Comprehensive Protocols for Detecting Xenotransplantation-Relevant Viruses
Abstract
1. Introduction
2. Experimental Design
2.1. Samples from the Animals
2.2. Materials
- Pancoll human medium (PAN-Biotech GmbH, Aidenbach, Germany).
- DNA and RNA isolation kits (QIAGEN, Hilden, Germany):
- 2.1
- DNeasy Blood & Tissue kit;
- 2.2
- QIAamp DNA FFPE Tissue Kit;
- 2.3
- RNeasy DSP FFPE Kit;
- 2.4
- RNase-Free DNase Set;
- 2.5
- RNeasy Plus Mini kit.
- Ethanol, methanol (both from Carl Roth, Karlsruhe, Germany).
- DreamTaq DNA polymerase (Thermo Fisher Scientific, Waltham, MA, USA).
- PVDF membrane (0.2 µm; Carl Roth, Karlsruhe, Germany).
2.3. Equipment
- Microcentrifuge Heraeus FRESCO 21 (Thermo Fisher Scientific, Waltham, MA, USA);
- NanoDrop Spectrophotometer (Peqlab Biotechnologie GmbH, Erlangen, Germany);
- Biometra TRIO cycler (Analytik Jena, Jena, Germany);
- qTOWER3 G qPCR cycler (Analytik Jena, Jena, Germany);
- Gel electrophoresis chamber (Bio-Rad, Hercules, CA, USA);
- Semi-dry electro blotter (Peqlab Biotechnologie GmbH, Erlangen, Germany);
- Imaging device (Peqlab Biotechnologie GmbH).
3. Procedure
3.1. Nucleic Acid Isolation
3.1.1. Isolation of PBMCs from Blood Samples
- Place 5 mL of lymphocyte separation medium (Pancoll human medium, PAN-Biotech GmbH, Aidenbach, Germany) in a 15 mL reaction tube. Hold the tube at a 45° angle.
CRITICAL STEP: Slowly layer 5 mL of diluted blood (1:1 ratio of media to blood) on top using the pipetting aid, release very slowly and close the lid.
CRITICAL STEP: Centrifuge at 900× g, 25 min, 21 °C (switch off the brake).
- 4.
- The following layers are obtained after centrifugation (from bottom to top):
- -
- Erythrocytes;
- -
- PBMCs, white milky layer;
- -
- Plasma, yellow liquid.
- 5.
- If desired, transfer the plasma without disturbing the PBMC layer to a 1.5 mL reaction tube and store it at −20 °C.
- 6.
- Carefully aspirate the PBMC layer with a pipette and transfer it into a new 15 mL reaction tube.
- 7.
- Wash the PBMC fraction with PBS.
- 8.
- Centrifuge at 350× g, 10 min, 21 °C with brakes.
- 9.
- Discard the supernatant and resuspend the PBMCs (visible as pellet) in PBS.
- 10.
- Repeat the PBS wash step.
- 11.
- Resuspend the PBMC pellet in an appropriate volume of PBS, aliquot in 1.5 mL reaction tubes and store them at −20 °C.
3.1.2. DNA Isolation from Blood Samples and Cell Cultures
3.1.3. DNA Isolation from Tissue Samples
3.1.4. Nucleic Acid Extraction from Formalin-Fixed Paraffin-Embedded (FFPE) Tissue
3.1.5. Nucleic Acid Extraction from Pig Skin
3.1.6. Nucleic Acid Quality Control and Quantification
- Open the NanoDrop software (Nano Drop 1000, version 3.8.1) on the connected computer.
- Initialize the instrument by loading 2 μL of nuclease-free water.
- Perform a blank measurement using 2 μL of elution buffer.
- Load 2 μL of the DNA sample and measure its concentration.
- Record the DNA/RNA concentration as well as the 260/280 and 260/230 ratios to evaluate sample purity.
3.1.7. RNA Isolation
3.2. PCR Methods
3.2.1. Primers and Probes Required
3.2.2. Conventional PCR
3.2.3. Real-Time PCR
- Perform the following steps in a clean master-mix-only room.
- Clean the surface with DNase/RNase solution. Use filter tips all the time.
- Prepare the primer–probe mix for the virus to be detected (Table A1).
- Also prepare primer–probe mix for the internal control, i.e., GAPDH gene (Table A1).
- Prepare a master mix in a new vial by adding SensiFAST probe no-ROX mix followed by virus primer–probe and GAPDH primer–probe, and top up the volume with nuclease-free water (Table A2).
- Vortex the master mix vial and give it a short spin.
- From now on, perform steps in a clean hood. Take a 96-well qPCR plate. Dispense 16 µL of master mix in each well, followed by 4 µL (100 ng) of DNA sample, making a total volume of 20 µL per well (prepare triplicates for each sample).
- Also include a positive control, which is gene block DNA, and a no-template control (NTC), which is nuclease-free water.
- Seal the plate carefully using the plastic sheet provided.
- Give the plate a short spin in the centrifuge to spin down the liquid and to remove any air bubbles.
- Place the plate in a qTOWER3 G qPCR cycler (Analytik Jena, Jena, Germany)
- Set up the protocol for the respective virus (Table A3) and run it. At the end, observe the graph and calculate the Ct (threshold cycle) value.
