Compendium of Agrobacterium-Mediated Tissue Culture Transformation Methods of Various Solanaceae Species
Abstract
1. Introduction
2. Experimental Design
2.1. Chemicals
- Murashige & Skoog (MS) Salts (Duchefa, Netherlands; Cat. no.: M0221).
- Daishin Agar (Duchefa, Netherlands, Cat. no.: D1004; CAS: 9002–18–0). Note: density ≥ 1000 g/cm2.
- Naphtalenacetic acid (Duchefa, Netherlands; Cat. no.: N0903; CAS: 86–87–3).
- Acetosyringone (Merck, Germany; Cat. no.: D134406, CAS: 2478–38–8).
- Amphotericin B fungicide (Merck, Germany; Cat. no.: A2411; CAS: 1397–89–3).
- Cefotaxime sodium anti-bacterial (Duchefa, Netherlands; Cat. no.: C0111, stored at 4 °C).
- Basta herbicide (CAS: 77182–82–2, 183 g/L Glufosinate; liquid, stored at RT). Note: as applicable, check for regional law restrictions.
- 6-Benzylaminopurine (Duchefa, Netherlands; Cat. no.: B0904; CAS: 1214–39–7).
- Indole-3-acetic acid (Merck, Germany; Cat. no.: I2886; CAS: 87–51–4).
- Gibberellic acid 3 (GA3) (Duchefa, Netherlands; Cat. no.: G0907; CAS: 77–06–5).
- Hygromycin B (Duchefa, Netherlands; Cat. no.: H0192; CAS: 31282–04–9).
- Kanamycin monosulfate (Duchefa, Netherlands; Cat. no.: K0126, CAS: 25389–94–0).
- Ticarcillindisodium/potassium-clavulanate (Duchefa, Netherlands; Cat. no: T0190).
- trans-Zeatin (Duchefa, Netherlands; Cat. No.: Z0917, CAS: 1637–39–4).
- Vancomycin hydrochloride (Duchefa, Netherlands; Cat. no.: V0155, CAS: 1404–93–9).
- Zeatin riboside (Duchefa, Netherlands; Cat. no.: Z0937, CAS: 6025–53–2).
2.2. Equipment
- Standard laboratory equipment:Petri dishes, pH meter, hot plate with magnetic stirrer, analytical balance, refrigerator, freezer, water purification system, autoclave, spatulas, forceps, laminar flow hood, and plant cultivation chambers (soil-free).
- Canning Jars type “Sturz-Gläser” ¼ Liter (height: 7 cm; top-diameter: 10.5 cm) and ½ (height: 11 cm; top-diameter: 10.5 cm) Liter (Weck, Bonn, Germany): Baked-sterilized: 180 °C for 10 h (Binder Heating Chamber).
- Razor-blade holder and beard-shaving quality razor blades (a very sharp edge is essential).
- Tea ball sieves wrapped in aluminum foil and autoclaved.
3. Procedures
3.1. Cleanliness and Working Aseptically
3.2. Sterilization of Glass- and Metalware
3.3. Working with Agrobacteria
3.4. Climate Chamber Conditions
3.5. Stable Tissue Culture Transformation of Solanum Lycopersicum (Tomato) ‘Moneymaker’
3.5.1. Seed Sterilization (1 h)
- Swirling for 3 min by hand in 200 mL 70% ethanol.
- Swirling for 10 min by hand in 200 mL 1.25% sodium-hypochlorite containing a few drops of 0.001% Triton X-100 (in sterile water).
- Rinsing by passaging in 3 × 200 mL for 30–60 s each in sterile double-distilled or ion-exchanged deionized water.
- Remove the seeds from the tea sieves by depositing them into a dry, sterile jar.
- Let the seeds dry under the laminar flow hood overnight.
- Store the dried, sterile seeds in the same jar for two days in the dark at 4 °C.
