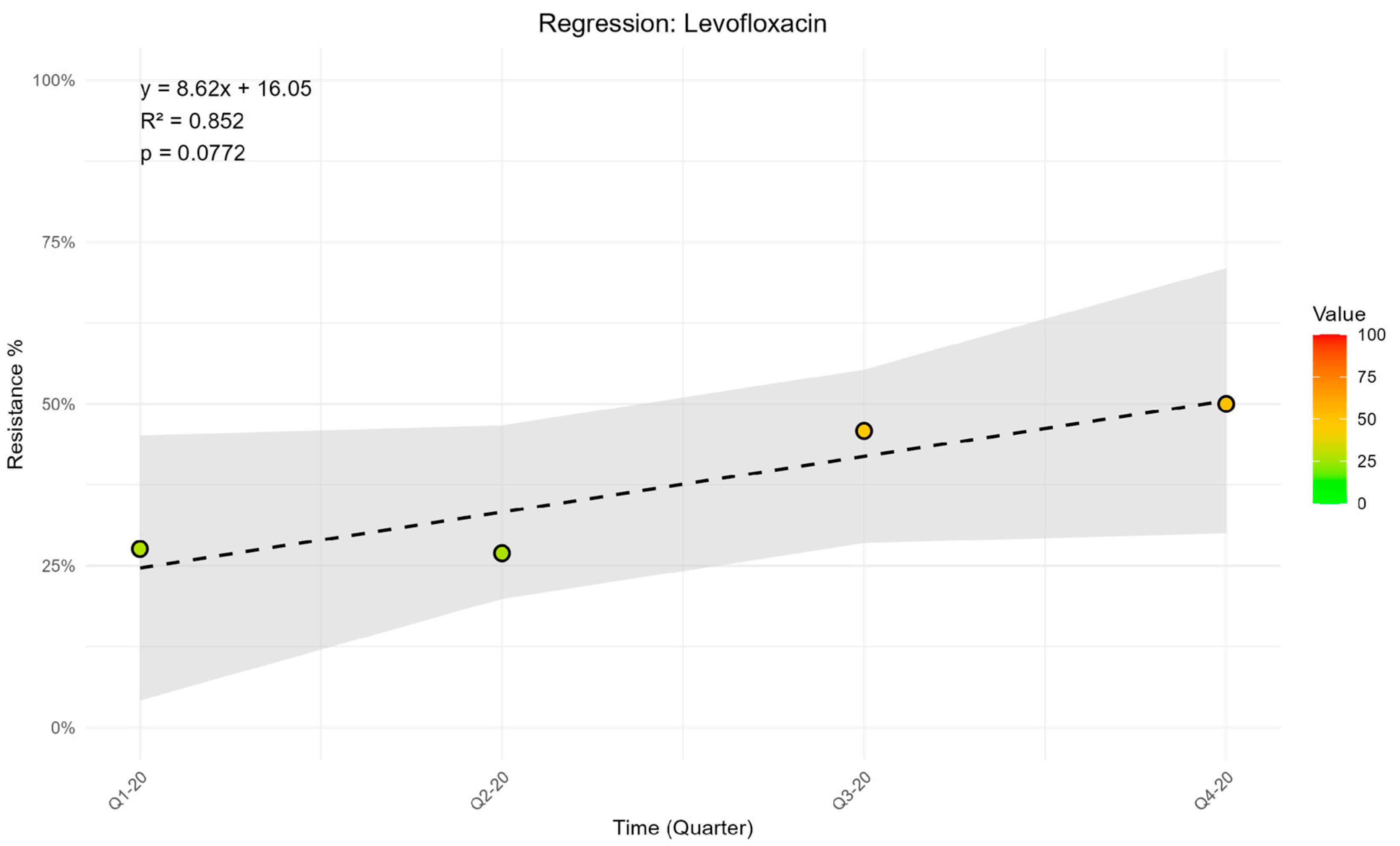
 CRITICAL STEP. To import the data into R, the WHONET output must be saved in .csv or another compatible database format. In this protocol, a worksheet was structured with time intervals arranged in rows and the antimicrobial agents investigated listed in columns. The resistance values retrieved for each molecule within the corresponding time interval were entered into the table cells.