3.3. Western Blot Analysis
3.3.1. SDS-PAGE
3.3.2. Western Blot Analysis PCMV/PRV
3.3.3. Western Blot Analysis PLHV
3.3.4. Western Blot Analysis PERV
3.3.5. Western Blot Analysis HEV
4. Expected Results
4.1. Results of PCR Analysis
4.2. Results of Western Blot Analysis
4.2.1. PCMV/PRV
4.2.2. PLHV
4.2.3. PERV
4.2.4. HEV
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Compound | Amount (µL) |
|---|---|
| FAM mix for viruses PCMV/PRV, PLHV-1, PLHV-2, PCV1, PCV2, PCV3, PCV4, PPV1, TTSuV1, TTSuV2, HEV | |
| Fwd | 10 |
| Rev | 10 |
| probe | 2.5 |
| H2O | 77.5 |
| total | 100 |
| FAM mix for viruses PLHV-3, APPV, PERVpol | |
| Fwd | 10 |
| Rev | 10 |
| probe | 5 |
| H2O | 75 |
| total | 100 |
| FAM mix for PRE-1 | |
| Fwd | 10 |
| Rev | 10 |
| probe | 2 |
| H2O | 78 |
| total | 100 |
| HEX mix for hGAPDH, pGAPDH | |
| Fwd | 2.5 |
| Rev | 2.5 |
| probe | 2.5 |
| H2O | 92.5 |
| total | 100 |
| Compound | µL |
|---|---|
| 2× SensiFast Probe No-ROX Mix | 10 |
| FAM mix for virus | 1.8 |
| HEX mix for GAPDH | 1.8 |
| H2O | 2.4 |
| Total | 16 |
| DNA template | 4 |
| Total reaction mix | 20 |
| Virus | Time | Temperature | Number of Cyles |
|---|---|---|---|
| PCMV/PRV | |||
| Inactivation | 2 min | 50 °C | |
| Initial Denaturation | 10 min | 95 °C | |
| Denaturation | 15 s | 95 °C | 45 cycles |
| Annealing/Extension | 1 min | 60 °C | |
| PLHV-1, PLHV-2, PPV1, PCV3 | |||
| Inactivation | 5 min | 95 °C | |
| Denaturation | 15 s | 95 °C | 45 cycles |
| Annealing | 1 min | 56 °C | |
| Extension | 30 s | 72 °C | |
| PLHV-3 | |||
| Inactivation | 10 min | 90 °C | |
| Denaturation | 30 s | 90 °C | 45 cycles |
| Annealing/Extension | 30 s | 59 °C | |
| PCV-1,2,4 | |||
| Initial Denaturation | 30 s | 95 °C | |
| Denaturation | 5 s | 95 °C | 45 cycles |
| Annealing/Extension | 1 min | 60 °C | |
| HEV | |||
| Reverse Transcription | 30 min | 50 °C | |
| Inactivation | 15 min | 95 °C | |
| Denaturation | 10 s | 95 °C | 45 cycles |
| Annealing | 20 s | 55 °C | |
| Extension | 15 s | 72 °C | |
| APPV | |||
| Reverse Transcription | 30 min | 50 °C | |
| Inactivation | 15 min | 95 °C | |
| Denaturation | 30 s | 95 °C | 40 cycles |
| Annealing | 30 s | 56 °C | |
| Extension | 30 s | 72 °C | |
| SARS-CoV-2 | |||
| Reverse Transcription | 10 min | 55 °C | |
| Inactivation | 3 min | 95 °C | |
| Denaturation | 15 s | 95 °C | 45 cycles |
| Annealing/Extension | 30 s | 58 °C | |
| TTSuV1, TTSuV2 | |||
| Activation | 2 min | 50 °C | |
| Denaturation | 10 min | 95 °C | |
| Annealing | 15 s | 60 °C | 45 cycles |
| Extension | 15 s | 60 °C | |
| PERV-C | |||
| Initial Denaturation | 10 min | 95 °C | |
| Denaturation | 15 s | 95 °C | 45 cycles |
| Annealing | 30 s | 55 °C | |
| Extension | 30 s | 72 °C | |
| Final Extension | 5 min | 72 °C | |
| PERVpol | |||
| Inactivation | 5 min | 95 °C | |
| Denaturation | 15 s | 95 °C | 45 cycles |
| Annealing | 30 s | 62 °C | |
| Extension | 30 s | 72 °C | |
| PRE-1 | |||
| Inactivation | 5 min | 95 °C | |
| Denaturation | 15 s | 95 °C | 45 cycles |
| Annealing | 30 s | 60 °C | |
| Extension | 30 s | 72 °C |
Appendix B
| Solution | Ingredients | Remarks |
|---|---|---|
| 6× SDS-PAGE Sample Buffer | ||
| 0.375 g | Tris | pH 6.8 |
| 12% | SDS | |
| 60% | Glycerol | |
| 0.6 M | DTT | |
| 0.06% | Bromophenol blue | |
| 10× SDS-PAGE Running Buffer (1 L) | ||
| 30.2 g | Tris | |
| 144 g | Glycine | |
| 10 g | SDS | |
| 10× Transfer Buffer | 1× Transfer Buffer | |
| 72 g | Glycine | 1× 10× Transfer buffer |
| 15.1 g | Tris | 20% Methanol |