3.5.2. Preparing Cotyledon Material for Transformation
- Climate chamber conditions: see Section 3.4.
- Sowing the seeds (1 to 3 h)
 CRITICAL STEP The seeds should germinate in absolute darkness. The cotyledons should be about 1 cm long, still etiolated, and the first true leaves not yet developed (see also [19,20]). See Figure 3 and Figure A2.
CRITICAL STEP The seeds should germinate in absolute darkness. The cotyledons should be about 1 cm long, still etiolated, and the first true leaves not yet developed (see also [19,20]). See Figure 3 and Figure A2.- 3.
- Isolating the cotyledons for transformation (3 h/construct plus 2 days)
- Apply a few drops of the Tomato Liquid Germination Medium (ToLGM, Section 5.3.3) onto a sterile Petri dish so that the plant material does not dry out while cutting it.
- Excise the cotyledons from the seedlings by cutting directly below the leaves and discarding the rest.
- Remove the top and base of the leaf and make a small incision along the long axis near the center of each leaf (Figure 3).
 CRITICAL Do not squeeze or crush the cotyledons.
CRITICAL Do not squeeze or crush the cotyledons.- Transfer the prepped cotyledons onto the Tomato Conditioning Medium (ToCM, Section 5.3.4) with the adaxial (upper) surface down, in full contact with the medium. Usually 10 Petri dishes are enough, with each dish holding about 20 cotyledons with enough space between them and the walls (Figure 3). If a Petri dish does become contaminated, discard it immediately.OPTIONAL: If possible, use only cotyledons that are not rolled, as they are very difficult to lay out on the selection plates and need to have good contact with the media to be selected properly.
- Incubate the cotyledons for 2 days in absolute darkness at 22 °C.
3.5.3. Preparing the Agrobacterium Suspension for Tomato
- Plate growth of Agrobacteria (2 h)Streak out the Agrobacteria on YEB-plates (Section 5.1.1) with the appropriate selection antibiotics roughly at the same time you start sowing the seeds (Section 3.5.2 step 2). Re-streak them two days before cutting the cotyledons (Section 3.5.2 step 3). This should fall on the Monday after the Friday you started cutting.
- 2.
- Pre-culture liquid growth of Agrobacteria (1 day)Inoculate 3 mL low-salt LB (ls-LB, Section 5.1.2) with one single colony with the appropriate selection antibiotics. Shake overnight at 180 rpm at 28 °C. This should fall on the Wednesday you started cutting the cotyledons on (Section 3.5.2 step 3).
- 3.
- Agrobacteria infection culture (1 day)One day before co-cultivation (Section 3.5.4), inoculate 1 mL of the pre-culture (Section 3.5.3 step 2) into 100 mL bacterial growth medium (BGM; Section 5.1.3) with the appropriate antibiotics. Shake overnight at 180 rpm at 28 °C. This should fall on the Thursday after you cut the cotyledons on (Section 3.5.2 step 3).
3.5.4. Transformation: Infection and Co-Cultivation of Plant Material with Agrobacteria
- Bring the Agrobacteria to the infection concentration (2 h)
- Centrifuge the infection culture (Section 3.5.3 step 3) for 10 min at 3000× g, RT.
- Resuspend the bacteria in 10 mM MgSO4 with 200 µM acetosyringone and adjust the OD600 to 1.0, in a final volume of 50 mL.
- 2.
- Co-cultivate Agrobacteria with the wounded cotyledon tissue (2 days)
- Dispense about 1 drop to each cotyledon with a 1000 µL pipette. Total volume should roughly be 2 mL per Petri dish.
- Incubate the co-cultivation for 2 days in the dark at 22 °C.
3.5.5. Selection of Transformed Tissue
- Climate chamber conditions: see Section 3.4
- Initial transfer to selection medium (3 days)
- Transfer the cotyledons adaxial (upper) side up to small canning jars (1/4 L) with the Tomato Selection Medium (ToSM, Section 5.3.5). This should fall on the Monday after co-cultivation (Section 3.5.4 step 2).