CRITICAL STEP. To import the data into R, the WHONET output must be saved in .csv or another compatible database format. In this protocol, a worksheet was structured with time intervals arranged in rows and the antimicrobial agents investigated listed in columns. The resistance values retrieved for each molecule within the corresponding time interval were entered into the table cells.3.6. Data Analysis in R
4. Expected Results
4.1. WHOnet Outputs
4.2. R Outputs
5. Discussion
5.1. Strengths and Limitations
5.2. Enhancing Infection Management Through Data-Driven Protocols and Telemedicine
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AMR | Antimicrobial resistance |
| CLSI | Clinical and Laboratory Standards Institute |
| DTR | Difficult-to-Treat Resistant |
| EARS-NET | European Antimicrobial Resistance Surveillance Network |
| ESAC-NET | European Surveillance of Antimicrobial Consumption Network |
| EUCAST | European Committee on Antimicrobial Susceptibility Testing |
| JANIS | Japan Nosocomial Infections Surveillance |
| MDR | Multi-Drug Resistant |
| MIC | Minimum Inhibitory Concentration |
| PDR | Pan Drug Resistant |
| UN | United Nations |
| WHO | World Health Organization |
| XDR | Extensively Drug Resistant |
| %RIS | Percentage of Resistant, Intermediate and Susceptible |
Appendix A
=IFERROR( INDEX('01.%RIS'!$C$9:$AU$90; MATCH (1; ('01.%RIS'!$F$9:$F$90=data_to_R!C$1)* ('01.%RIS'!$J$9:$J$90=data_to_R!$A2) ;0) ;15) ;"")
-
Target Area –> INDEX('01.%RIS'!$C$9:$AU$90;; ... ; 15)
-
Identification of data source: from range C9:AU90 in the sheet '01.%RIS';
-
The row found from the MATCH(...) formula;
-
The column found from the 15th column of that range (the column contains resistance data matched for organism, molecules and quarter).
-
-
Row Lookup –> MATCH(1; (...)*(...); 0)
-
MATCH(1; ...) looks for the first 1 (TRUE) in an array of 1s and 0s created by the following multiplication;
-
(...)*(...) multiplication defines the conditions to satisfy in order to make the relationship TRUE and therefore allow the data to be transposed into that cell where the formula was written;
-
(...) the brackets of point b contain the description of the criteria, in our formula they are:
-
'01.%RIS'!$F$9:$F$90=data_to_R!C$1 this equation searches among all antibiotics in the list F9:F90, the antibiotic in cell C1;
-
'01.%RIS'!$J$9:$J$90=data_to_R!$A2 this equation searches among all quarters in the list J9:J90, the quarter in cell A2;
-
-
If both conditions are satisfied, the row is correctly identified for the INDEX (...) function. If both conditions are not satisfied, an error message is returned.
-
-
INDEX(...; ...; 15)
-
Once the matching by row and by column is found, the formula extracts the value;
-
-
IFERROR(..., "")
-
If no match is found (i.e., the combination does not exist), the formula returns an empty string ("") instead of an error.
-
| Reference | Descriptor |
|---|---|
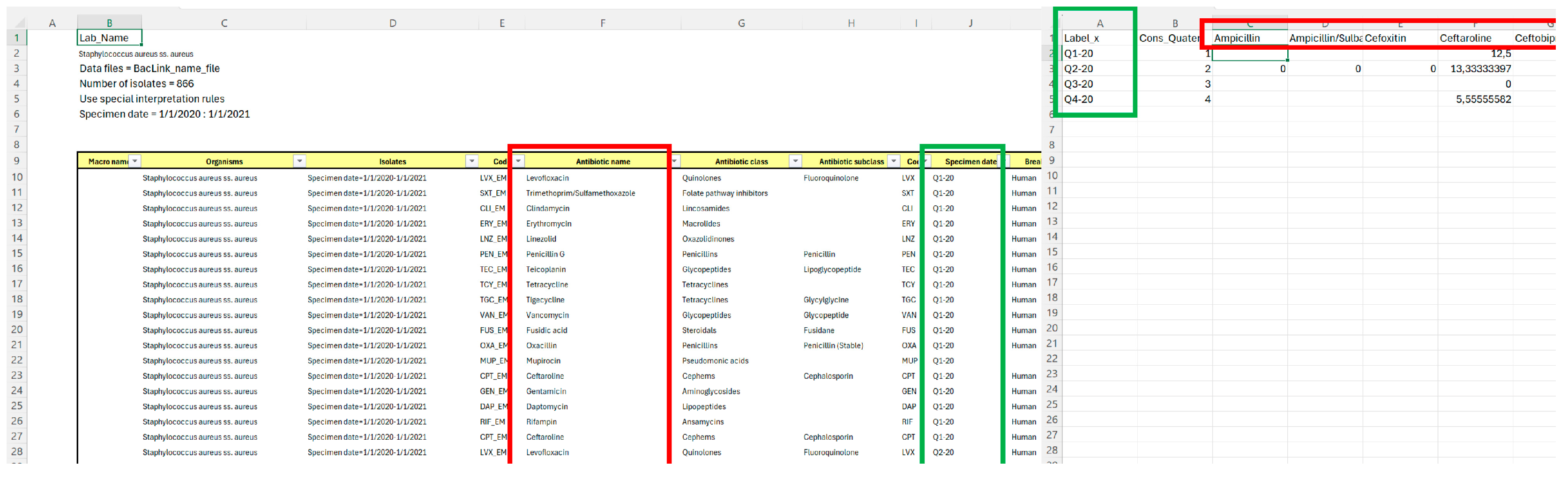
| ‘01.%RIS’ | This Excel spreadsheet is generated by WHONET during the results export process described in step 3.5. By selecting the “Save table” option shown in Figure 2, the software creates a .csv file on the researcher’s computer containing the displayed information. (Figure A1, left side) |