| 2.5 g | SDS | Top up with water |
| 10× Tris-buffered saline (TBS) (1 L) | pH 7.5 | |
| 24.2 g | Tris | |
| 87.6 g | Sodium chloride | |
| TBS-T | ||
| 0.05% Tween 20 in 1× TBS | ||
| Blocking buffer | ||
| 5% milk powder in TBS-T | ||
References
- Lu, T.; Yang, B.; Wang, R.; Qin, C. Xenotransplantation: Current Status in Preclinical Research. Front. Immunol. 2020, 10, 3060. [Google Scholar] [CrossRef] [PubMed]
- Längin, M.; Mayr, T.; Reichart, B.; Michel, S.; Buchholz, S.; Guethoff, S.; Dashkevich, A.; Baehr, A.; Egerer, S.; Bauer, A.; et al. Consistent success in life-supporting porcine cardiac xenotransplantation. Nature 2018, 564, 430–433. [Google Scholar] [CrossRef]
- Singh, A.K.; Goerlich, C.E.; Zhang, T.; Lewis, B.; Hershfeld, A.; Braileanu, G.; Kurvi, K.; Rice, K.; Sentz, F.; Mudd, S.; et al. Genetically engineered pig heart transplantation in non-human primates. Commun. Med. 2025, 5, 6. [Google Scholar] [CrossRef] [PubMed]
- Moazami, N.; Stern, J.M.; Khalil, K.; Kim, J.I.; Narula, N.; Mangiola, M.; Weldon, E.P.; Kagermazova, L.; James, L.; Lawson, N.; et al. Pig-to-human heart xenotransplantation in two recently deceased human recipients. Nat. Med. 2023, 29, 1989–1997. [Google Scholar] [CrossRef]
- Cooper, D.K.C.; Kobayashi, T. Xenotransplantation experiments in brain-dead human subjects-A critical appraisal. Am. J. Transplant. 2024, 24, 520–525. [Google Scholar] [CrossRef]
- Cooper, D.K.; Matsumoto, S.; Abalovich, A.; Itoh, T.; Mourad, N.I.; Gianello, P.R.; Wolf, E.; Cozzi, E. Progress in Clinical Encapsulated Islet Xenotransplantation. Transplantation 2016, 100, 2301–2308. [Google Scholar] [CrossRef]
- Mohiuddin, M.M.; Singh, A.K.; Scobie, L.; Goerlich, C.E.; Grazioli, A.; Saharia, K.; Crossan, C.; Burke, A.; Drachenberg, C.; Oguz, C.; et al. Graft dysfunction in compassionate use of genetically engineered pig-to-human cardiac xenotransplantation: A case report. Lancet 2023, 402, 397–410. [Google Scholar] [CrossRef]
- Loupy, A.; Goutaudier, V.; Giarraputo, A.; Mezine, F.; Morgand, E.; Robin, B.; Khalil, K.; Mehta, S.; Keating, B.; Dandro, A.; et al. Immune response after pig-to-human kidney xenotransplantation: A multimodal phenotyping study. Lancet 2023, 402, 1158–1169. [Google Scholar] [CrossRef]
- Montgomery, R.A.; Stern, J.M.; Lonze, B.E.; Tatapudi, V.S.; Mangiola, M.; Wu, M.; Weldon, E.; Lawson, N.; Deterville, C.; Dieter, R.A.; et al. Results of Two Cases of Pig-to-Human Kidney Xenotransplantation. N. Engl. J. Med. 2022, 386, 1889–1898. [Google Scholar] [CrossRef]
- Locke, J.E.; Kumar, V.; Anderson, D.; Porrett, P.M. Normal graft function after pig-to-human kidney xenotransplant. JAMA Surg. 2023, 158, 1106–1108. [Google Scholar] [CrossRef] [PubMed]
- Yamada, K.; Tasaki, M.; Sekijima, M.; Wilkinson, R.A.; Villani, V.; Moran, S.G.; Cormack, T.A.; Hanekamp, I.M.; Hawley, R.J.; Arn, J.S.; et al. Porcine cytomegalovirus infection is associated with early rejection of kidney grafts in a pig to baboon xenotransplantation model. Transplantation 2014, 98, 411–418.3. [Google Scholar] [CrossRef]
- Sekijima, M.; Waki, S.; Sahara, H.; Tasaki, M.; Wilkinson, R.A.; Villani, V.; Shimatsu, Y.; Nakano, K.; Matsunari, H.; Nagashima, H.; et al. Results of life-supporting galactosyltransferase knockout kidneys in cynomolgus monkeys using two different sources of galactosyltransferase knockout Swine. Transplantation 2014, 98, 419–426. [Google Scholar] [CrossRef]