CRITICAL If you are selecting with Kan, use 35 mg/L at this point. For other antibiotics, see the Reagent Setup Section.
- Incubate for 3 days in the climate chamber.
- 3.
- Sub-culturing for shoot and callus isolation (8 to 10 weeks)
- Transfer the cotyledons to fresh ToSM in small canning jars (1/4 L) once a week.
CRITICAL If you are selecting with Kan, use 50 mg/L at this point for two weeks, and thereafter 100 mg/L Kan.
- After a couple of weeks, the first callus and some with shoots should form.
- Once identifiable, place callus and shoots in direct contact with ToSM.
 CRITICAL Shoots are to be cut near the base; avoid any old or callus-like tissues.
CRITICAL Shoots are to be cut near the base; avoid any old or callus-like tissues.3.5.6. Regeneration of Transgenic Tomato Plants (2 to 4 Weeks)
 CRITICAL If you are selecting with Kan, use 20 mg/L. OPTIONAL: If you have any trouble with rooting, try leaving out the selection antibiotic.
CRITICAL If you are selecting with Kan, use 20 mg/L. OPTIONAL: If you have any trouble with rooting, try leaving out the selection antibiotic.- The callus should be kept, as more shoots will form over time. You can discard calli that lose their vitality (yellow or brown). This is useful in case it is difficult to readily identify transgenic plants.
- Do not let the shoots touch the top of the jars to maintain axenic culture. You may remove the top of the plants and transfer the cuttings to fresh ToRM.
- You can bring the plants to the greenhouse as soon as the roots are well visible and moderately extensive (~1 to 3 cm long with lateral roots, see Figure A3).
- Tomato plants take up a lot of space and take time to flower and set seed; therefore, we advise that you test the transgenic status of each plant as soon as possible in order to preserve greenhouse space and assay if the transformation vector was performing as desired.
3.6. Stable Tissue Culture Transformation of Solanum Tuberosum (Potato)
3.6.1. Plant Material (Continuous, Every Four Weeks)
- You will need about 25 sterile plants, three to four weeks old
3.6.2. Preparing the Agrobacterium Suspension for Potato
- Plate growth of Agrobacteria (2 days)
- 2.
- Pre-culture liquid growth of Agrobacteria (1 day)
- 3.
- Agrobacteria infection culture (2 h)
3.6.3. Transformation: Infection and Co-Cultivation of Plant Material with Agrobacteria (2 h/Construct Plus 2 to 3 Days)
- Dispense 10 mL of liquid MS Medium (PoMS, Section 5.4.2) into 8 Petri dishes (9 cm Ø).
- Excise leaves from the potato tissue culture, immediately remove the petiole and a bit of the leaf base, and apply two tiny transverse cuts to the mid-vein of the abaxial surface.
- Plan for two hours per construct for leaf excision.
- Place the cut leaf in the liquid PoGM (Section 5.4.3), adaxial side facing down, directly in contact with the medium.
 CRITICAL The leaves are very sensitive to any kind of injury, i.e., being burned or handled too harshly. Therefore, never squeeze the tissue too hard, and let the forceps cool down to room temperature after flame sterilization. Use very sharp razor blades to avoid microscopic bludgeoning damage. Finally, use healthy, vibrant, green leaves from the top of the plants.
CRITICAL The leaves are very sensitive to any kind of injury, i.e., being burned or handled too harshly. Therefore, never squeeze the tissue too hard, and let the forceps cool down to room temperature after flame sterilization. Use very sharp razor blades to avoid microscopic bludgeoning damage. Finally, use healthy, vibrant, green leaves from the top of the plants.- You should prepare around 100 leaves in total, roughly 12 to 15 leaves per Petri dish (Figure A4).
- Apply 50 µL of the Agrobacterium infection culture (Section 3.6.2 step 3) directly to the Petri dish and shake lightly for 3 to 5 min on a shaker.