| data_to_R | This sheet must be created by the researcher, preferably in the same file as sheet ‘01.%RIS’. Column A contains the x-axis labels of the graphs (quarters), while column B contains the quartile count, useful for regression analysis (1, 2, 3, 4). Row 1, for columns C and later, contains the names of the antimicrobials being studied (in this protocol, only levofloxacin was selected for demonstration purposes). (Figure A1, right side). The structure of this sheet is presented in Table 1. |
| $C$9:$AU$90 | The range refers to all data exported from WHONET in .csv format. In this protocol, it includes the time variable (quarters, located in column J of sheet ‘01.%RIS’) and the antimicrobial variable (antibiotic names, located in column F of sheet ‘01.%RIS’). |
| $F$9:$F$90 | The range refers to the column containing data on the tested antibiotics in the ‘01.%RIS’ sheet. These values are matched with the headers in the data_to_R sheet using the formula described in Appendix A, which extracts and translates the corresponding data for each antibiotic and quarter. |
| $J$9:$J$90 | The range refers to the column containing data on specific quarter in the ‘01.%RIS’ sheet. These values are matched with the headers in the data_to_R sheet using the formula described in Appendix A, which extracts and translates the corresponding data for each antibiotic and quarter. |
| C$1 | This part of the formula locks the selection to row 1 of the data_to_R sheet for the formula described above, enabling the formula to be dragged across the columns of the data_to_R sheet where the antimicrobial names are listed in the headers. |
| $A2 | This part of the formula locks the selection to column A of the data_to_R sheet for the formula described above, enabling the formula to be dragged across the rows of the data_to_R sheet where quarters are listed in the labels. |
Appendix B
- “…path of your data file as organized in Appendix A…” ->
- ○
- “C:/Users/User_Name/Desktop/Folder_Name/file.xlsx”
- “the name of the sheet which contains the data to import in R” ->
- ○
- “data_to_import_into_R”
- readxl,
- ggplot2,
- broom,
- scales.
library(readxl) # Loads functions to read Excel (.xls and .xlsx) files into R.
library(ggplot2) # Loads tools to create advanced and customizable data visualizations using the grammar of graphics.
library(broom) # Provides functions to convert statistical analysis objects into tidy data frames for easy analysis and visualization.
library(scales) # Ensure 'scales' is loaded for rescale
# Read data
data <- read_excel("…path of your data file as organized in Appendix A…", sheet = "the name of the sheet which contains the data to import in R")
x_labels <- data[[1]]
time <- data[[2]] # This 'time' variable contains all original values from data[[2]]
colnames(data)[2] <- "Time"
# --- Plot Export Settings ---
# Define the output folder. It's good practice to create it if it doesn't exist.
# We will use a subfolder "Regression_Plots" within your Excel file's directory.
# Make sure the base path for your Excel file is correct for your system.
# Extract the directory of the Excel file
excel_dir <- dirname("…path of your data file as organized in Appendix A…")
output_dir <- file.path(excel_dir, "Regression_Plots") # The R output data will be placed in a folder called "Regression_Plots" in the same path as your datasheet.