- Denner, J.; Längin, M.; Reichart, B.; Krüger, L.; Fiebig, U.; Mokelke, M.; Radan, J.; Mayr, T.; Milusev, A.; Luther, F.; et al. Impact of porcine cytomegalovirus on long-term orthotopic cardiac xenotransplant survival. Sci. Rep. 2020, 10, 17531. [Google Scholar] [CrossRef] [PubMed]
- Krüger, L.; Längin, M.; Reichart, B.; Fiebig, U.; Kristiansen, Y.; Prinz, C.; Kessler, B.; Egerer, S.; Wolf, E.; Abicht, J.M.; et al. Transmission of Porcine Circovirus 3 (PCV3) by Xenotransplantation of Pig Hearts into Baboons. Viruses 2019, 11, 650. [Google Scholar] [CrossRef] [PubMed]
- Gazda, L.S.; Collins, J.; Lovatt, A.; Holdcraft, R.W.; Morin, M.J.; Galbraith, D.; Graham, M.; Laramore, M.A.; Maclean, C.; Black, J.; et al. A comprehensive microbiological safety approach for agarose encapsulated porcine islets intended for clinical trials. Xenotransplantation 2016, 23, 444–463. [Google Scholar] [CrossRef]
- Meier, R.P.H.; Pierson, R.N., 3rd; Fishman, J.A.; Buhler, L.H.; Bottino, R.; Ladowski, J.M.; Ekser, B.; Wolf, E.; Brenner, P.; Ierino, F.; et al. International Xenotransplantation Association (IXA) Position Paper on Kidney Xenotransplantation. Xenotransplantation 2025, 32, e70003. [Google Scholar] [CrossRef]
- Meng, X.J. Swine hepatitis E virus: Cross-species infection and risk in xenotransplantation. Curr. Top. Microbiol. Immunol. 2003, 278, 185–216. [Google Scholar]
- Morozov, V.A.; Morozov, A.V.; Rotem, A.; Barkai, U.; Bornstein, S.; Denner, J. Extended Microbiological Characterization of Gottingen Minipigs in the Context of Xenotransplantation: Detection and Vertical Transmission of Hepatitis E Virus. PLoS ONE 2015, 10, e0139893. [Google Scholar] [CrossRef]
- Jhelum, H.; Kaufer, B.; Denner, J. Application of Methods Detecting Xenotransplantation-Relevant Viruses for Screening German Slaughterhouse Pigs. Viruses 2024, 16, 1119. [Google Scholar] [CrossRef]
- Morozov, V.A.; Abicht, J.M.; Reichart, B.; Mayr, T.; Guethoff, S.; Denner, J. Active replication of porcine cytomegalovirus (PCMV) following transplantation of a pig heart into a baboon despite undetected virus in the donor pig. Ann. Virol. Res. 2016, 2, 1018. [Google Scholar]
- Morozov, V.A.; Morozov, A.V.; Denner, J. New PCR diagnostic systems for the detection and quantification of porcine cytomegalovirus (PCMV). Arch. Virol. 2016, 161, 1159–1168. [Google Scholar] [CrossRef] [PubMed]
- Halecker, S.; Hansen, S.; Krabben, L.; Ebner, F.; Kaufer, B.; Denner, J. How, where and when to Screen for Porcine Cytomegalovirus (PCMV) in Donor Pigs for Xenotransplantation. Sci. Rep. 2022, 12, 21545. [Google Scholar] [CrossRef]
- Fishman, J.A. Infectious disease risks in xenotransplantation. Am. J. Transplant. 2018, 18, 1857–1864. [Google Scholar] [CrossRef]
- Denner, J. Porcine Lymphotropic Herpesviruses (PLHVs) and Xenotranplantation. Viruses 2021, 13, 1072. [Google Scholar] [CrossRef]
- Karuppannan, A.K.; Opriessnig, T. Possible risks posed by single-stranded DNA viruses of pigs associated with xenotransplantation. Xenotransplantation 2018, 25, e12453. [Google Scholar] [CrossRef]
- Jiang, M.; Xie, Z.; Fan, Q.; Tang, G.; Pan, N.; Zeng, H.; Xu, Y.; Zhang, C.; Luo, M.; Li, F. Epidemiological characteristics of Circovirus Human infection in hospitalized patients in a representative infectious disease hospital in Guangzhou, China, from Feb 2023 to Dec 2024. Diagn. Microbiol. Infect. Dis. 2025, 113, 116962. [Google Scholar] [CrossRef]
- Russell, G.C.; Stewart, J.P.; Haig, D.M. Malignant catarrhal fever: A review. Vet. J. 2009, 179, 324–335. [Google Scholar] [CrossRef]