- Incubate the co-culture plates in the dark at 22 °C for 2 to 3 days.
3.6.4. Selection of Transformed Tissue (8 to 10 Weeks)
- Transfer the leaves to the Potato Callus Induction Medium (PoCIM, Section 5.4.4), adaxial side facing down, in direct contact with the medium (Figure A5).
- After one week, transfer the leaves to the Potato Selection Medium (PoSM, Section 5.4.5), adaxial side facing down, in direct contact with the medium.
 WARNING Leaves and the jars with contaminated with fungi are best just discarded. OPTIONAL: Uninfected leaves adjacent to infected material can be transferred to PoSM with fungicide amphotericin (Section 5.2.1) and sealed with Parafilm.
WARNING Leaves and the jars with contaminated with fungi are best just discarded. OPTIONAL: Uninfected leaves adjacent to infected material can be transferred to PoSM with fungicide amphotericin (Section 5.2.1) and sealed with Parafilm. WARNING: Leaves with a strong Agrobacterium infection should also be discarded. OPTIONAL: As above, try removing uninfected material to fresh jars, and you can try washing infected leaves with 10 mM MgSO4 plus 250 mg/L cefotaxime (Section 5.2.2) and isolate them from healthy tissue in their own, separate jars. If necessary, this can be performed every few weeks; washing is more effective than just adding it to the medium.
WARNING: Leaves with a strong Agrobacterium infection should also be discarded. OPTIONAL: As above, try removing uninfected material to fresh jars, and you can try washing infected leaves with 10 mM MgSO4 plus 250 mg/L cefotaxime (Section 5.2.2) and isolate them from healthy tissue in their own, separate jars. If necessary, this can be performed every few weeks; washing is more effective than just adding it to the medium.- Transfer to fresh medium every 10 days until callus appears or all the tissue dies. OPTIONAL: As long as there are no shoots, you may perform this and all of the above steps in Petri dishes if desired.
- 2.
- Callus isolation and cultivation
- As soon as you can clearly identify callus forming, you should move the callus to separate jars for further cultivation by cutting them free from any attached leaf tissue.
- Continue to transfer the callus every 10 days and expect shoot formation in a few weeks.
3.6.5. Regeneration of Transgenic Potato Plants (2 to 4 Weeks)
- After a couple of weeks, the first shoots should form. The shoots will be delicate and thin, but vital (Figure A6). Fresh shoots should not be allowed to touch the top of the jars to maintain axenic culture. They should be excised and moved to new jars. As they get taller, move them to larger jars (1/2 L).
 CRITICAL Shoots are to be cut near the base; avoid any old or callus-like tissues.
CRITICAL Shoots are to be cut near the base; avoid any old or callus-like tissues.- After approximately 2 months, shoots that are green and have at least one to two leaves can be cut and transferred to the Potato Rooting Medium (PoRM, Section 5.4.6). Keep in mind that potato shoots coming out of the calli will be thin and delicate (Figure A6).
- You can bring the plants to the greenhouse as soon as the roots are visible and minimally extensive. As with the shoots, the roots will be thin and fine.
- The callus should be kept, as more shoots will form over time. You can discard calli that lose their vitality (yellow or brown). This is useful in case it is difficult to readily identify transgenic plants.
3.7. Stable Tissue Culture Transformation of Nicotiana Benthamiana and Nicotiana Tabacum
3.7.1. Seed Preparation (2 Days)
- Surface-sterilize 10 to 25 seeds by placing them in Eppendorf tubes. Working under the laminar flow hood, passage the seeds by the following process:
- Add 1 mL 70% ethanol and place on a shaker for 3 min. Decant or remove by pipetting.
- Add 1 mL of 1.5% sodium hypochlorite containing a few drops of 0.001% Triton X-100 and place on a shaker for 8–10 min. Decant or remove by pipetting.