# Create the folder if it doesn't exist. 'recursive = TRUE' creates parent folders if needed.
if (!dir.exists(output_dir)) {
dir.create(output_dir, recursive = TRUE)
message(paste("Output folder created:", output_dir))
} else {
message(paste("Output folder already exists:", output_dir))
}
# Start the loop to analyze each variable
for (i in 3:ncol(data)) {
variable_name <- colnames(data)[i]
variable_values <- data[[i]]
# Remove NAs for the model (this dataframe will be used ONLY for FITTING the model)
df_model <- data.frame(time = time, y = variable_values)
df_model <- na.omit(df_model)
# Regression checks
if (nrow(df_model) < 2 || sd(df_model$y) == 0) {
message(paste("Variable skipped:", variable_name, "- insufficient data for regression"))
next
}
# Linear model
model <- lm(y ~ time, data = df_model)
coeff <- coef(model)
summary_model <- summary(model)
r2 <- summary_model$r.squared
if (nrow(coef(summary_model)) >= 2) {
p_value <- coef(summary_model)[2, 4]
formula_text <- paste0(
"y = ", round(coeff[2], 2), "x + ", round(coeff[1], 2),
"\nR\u00b2 = ", round(r2, 3), # Used \u00b2 for the squared symbol
"\np = ", format.pval(p_value, digits = 3, eps = 0.001)
)
} else {
formula_text <- "Invalid Model"
}
# Dataframe for points (uses the full original 'time' variable)
df_plot <- data.frame(
Time = time, # 'time' here is the global variable, includes all original values
Label = factor(x_labels, levels = unique(x_labels)),
Value = variable_values
)
# **NEW: Creating a dataframe for prediction over the entire 'time' range**
# Use all unique and sorted 'time' values from the original dataset
pred_time_values <- sort(unique(time))
pred_df <- data.frame(time = pred_time_values)
# Perform prediction of values and confidence interval
predictions <- predict(model, newdata = pred_df, interval = "confidence")
pred_data <- cbind(pred_df, as.data.frame(predictions))
colnames(pred_data) <- c("time", "fit", "lwr", "upr") # Rename columns for clarity
# Plot
p <- ggplot() +
# **NEW: Add geom_ribbon for the extended confidence interval**
geom_ribbon(
data = pred_data, # Use the new 'pred_data' dataframe
aes(x = time, ymin = lwr, ymax = upr),
fill = "grey80",
alpha = 0.5 # Makes the interval slightly transparent
) +
# **NEW: Add geom_line for the extended regression line**
geom_line(
data = pred_data, # Use the new 'pred_data' dataframe
aes(x = time, y = fit),
color = "black",
linetype = "dashed",
linewidth = 0.8 # Sets the line thickness (use 'linewidth' for ggplot2 >= 3.4.0)
) +
# Colorized points on the X-axis which will now use 'Time' (numeric from df_plot)
geom_point(
data = df_plot,
aes(x = Time, y = Value, fill = Value), # 'Time' is numeric
shape = 21,
color = "black",
size = 3,
stroke = 1,
na.rm = TRUE
) +
# Ensure the X-axis is continuous but with desired discrete labels
scale_x_continuous(
breaks = unique(df_plot$Time), # Numeric positions for breaks
labels = unique(df_plot$Label) # Discrete labels for breaks
) +
scale_y_continuous(
limits = c(0, 100),
labels = function(x) paste0(x, "%")
) +
scale_fill_gradientn(
colours = c("green", "goldenrod1", "red"),
values = scales::rescale(c(0, 50, 100)),
limits = c(0, 100),
name = "Value"
) +
ggtitle(paste("Regression:", variable_name)) +
xlab("Time (Quarter)") +
ylab("Resistance %") +
annotate(
"text",
x = min(pred_data$time, na.rm = TRUE), # Use the minimum 'time' value from prediction
y = 100,
label = formula_text,
hjust = 0, vjust = 1, # Top-left alignment
size = 4, color = "black"
) +
theme_minimal() +
theme(
axis.text.x = element_text(angle = 45, hjust = 1), # Rotate X-axis labels
plot.title = element_text(hjust = 0.5) # Center the plot title