- Denner, J. Porcine Endogenous Retroviruses and Xenotransplantation, 2021. Viruses 2021, 13, 2156. [Google Scholar] [CrossRef]
- Yang, L.; Güell, M.; Niu, D.; George, H.; Lesha, E.; Grishin, D.; Aach, J.; Shrock, E.; Xu, W.; Poci, J.; et al. Genome-wide inactivation of porcine endogenous retroviruses (PERVs). Science 2015, 350, 1101–1104. [Google Scholar] [CrossRef]
- Niu, D.; Wei, H.J.; Lin, L.; George, H.; Wang, T.; Lee, I.H.; Zhao, H.Y.; Wang, Y.; Kan, Y.; Shrock, E.; et al. Inactivation of porcine endogenous retrovirus in pigs using CRISPR-Cas9. Science 2017, 357, 1303–1307. [Google Scholar] [CrossRef] [PubMed]
- Jhelum, H.; Bender, M.; Reichart, B.; Mokelke, M.; Radan, J.; Neumann, E.; Krabben, L.; Abicht, J.M.; Kaufer, B.; Längin, M.; et al. Evidence for Microchimerism in Baboon Recipients of Pig Hearts. Viruses 2023, 15, 1618. [Google Scholar] [CrossRef] [PubMed]
- Denner, J. Monitoring for PERV Following Xenotransplantation. Transpl. Int. 2024, 37, 13491. [Google Scholar] [CrossRef] [PubMed]
- Krüger, L.; Kristiansen, Y.; Reuber, E.; Möller, L.; Laue, M.; Reimer, C.; Denner, J. A Comprehensive Strategy for Screening for Xenotransplantation-Relevant Viruses in a Second Isolated Population of Gottingen Minipigs. Viruses 2019, 12, 38. [Google Scholar] [CrossRef] [PubMed]
- Postel, A.; Hansmann, F.; Baechlein, C.; Fischer, N.; Alawi, M.; Grundhoff, A.; Derking, S.; Tenhündfeld, J.; Pfankuche, V.M.; Herder, V.; et al. Presence of atypical porcine pestivirus (APPV) genomes in newborn piglets correlates with congenital tremor. Sci. Rep. 2016, 6, 7735. [Google Scholar] [CrossRef] [PubMed]
- Opriessnig, T.; Huang, Y.W. SARS-CoV-2 does not infect pigs, but this has to be verified regularly. Xenotransplantation 2022, 29, e12772. [Google Scholar] [CrossRef]
- Denner, J. Sensitive detection systems for infectious agents in xenotransplantation. Xenotransplantation 2020, 18, e12594. [Google Scholar] [CrossRef]
- Denner, J. Limited availability of methods for the detection of xenotransplantation-relevant viruses in veterinary laboratories. Xenotransplantation 2024, 31, e12851. [Google Scholar] [CrossRef]
- Jhelum, H.; Kunec, D.; Papatsiros, V.; Kaufer, B.B.; Denner, J. Reliable Polymerase Chain Reaction Methods for Screening for Porcine Endogenous Retroviruses-C (PERV-C) in Pigs. Viruses 2025, 17, 164. [Google Scholar] [CrossRef]
- Morozov, V.A.; Heinrichs, G.; Denner, J. Effective Detection of Porcine Cytomegalovirus Using Non-Invasively Taken Samples from Piglets. Viruses 2017, 9, 9. [Google Scholar] [CrossRef] [PubMed]
- Denner, J. Commentary: Oral fluid testing can be used to monitor xenotransplant donor herds for porcine cytomegalovirus/roseolovirus status. Front. Vet. Sci. 2025, 12, 1571657. [Google Scholar] [CrossRef]
- Fiebig, U.; Abicht, J.M.; Mayr, T.; Längin, M.; Bähr, A.; Guethoff, S.; Falkenau, A.; Wolf, E.; Reichart, B.; Shibahara, T.; et al. Distribution of Porcine Cytomegalovirus in Infected Donor Pigs and in Baboon Recipients of Pig Heart Transplantation. Viruses 2018, 10, 66. [Google Scholar] [CrossRef]
- Halecker, S.; Krabben, L.; Kristiansen, Y.; Krüger, L.; Möller, L.; Becher, D.; Laue, M.; Kaufer, B.; Reimer, C.; Denner, J. Rare isolation of human-tropic recombinant porcine endogenous retroviruses PERV-A/C from Göttingen minipigs. Virol. J. 2022, 19, 30. [Google Scholar] [CrossRef] [PubMed]
- Mueller, N.J.; Barth, R.; Yamamoto, S.; Kitamura, H.; Patience, C.; Yamada, K.; Cooper, D.K.C.; Sachs, D.H.; Kaur, A.; Fishman, J.A. Activation of Cytomegalovirus in Pig-to-Primate Organ Xenotransplantation. J. Virol. 2002, 76, 4866–4872. [Google Scholar] [CrossRef]