- Wash with 1 mL for 1 min in sterile double-distilled or ion-exchanged deionized water. Decant or remove by pipetting.
- ○
- Repeat this wash step two more times.
- Resuspend the seeds in 0.1% agarose (Section 5.5.1) and distribute on Nicotiana Growth Medium (NicoGM, Section 5.5.2) in jars or Petri dishes. You may find with practice that you can dispense the seeds just in pure, sterile water.
- Place the jars in the dark and cold (4 °C) for 2 days.
- Bring the vernalized seeds to your cultivation chamber.
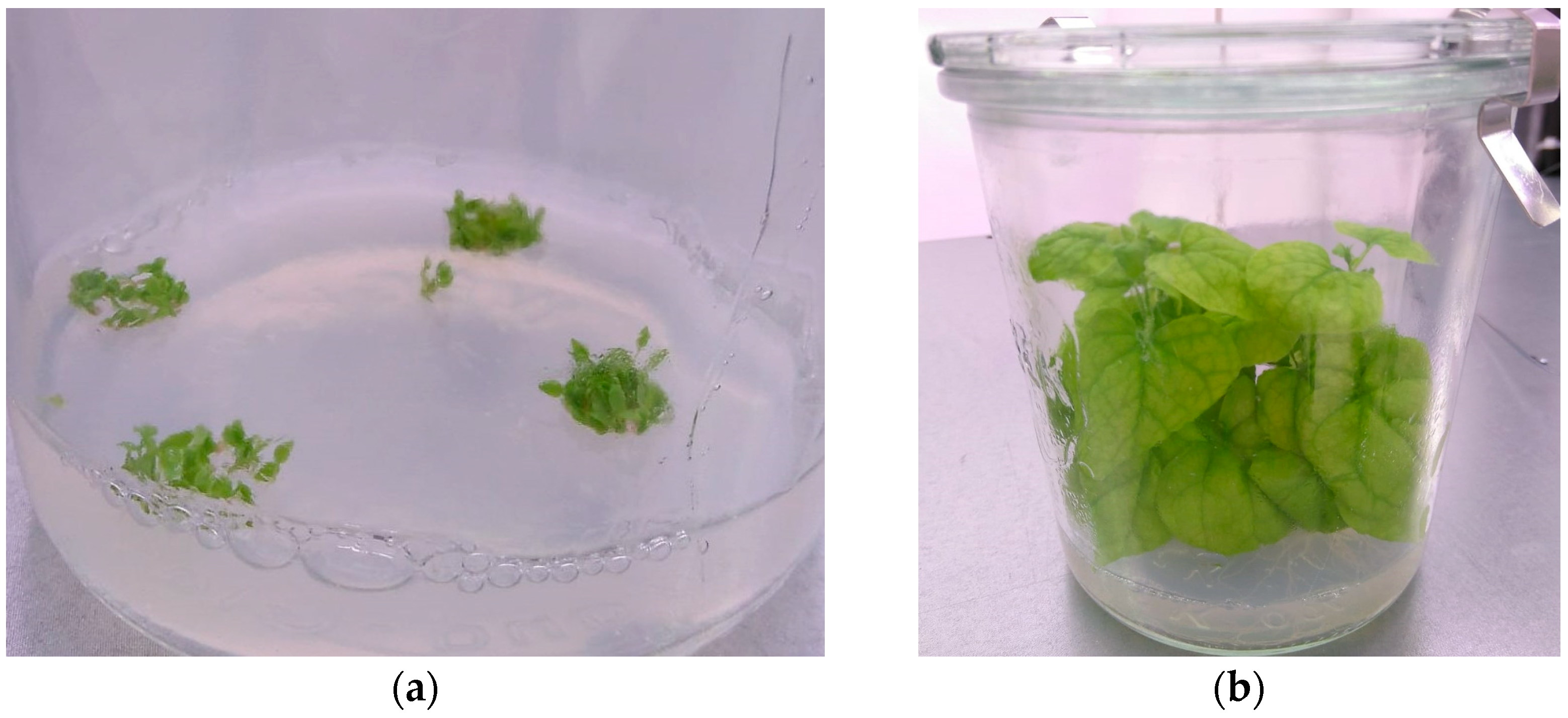
- Grow N. benthamiana for 7–8 weeks; you need 6 plants per transformation (example in Figure 5).