)
print(p) # Display the plot on screen
# --- Export the plot as an image (PNG) ---
# Create a safe filename for the variable
safe_variable_name <- gsub("[^A-Za-z0-9_\\-]", "_", variable_name) # Removes invalid characters
filename <- paste0(safe_variable_name, "_regression_plot.png")
filepath <- file.path(output_dir, filename)
ggsave(
filename = filepath,
plot = p,
width = 10, # Plot width in inches
height = 6, # Plot height in inches
dpi = 300 # Resolution in DPI (Dots Per Inch) - 300 is standard for high quality
)
message(paste("Plot saved:", filepath))
}
References
- Shutt, A.E.; Ashiru-Oredope, D.; Price, J.; Padoveze, M.C.; Shafiq, N.; Carter, E.; Ghataure, A.; Shariq, S.; Holmes, A.H.; Charani, E. The intersection of the social determinants of health and antimicrobial resistance in human populations: A systematic review. BMJ Glob. Health 2025, 10, e017389. [Google Scholar] [CrossRef]
- Morrison, L.; Zembower, T.R. Antimicrobial Resistance. Gastrointest. Endosc. Clin. N. Am. 2020, 30, 619–635. [Google Scholar] [CrossRef]
- Georges, R.-N.; Doumèche, B. Understanding current challenges of antibiotic resistance to find new solutions for 2050: Scientific, medical and political issues. Arch. Microbiol. 2025, 207, 162. [Google Scholar] [CrossRef]
- OECD. Tackling Wasteful Spending on Health. 10 January 2017. Available online: https://www.oecd.org/en/publications/tackling-wasteful-spending-on-health_9789264266414-en.html (accessed on 30 June 2025).
- Cassini, A.; Högberg, L.D.; Plachouras, D.; Quattrocchi, A.; Hoxha, A.; Simonsen, G.S.; Colomb-Cotinat, M.; Kretzschmar, M.E.; Devleesschauwer, B.; Cecchini, M.; et al. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: A population-level modelling analysis. Lancet Infect. Dis. 2019, 19, 56–66. [Google Scholar] [CrossRef]
- Kadeřábková, N.; Mahmood, A.J.S.; Mavridou, D.A.I. Antibiotic susceptibility testing using minimum inhibitory concentration (MIC) assays. npj Antimicrob. Resist. 2024, 2, 37. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez-Martín, C.B.; Martínez-Martínez, S.; Petrocchi-Rilo, M. Analysis of Susceptibility or Resistance to Antimicrobial Agents. In Streptococcus suis: Methods in Molecular Biology; Humana: New York, NY, USA, 2024; Volume 2815, pp. 51–71. [Google Scholar] [CrossRef]
- Suzuki, H.; Perencevich, E.N.; Nair, R.; Livorsi, D.J.; Goto, M. Excess Length of Acute Inpatient Stay Attributable to Acquisition of Hospital-Onset Gram-Negative Bloodstream Infection with and without Antibiotic Resistance: A Multistate Model Analysis. Antibiotics 2020, 9, 96. [Google Scholar] [CrossRef] [PubMed]
- Vedovetto, A.; Soriani, N.; Merlo, E.; Gregori, D. The burden of inappropriate emergency department pediatric visits: Why Italy needs an urgent reform. Health Serv. Res. 2014, 49, 1290–1305. [Google Scholar] [CrossRef]
- Cuningham, W.; Anderson, L.; Bowen, A.C.; Buising, K.; Connors, C.; Daveson, K.; Martin, J.; McNamara, S.; Patel, B.; James, R.; et al. Antimicrobial stewardship in remote primary healthcare across northern Australia. PeerJ 2020, 8, e9409. [Google Scholar] [CrossRef]
- Klein, E.Y.; Van Boeckel, T.P.; Martinez, E.M.; Pant, S.; Gandra, S.; Levin, S.A.; Goossens, H.; Laxminarayan, R. Global increase and geographic convergence in antibiotic consumption between 2000 and 2015. Proc. Natl. Acad. Sci. USA 2018, 115, E3463–E3470. [Google Scholar] [CrossRef]
- Van Boeckel, T.P.; Gandra, S.; Ashok, A.; Caudron, Q.; Grenfell, B.T.; Levin, S.A.; Laxminarayan, R. Global antibiotic consumption 2000 to 2010: An analysis of national pharmaceutical sales data. Lancet Infect. Dis. 2014, 14, 742–750. [Google Scholar] [CrossRef] [PubMed]