- Chmielewicz, B.; Goltz, M.; Franz, T.; Bauer, C.; Brema, S.; Ellerbrok, H.; Beckmann, S.; Rziha, H.-J.; Lahrmann, K.-H.; Romero, C.; et al. A novel porcine gammaherpesvirus. Virology 2003, 308, 317–329. [Google Scholar] [CrossRef]
- McMahon, K.J.; Minihan, D.; Campion, E.M.; Loughran, S.T.; Allan, G.; McNeilly, F.; Walls, D. Infection of pigs in Ireland with lymphotropic gamma-herpesviruses and relationship to postweaning multisystemic wasting syndrome. Vet. Microbiol. 2006, 116, 60–66. [Google Scholar] [CrossRef]
- Chen, N.; Xiao, Y.; Li, X.; Li, S.; Xie, N.; Yan, X.; Li, X.; Zhu, J. Development and application of a quadruplex real-time PCR assay for differential detection of porcine circoviruses (PCV1 to PCV4) in Jiangsu province of China from 2016 to 2020. Transbound. Emerg. Dis. 2021, 68, 1615–1624. [Google Scholar] [CrossRef]
- Palinski, R.; Piñeyro, P.; Shang, P.; Yuan, F.; Guo, R.; Fang, Y.; Byers, E.; Hause, B.M. A Novel Porcine Circovirus Distantly Related to Known Circoviruses Is Associated with Porcine Dermatitis and Nephropathy Syndrome and Reproductive Failure. J. Virol. 2017, 91, e01879. [Google Scholar] [CrossRef] [PubMed]
- Opriessnig, T.; Shen, H.G.; Pal, N.; Ramamoorthy, S.; Huang, Y.W.; Lager, K.M.; Beach, N.M.; Halbur, P.G.; Meng, X.J. A Live-Attenuated Chimeric Porcine Circovirus Type 2 (PCV2) Vaccine Is Transmitted to Contact Pigs but Is Not Upregulated by Concurrent Infection with Porcine Parvovirus (PPV) and Porcine Reproductive and Respiratory Syndrome Virus (PRRSV) and Is Efficacious in a PCV2b-PRRSV-PPV Challenge Model. Clin. Vaccine Immunol. 2011, 18, 1261–1268. [Google Scholar] [CrossRef] [PubMed]
- Xiao, C.T.; GimeFnez-Lirola, L.; Huang, Y.W.; Meng, X.J.; Halbur, P.G.; Opriessnig, T. The prevalence of Torque teno sus virus (TTSuV) is common and increases with the age of growing pigs in the United States. J. Virol. Methods 2012, 183, 40–44. [Google Scholar] [CrossRef]
- Jothikumar, N.; Cromeans, T.L.; Robertson, B.H.; Meng, X.J.; Hill, V.R. A broadly reactive one-step real-time RT-PCR assay for rapid and sensitive detection of hepatitis E virus. J. Virol. Methods 2006, 131, 65–71. [Google Scholar] [CrossRef]
- Dagotto, G.; Mercado, N.B.; Martinez, D.R.; Hou, Y.J.; Nkolola, J.P.; Carnahan, R.H.; Crowe, J.E.; Baric, R.S.; Barouch, D.H. Comparison of subgenomic and total RNA in SARS-CoV-2- challenged rhesus macaques. J. Virol. 2021, 95, e02370-20. [Google Scholar] [CrossRef]
- Duvigneau, J.; Hartl, R.; Groiss, S.; Gemeiner, M. Quantitative simultaneous multiplex real-time PCR for the detection of porcine cytokines. J. Immunol. Methods 2005, 306, 16–27. [Google Scholar] [CrossRef]
- Behrendt, R.; Fiebig, U.; Norley, S.; Gürtler, L.; Kurth, R.; Denner, J. A neutralization assay for HIV-2 based on measurement of provirus integration by duplex real-time PCR. J. Virol. Methods 2009, 159, 40–46. [Google Scholar] [CrossRef]
- Walker, J.A.; Hughes, D.A.; Anders, B.A.; Shewale, J.; Sinha, S.K.; Batzer, M.A. Quantitative intra-short interspersed element PCR for species-specific DNA identification. Anal. Biochem. 2003, 316, 259–269. [Google Scholar] [CrossRef]
- Takeuchi, Y.; Patience, C.; Magre, S.; Weiss, R.A.; Banerjee, P.T.; Le Tissier, P.; Stoye, J.P. Host range and interference studies of three classes of pig endogenous retrovirus. J. Virol. 1998, 72, 9986–9991. [Google Scholar] [CrossRef] [PubMed]