3.7.2. Preparing the Agrobacterium Suspension for Nicotiana
- Plate growth of Agrobacteria (2 days)
- 2.
- Pre-culture liquid growth of Agrobacteria (1 day)
- 3.
- Agrobacteria infection culture (1 day)
3.7.3. Infection and Co-Cultivation of Plant Leaves with Agrobacterium (2 Days)
- Collect the bacteria from the infection culture (Section 3.7.2 step 3) by centrifugation at 3000× g for 5 to 10 min and resuspend the pellet in sterile 10 mM MgCl2 without antibiotics; adjust the OD600 to 1.0, in a final volume of 100 mL. This should now fall on a Wednesday.
- Place ~30 mL of the suspension into 2 to 3 sterile Petri dishes (Ø 9 cm). You will be dipping the leaf explants into the Agrobacterium solution.
- Cut the leaves into about 30–35 square pieces per sterile Petri dish, avoiding the central vein:
- For N. tabacum 0.5 × 0.5 cm squares
- For N. benthamiana 1 × 1 cm squares
 CRITICAL Use green and healthy leaves. Do not let the leaf pieces overlap.
CRITICAL Use green and healthy leaves. Do not let the leaf pieces overlap. CRITICAL The leaves are sensitive to any kind of injury; if burned, singed, or handled too harshly, the tissue will die. Therefore, never squeeze the tissue and make sure to let the forceps cool down to RT after flame sterilization. Try to use very sharp razor blades to mitigate bludgeoning damage.
CRITICAL The leaves are sensitive to any kind of injury; if burned, singed, or handled too harshly, the tissue will die. Therefore, never squeeze the tissue and make sure to let the forceps cool down to RT after flame sterilization. Try to use very sharp razor blades to mitigate bludgeoning damage.- 4.
- Incubate the explants abaxial-side down in the bacteria suspension for 3 min with gentle shaking.
- 5.
- Transfer the explants to Nicotiana Growth Medium (NicoGM, 5.5.2), abaxial surface down, in contact with the medium.
- 6.
- About 100 pieces are enough for one transformation (about 3 Petri dishes).
- 7.
- Incubate them for 2 days in the dark at RT.
- 8.
- At any time during the procedure, if you see excessive Agrobacteria growth on the explant tissue, exchange the plates with fresh ones before proceeding.
3.7.4. Selection of Transformed Callus Tissue (Typically 3 to 6 Weeks)
- Transfer leaves to Nicotiana Selection Medium (NicoSM, Section 5.5.3) in small jars. This should fall on a Friday after co-cultivation (Section 3.7.3 step 7).
 CAUTION At the beginning, leaves with strong Agrobacterium infection should be outright discarded or washed in 10 mM MgSO4 containing 250 mg/L cefotaxime and incubated separately on NicoSM plates, away from the other explants. If necessary, this can be performed every few weeks; washing is more effective than just adding it to the medium.
CAUTION At the beginning, leaves with strong Agrobacterium infection should be outright discarded or washed in 10 mM MgSO4 containing 250 mg/L cefotaxime and incubated separately on NicoSM plates, away from the other explants. If necessary, this can be performed every few weeks; washing is more effective than just adding it to the medium.- 2.
- The leaves must be transferred to fresh NicoSM medium every 7 to 10 days.