- Polacek, C.; Timbrook, T.T.; Cui, C.; Heins, Z.; Rosenthal, N.A. Prevalence and healthcare burden of inappropriate antimicrobial treatment in patients at high risk of complications from acute respiratory infections: A scoping review. Front. Med. 2025, 12, 1533797. [Google Scholar] [CrossRef]
- Licata, F.; Quirino, A.; Pepe, D.; Matera, G.; Bianco, A.; Collaborative Group. Antimicrobial Resistance in Pathogens Isolated from Blood Cultures: A Two-Year Multicenter Hospital Surveillance Study in Italy. Antibiotics 2020, 10, 10. [Google Scholar] [CrossRef]
- Ribeiro, C.d.S.; van de Burgwal, L.H.; Regeer, B.J. Overcoming challenges for designing and implementing the One Health approach: A systematic review of the literature. One Health 2019, 7, 100085. [Google Scholar] [CrossRef] [PubMed]
- McSorley, J.C. Analysis of ESAC-Net/EARS-Net Data from 29 EEA Countries for Spatiotemporal Associations Between Antimicrobial Use and Resistance-Implications for Antimicrobial Stewardship? Antibiotics 2025, 14, 399. [Google Scholar] [CrossRef]
- Kajihara, T.; Yahara, K.; Hirabayashi, A.; Shibayama, K.; Sugai, M. Japan Nosocomial Infections Surveillance (JANIS): Current Status, International Collaboration, and Future Directions for a Comprehensive Antimicrobial Resistance Surveillance System. Jpn. J. Infect. Dis. 2021, 74, 87–96. [Google Scholar] [CrossRef]
- Oplustil, C.P.; Nunes, R.; Mendes, C.; RESISTNET Group. Multicenter evaluation of resistance patterns of Klebsiella pneumoniae, Escherichia coli, Salmonella spp. and Shigella spp. isolated from clinical specimens in Brazil: RESISTNET Surveillance Program. Braz. J. Infect. Dis. 2001, 5, 8–12. [Google Scholar] [CrossRef]
- Do, P.C.; Assefa, Y.A.; Batikawai, S.M.; Reid, S.A. Strengthening antimicrobial resistance surveillance systems: A scoping review. BMC Infect. Dis. 2023, 23, 593. [Google Scholar] [CrossRef]
- Delpy, L.; Astbury, C.C.; Aenishaenslin, C.; Ruckert, A.; Penney, T.L.; Wiktorowicz, M.; Ciss, M.; Benko, R.; Bordier, M. Integrated surveillance systems for antibiotic resistance in a One Health context: A scoping review. BMC Public Health 2024, 24, 1717. [Google Scholar] [CrossRef] [PubMed]
- WHONET. Software. Available online: https://whonet.org/software.html (accessed on 4 June 2025).
- Stelling, J.; Yih, W.K.; Galas, M.; Kulldorff, M.; Pichel, M.; Terragno, R.; Tuduri, E.; Espetxe, S.; Binsztein, N.; O’Brien, T.F.; et al. Automated use of WHONET and SaTScan to detect outbreaks of Shigella spp. using antimicrobial resistance phenotypes. Epidemiol. Infect. 2010, 138, 873–883. [Google Scholar] [CrossRef]
- O’Brien, T.F.; Stelling, J.M. WHONET: Removing obstacles to the full use of information about antimicrobial resistance. Diagn. Microbiol. Infect. Dis. 1996, 25, 162–168. [Google Scholar] [CrossRef] [PubMed]
- Roberts, S.C.; Martinello, R.A. Can Patient Safety Be Improved by Using WHONET-SaTScan to Identify Outbreaks? NEJM Evid. 2024, 3, EVIDe2400086. [Google Scholar] [CrossRef]
- Peñalva, G.; Crespo-Robledo, P.; Molvik, M.; López-Navas, A.; Kacelnik, O.; Cisneros, J.M.; EU-JAMRAI WP7.4.1 group. A step forward in antibiotic use and resistance monitoring: A quarterly surveillance system pilot in 11 European Union/European Economic Area countries, September 2017 to May 2020. Eurosurveillance 2022, 27, 2200082. [Google Scholar] [CrossRef]
- WHONET. BacLink: Exporting data from an LIS. 2022. Available online: https://whonet.org/WebDocs/BacLink.4_Data_exports_Exporting_data_from_Laboratory_information_systems.pdf (accessed on 4 June 2025).