- Plotzki, E.; Keller, M.; Ehlers, B.; Denner, J. Immunological methods for the detection of porcine lymphotropic herpesviruses (PLHV). J. Virol. Methods 2016, 233, 72–77. [Google Scholar] [CrossRef] [PubMed]
- Kaulitz, D.; Fiebig, U.; Eschricht, M.; Wurzbacher, C.; Kurth, R.; Denner, J. Generation of neutralising antibodies against porcine endogenous retroviruses (PERVs). Virology 2011, 411, 78–86. [Google Scholar] [CrossRef]
- Jhelum, H.; Papatsiros, V.; Papakonstantinou, G.; Krabben, L.; Kaufer, B.; Denner, J. Screening for Viruses in Indigenous Greek Black Pigs. Microorganisms 2024, 12, 315. [Google Scholar] [CrossRef] [PubMed]
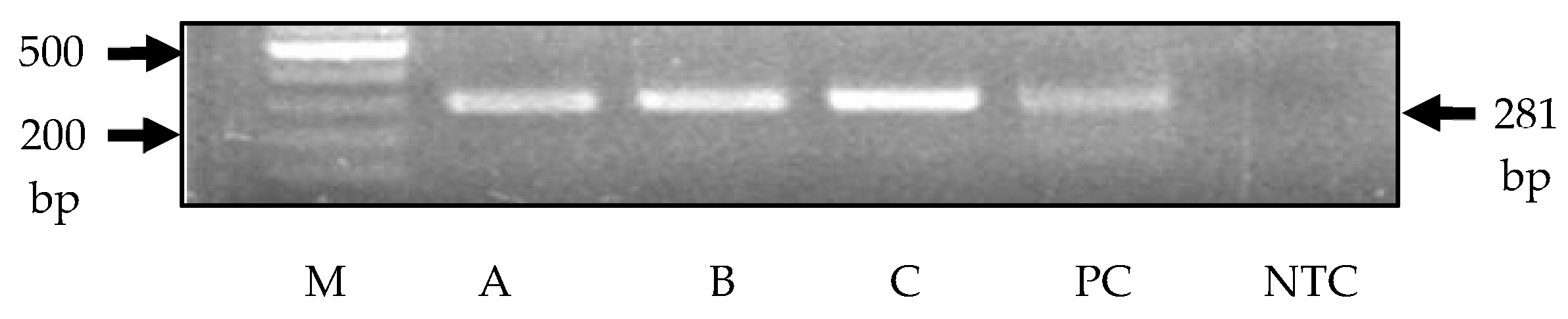
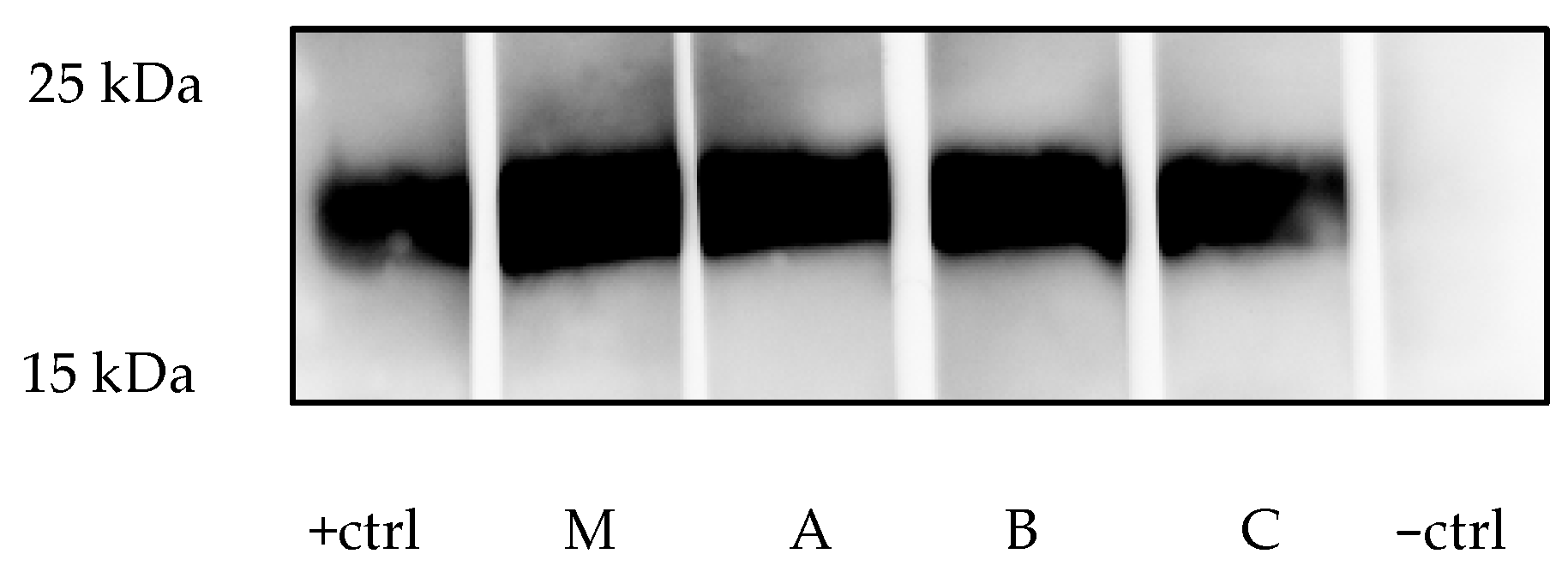
| Virus | Primer/Probe | Sequence (5′ to 3′) | Reference |
|---|---|---|---|
| PCMV/PRV | PCMV-Fwd | ACTTCGTCGCAGCTCATCTGA | Müller et al., 2002 [43] |
| PCMV-Rev | GTTCTGGGATTCCGAGGTTG | ||
| PCMV-Probe | 6FAM-CAGGGCGGCGGTCGAGCTC-BHQ | ||
| PLHV-1 | PLHV-1 (1125)-Fwd | CTCACCTCCAAATACAGCGA | Chmielewicz et al., 2003 [44] |
| PLHV-1 (1125) Rev | GCTTGAATCGTGTGTTCCATAG | ||
| PLHV-1 (1125) probe | 6FAM-CTGGTCTACTGAATCGCCGCTAACAG-TAMRA | ||
| PLHV-2 | PLHV-2 (1155s) | GTCACCTGCAAATACACAGG | Chmielewicz et al., 2003 [44] |
| PLHV-2 (1155as) | GGCTTGAATCGTATGTTCCATAT | ||
| PLHV-2 (1155) probe | 6FAM-CTGGTCTACTGAAGCGCTGCCAATAG-TAMRA | ||
| PLHV-3 | PLHV-3 (210s)-Fwd | AACAGCGCCAGAAAAAAAGG | McMahon et al., 2006 [45] |
| PLHV-3 (210as)-Rev | GGAAAGGTAGAAGGTGAACCATAAAA | ||
| PLHV-3 (210)-probe | 6FAM-CCAAAGAGGAAAATC-MGB | ||
| PCV1 | PCV1 Fwd | AACCCCATAAGAGGTGGGTGTT | Chen et al., 2021 [46] |
| PCV1 Rev | TTCTACCCTCTTCCAAACCTTCCT | ||
| PCV1 probe | 6FAM-TCCGAGGAGGAGAAAAACAAAATACGGGA-BHQ1 | ||
| PCV2 | PCV2 Fwd | CTGAGTCTTTTTTATCACTTCGTAATGGT | Chen et al., 2021, [46] |
| PCV2 Rev | ACTGCGTTCGAAAACAGTATATACGA | ||
| PCV2 probe | ROX-TTAAGTGGGGGGTCTTTAAGATTAAATTCTCTGAATTGT-BHQ2 | ||
| PCV3 | PCV3 Fwd | AGTGCTCCCCATTGAACG | Palinski et al., 2017 [47] |
| PCV3 Rev | ACACAGCCGTTACTTCAC | ||
| PCV3 probe | 6FAM-ACCCCATGGCTCAACACATATGACC-BHQ1 | ||
| PCV4 | PCV4 Fwd | ATTATTAAACAGACTTTATTTGTGTCATCACTT | Chen et al., 2021 [46] |
| PCV4 Rev | ACAGGGATAATGCGTAGTGATCACT | ||
| PCV4 probe | 6FAM-ATACTACACTTGATCTTAGCCAAAAGGCTCGTTGA-BHQ1 | ||
| PPV-1 | PPV-1 Fwd | CAGAATCAGCAACCTCACCA | Opriessnig et al., 2011 [48] |
| PPV-1 Rev | GCTGCTGGTGTGTATGGAAG | ||
| PPV-1 probe | 6FAM-TGCAAGCTT/ZEN/AATGGTCGCACTAGACA-BHQ1 | ||
| TTSuV1 | TTSuV1-Fwd | CGAATGGCTGAGTTTATGCC | Xiao et al., 2012 [49] |
| TTSuV1-Rev | GATAGGCCCCTTGACTCCG | ||
| TTSuV1-probe | 6FAM-AACTGTCTA/ZEN/GCGACTGGGCGGGT-3IABkFQ | ||
| TTSuV2 | TTSuV2-Fwd | CGAATGGCTGAGTTTATGCC | Xiao et al., 2012 [49] |
| TTSuV2-Rev | GATAGGCCCCTTGACTCCG | ||
| TTSuV2-probe | 6FAM-AACAGAGCT/ZEN/GAGTGTCTAACCGCCTG-3IABkFQ | ||
| HEV | HEV-Fwd | GGTGGTTTCTGGGGTGAC | Jothikumar et al., 2006 [50] |
| HEV-Rev | AGGGGTTGGTTGGATGAA | ||
| HEV-probe | 6FAM-TGATTCTCAGCCCTTCGC-BHQ | ||
| APPV | APPV_5587-Fwd | CAGAGRAAAGGKCGAGTGGG | Postel et al., 2016 [34] |
| APPV_5703-Rev | ACCATAYTCTTGGGCCTGSAG | ||
| APPV_CT-59 probe | 6FAM-ACTACTATCCTTCGGGGGTAGTACCGA-BHQ1 | ||
| SARS-CoV-2 | ORF1ab.F | GGCCAATTCTGCTGTCAAATTA | Dagotta et al., 2021 [51] |
| ORF1ab.F | CAGTGCAAGCAGTTTGTGTAG | ||
| probe | 6FAM-ACAGATGTCTTGTGCTGCCGGTA-BHQ1 | ||
| pGAPDH | pGAPDH-Fwd | GATCGAGTTGGGGCTGTGACT | Duvigneau et al., 2005 [52] |
| pGAPDH-Rev | ACATGGCCTCCAAGGAGTAAGA | ||
| pGAPDH-probe | HEX-CCACCAACCCCAGCAAGAG-BHQ | ||
| hGAPDH | hGAPDH-Fwd | GGCGATGCTGGCGCTGAGTAC | Behrendt et al., 2009 [53] |