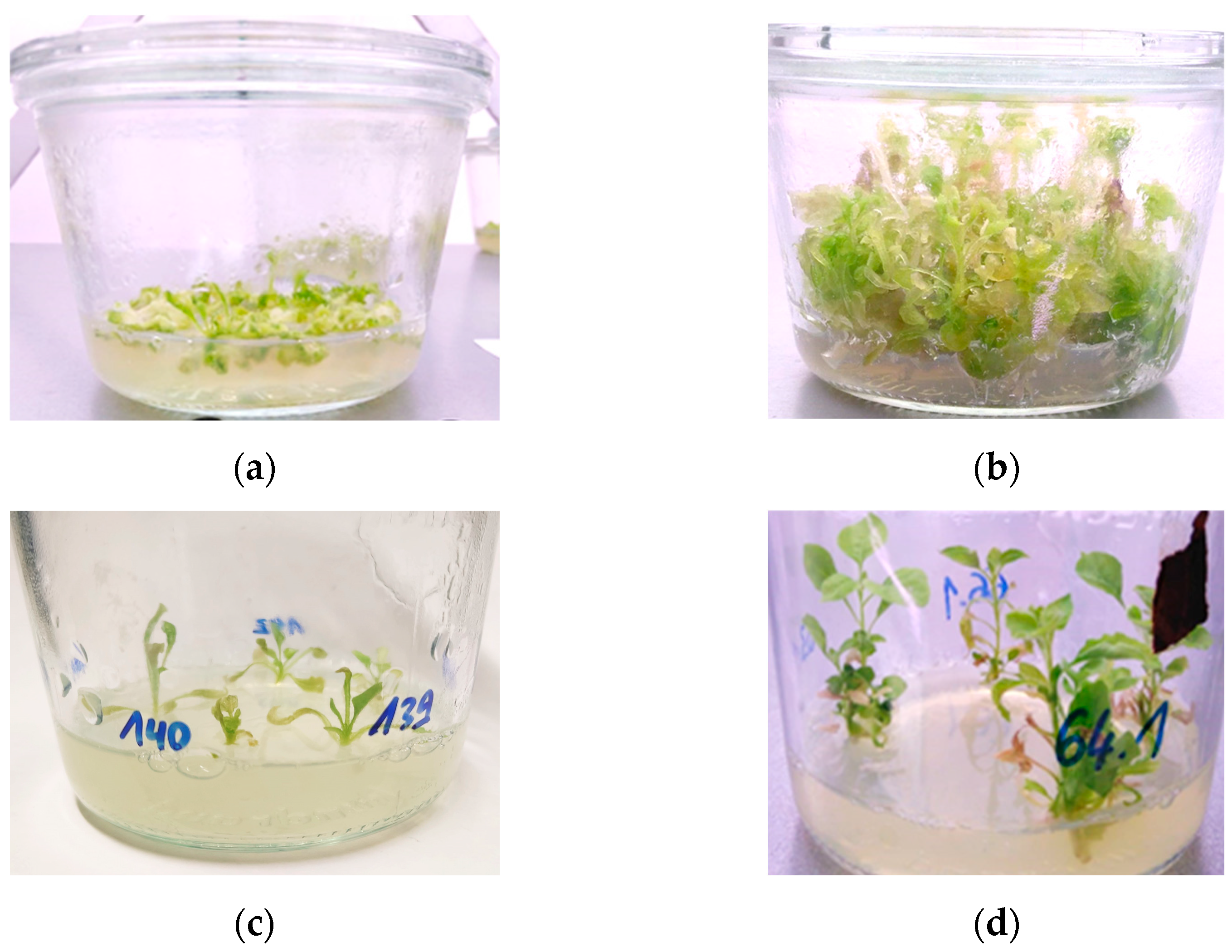
- As soon as you can clearly identify callus forming, you should move the callus to separate jars by excising it free from any attached leaf tissue. Maintain the callus on NicoSM every 10 days as well.