- Hackenberger, B.K. R software: Unfriendly but probably the best. Croat. Med J. 2020, 61, 66–68. [Google Scholar] [CrossRef]
- Vielhauer, J.; Mahajan, U.M.; Adorjan, K.; Benesch, C.; Oehrle, B.; Beyer, G.; Sirtl, S.; Johlke, A.-L.; Allgeier, J.; Pernpruner, A.; et al. Electronic data capture in resource-limited settings using the lightweight clinical data acquisition and recording system. Sci. Rep. 2024, 14, 19056. [Google Scholar] [CrossRef] [PubMed]
- de Lima, D.A.; Helito, C.P.; de Lima, L.L.; Clazzer, R.; Gonçalves, R.K.; de Camargo, O.P. How to Perform a Meta-Analysis: A Practical Step-By-Step Guide Using R Software and Rstudio. Acta Ortop. Bras. 2022, 30, e248775. [Google Scholar] [CrossRef] [PubMed]
- WHONET. Additional Installation Information. Available online: https://whonet.org/InstallationIssues.html (accessed on 4 June 2025).
- WHONET. Microbiology Laboratory Database Software. Available online: https://whonet.org/ (accessed on 4 June 2025).
- R: The R Project for Statistical Computing. Available online: https://www.r-project.org/ (accessed on 4 June 2025).
- Qiao, H.; Chen, Y.; Qian, C.; Guo, Y. Clinical data mining: Challenges, opportunities, and recommendations for translational applications. J. Transl. Med. 2024, 22, 185. [Google Scholar] [CrossRef] [PubMed]
- Pietrantonio, F.; Vinci, A.; Maurici, M.; Ciarambino, T.; Galli, B.; Signorini, A.; La Fazia, V.M.; Rosselli, F.; Fortunato, L.; Iodice, R.; et al. Intra- and Extra-Hospitalization Monitoring of Vital Signs—Two Sides of the Same Coin: Perspectives from LIMS and Greenline-HT Study Operators. Sensors 2023, 23, 5408. [Google Scholar] [CrossRef]
- Marra, A.R.; Langford, B.J.; Nori, P.; Bearman, G. Revolutionizing antimicrobial stewardship, infection prevention, and public health with artificial intelligence: The middle path. Antimicrob. Steward. Heal. Epidemiol. 2023, 3, e219. [Google Scholar] [CrossRef]

| Time Period Label | Time ID 1 | Antibiotic 1 | Antibiotic 2 | Antibiotic … |
|---|---|---|---|---|
| Period 1 | 1 | data | data | … |
| Period 2 | 2 | data | data | … |
| … | … | … | … | … |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ingravalle, F.; Vinci, A.; Ciotti, M.; Fontana, C.; Pica, F.; Sebastiani, E.; Donnoli, C.; Guido Rizzo, M.; Tedesco, D.; D’Arezzo, S.; et al. A Six-Step Protocol for Monitoring Antimicrobial Resistance Trends Using WHONET and R: Real-World Application and R Code Integration. Methods Protoc. 2025, 8, 115. https://doi.org/10.3390/mps8050115
Ingravalle F, Vinci A, Ciotti M, Fontana C, Pica F, Sebastiani E, Donnoli C, Guido Rizzo M, Tedesco D, D’Arezzo S, et al. A Six-Step Protocol for Monitoring Antimicrobial Resistance Trends Using WHONET and R: Real-World Application and R Code Integration. Methods and Protocols. 2025; 8(5):115. https://doi.org/10.3390/mps8050115
Chicago/Turabian StyleIngravalle, Fabio, Antonio Vinci, Marco Ciotti, Carla Fontana, Francesca Pica, Emanuele Sebastiani, Clara Donnoli, Martino Guido Rizzo, Dario Tedesco, Silvia D’Arezzo, and et al. 2025. "A Six-Step Protocol for Monitoring Antimicrobial Resistance Trends Using WHONET and R: Real-World Application and R Code Integration" Methods and Protocols 8, no. 5: 115. https://doi.org/10.3390/mps8050115
APA StyleIngravalle, F., Vinci, A., Ciotti, M., Fontana, C., Pica, F., Sebastiani, E., Donnoli, C., Guido Rizzo, M., Tedesco, D., D’Arezzo, S., Cicalini, S., Loiudice, M. T., & Maurici, M. (2025). A Six-Step Protocol for Monitoring Antimicrobial Resistance Trends Using WHONET and R: Real-World Application and R Code Integration. Methods and Protocols, 8(5), 115. https://doi.org/10.3390/mps8050115