| hGAPDH-Rev | TGGTTCACACCCATGACGA | ||
| hGAPDH-probe | HEX-CTTCACCACCATGGAGAAGGCTGGG-BHQ1 | ||
| PERVpol | PERV pol Fwd | CGACTG CCCCAAGGG TTC AA | Yang et al., 2015 [29] |
| PERV pol Rev | TCTCTCCTG CAA ATC TGG GCC | ||
| PERV pol probe | 6FAM-CACGTACTG GAG GAG GGTCACCTG -BHQ1 | ||
| PRE-1 | PRE-1 Fwd | GACTAGGAACCATGAGGTTGCG | Walker et al., 2003 [54] |
| PRE-1 Rev | AGCCTACACCACAGCCACAG | ||
| PRE-1 probe | FAM-TTTGATCCCTGGCCTTGCTCAGTGG-BHQ1 | ||
| PERV-C * | PERV-C Fwd PERV-C Rev | CTGACCTGGATTAGAACTGG ATGTTAGAGGATGGTCCTGG | Takeuchi et al., 1998 [55] |
| Virus | Method | Sensitivity (Copy Number per 100 ng DNA) | Sensitivity R2 | Reference |
|---|---|---|---|---|
| PCMV/PRV a | conventional PCR | 15 copies | Morozov et al., 2016 [21] | |
| nested PCR | 5 copies | |||
| real-time PCR | 2 copies | |||
| real-time PCR | 10 copies | 0.9964 | Jhelum et al., 2024 [58] | |
| HEV3 | real-time RT-PCR | 150–200 copies | Morozov et al., 2015 [18] | |
| real-time RT-PCR | 10 copies | 0.9962 | Jhelum et al., 2024 [58] | |
| PCV2 | real-time PCR | 1 copy | 0.9935 | Jhelum et al., 2024 [58] |
| PCV3 | real-time PCR | 10 copies | 0.9906 | Jhelum et al., 2024 [58] |
| PCV4 | real-time PCR | 100 copies | 0.9906 | Jhelum et al., 2024 [58] |
| PLHV-1 | real-time PCR | 1 copy | 0.9964 | Jhelum et al., 2024 [58] |
| PLHV-2 | real-time PCR | 1 copy | 0.9953 | Jhelum et al., 2024 [58] |
| PLHV-3 | real-time PCR | 1 copy | 0.9983 | Jhelum et al., 2024 [58] |
| PPV1 | real-time PCR | 10 copies | 0.9961 | Jhelum et al., 2024 [58] |
| Animal | PCMV/PRV | PLHV-1 | PLHV-2 | PLHV-3 | PCV1 | PCV2 | PCV3 | |||||||
| Virus ct | GAPDH ct | Virus ct | GAPDH ct | Virus ct | GAPDH ct | Virus ct | GAPDH ct | Virus ct | GAPDH ct | Virus ct | GAPDH ct | Virus ct | GAPDH ct | |
| A | n.d. | 21.66 | n.d. | 21.12 | n.d. | 21.35 | n.d. | 21.92 | 26.66 | 21.91 | n.d. | 21.30 | n.d. | 21.98 |
| B | n.d. | 21.62 | n.d. | 21.06 | n.d. | 21.81 | n.d. | 21.88 | n.d. | 21.54 | n.d. | 21.36 | n.d. | 21.63 |
| C | n.d. | 21.94 | n.d. | 21.78 | n.d. | 21.33 | n.d. | 21.19 | n.d. | 21.05 | n.d. | 21.55 | n.d. | 21.37 |
| Animal | PCV4 | PPV-1 | TTSuV1 | TTSuV2 | APPV | HEV | SARS-CoV-2 | |||||||
| Virus ct | GAPDH ct | Virus ct | GAPDH ct | Virus ct | GAPDH ct | Virus ct | GAPDH ct | Virus ct | GAPDH ct | Virus ct | GAPDH ct | Virus ct | GAPDH ct | |
| A | n.d. | 21.32 | n.d. | 21.62 | n.t. | n.t. | n.t. | n.t. | n.d. | 30.76 | n.d. | 30.02 | n.t. | n.t. |
| B | n.d. | 21.59 | n.d. | 21.41 | n.t. | n.t. | n.t. | n.t. | n.d. | 30.98 | n.d. | 30.50 | n.t. | n.t. |
| C | n.d. | 21.40 | n.d. | 21.04 | n.t. | n.t. | n.t. | n.t. | n.d. | 30.01 | n.d. | 30.46 | n.t. | n.t. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jhelum, H.; Kaufer, B.B.; Denner, J. Comprehensive Protocols for Detecting Xenotransplantation-Relevant Viruses. Methods Protoc. 2025, 8, 109. https://doi.org/10.3390/mps8050109
Jhelum H, Kaufer BB, Denner J. Comprehensive Protocols for Detecting Xenotransplantation-Relevant Viruses. Methods and Protocols. 2025; 8(5):109. https://doi.org/10.3390/mps8050109
Chicago/Turabian StyleJhelum, Hina, Benedikt B. Kaufer, and Joachim Denner. 2025. "Comprehensive Protocols for Detecting Xenotransplantation-Relevant Viruses" Methods and Protocols 8, no. 5: 109. https://doi.org/10.3390/mps8050109
APA StyleJhelum, H., Kaufer, B. B., & Denner, J. (2025). Comprehensive Protocols for Detecting Xenotransplantation-Relevant Viruses. Methods and Protocols, 8(5), 109. https://doi.org/10.3390/mps8050109