3.7.5. Regeneration of Transgenic Plants (Approximately 2 Months)
- Now the shoots should be large enough to be excised and moved to large jars with Nicotiana Rooting Medium (NicoRM, Section 5.5.4; Figure 6c).
- Move the plants to the greenhouse once they have several branches, leaves, and roots that are visible and moderately extensive (Figure 6d).
- The callus should be kept, as more shoots will form over time. You can discard calli that lose their vitality (yellow or brown). This is useful in case it is difficult to readily identify transgenic plants.
- Do not let the shoots touch the top of the jars to maintain axenic culture. You may remove the top (2 to 3 cm) of the plants and transfer the cuttings to fresh NicoRM.
4. Expected Results, Notes, and Discussion
5. Reagent Setup
5.1. Agrobacteria Media
5.1.1. YEB 1x
5.1.2. (Low-Salt) lsLB/LB 1x
5.1.3. Bacteria-Growth Medium (BGM) 1x
5.1.4. Acetosyringone 400 mM
5.1.5. Antibiotics for Bacteria
5.2. Hormones and Antibiotics for Plants
5.2.1. Amphotericin B Fungicide (ABF) 5 mg/mL
5.2.2. Cefotaxime Sodium (Cef) 250 mg/mL
5.2.3. Basta Herbicide (PPT) 20 g/L
5.2.4. 6-Benzylaminopurine (BAP) 1 mg/mL
5.2.5. Indole-3-Acetic Acid (IAA) 1 mg/mL
5.2.6. Gibberellic Acid (GA3) 20 µg/mL
5.2.7. Hygromycin B (Hyg) 12 mg/L or 15 mg/L
5.2.8. Naphtalenacetic Acid (NAA) 1 mg/mL
5.2.9. Kanamycin Monosulfate (Kan) 50 mg/mL
5.2.10. Ticarcillin Disodium and Clavulanate Potassium (15:1) (TiCla) 250 mg/mL
5.2.11. trans-Zeatin (t-Z) 1 mg/mL
5.2.12. Vancomycin Hydrochloride (Van) 125 mg/mL
5.2.13. Zeatin Riboside (ZR) 1.4 mg/mL
5.3. Plant Media for Tomato
5.3.1. NPT Vitamins Mix (NPT)
5.3.2. Tomato Medium (ToMe) 1 L
5.3.3. Tomato Liquid Germination Medium (ToLGM) 500 mL
5.3.4. Tomato Conditioning Medium (ToCM) 1 L
5.3.5. Tomato Selection Medium (toSM) 1 L
5.3.6. Tomato Rooting Medium (ToRM) 1 L
5.4. Plant Media for Potato
5.4.1. GmNPT Vitamins Mix (GmNPT) 200x
5.4.2. Potato MS Medium (PoMS/PoSMS) 1 L
5.4.3. Potato Growth Medium (PoGM) 1 L
5.4.4. Potato Callus Induction Medium (PoCIM) 1 L
5.4.5. Potato Selection Medium (PoSM) 1 L
5.4.6. Potato Rooting Medium (PoRM) 1 L
5.5. Plant Media for Nicotiana
5.5.1. 0.1%. Agarose
5.5.2. Nicotiana Growth Medium (NicoGM) 1 L
5.5.3. Nicotiana Selection Medium (NicoSM) 1L
5.5.4. Nicotiana Rooting Medium (NicoRM) 1 L
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A






References
- Sussex, I.M. The Scientific Roots of Modern Plant Biotechnology. Plant Cell 2008, 20, 1189–1198. [Google Scholar] [CrossRef] [PubMed]
- Loyola-Vargas, V.M.; Ochoa-Alejo, N. Plant Cell Culture Protocols; Methods in Molecular Biology; Walker, J.M., Ed.; Humana Press: Totowa, NJ, USA, 2018; Volume 1815, ISBN 978-1-4939-8593-7. [Google Scholar]
- Kumar, S.; Barone, P.; Smith, M. Transgenic Plants: Methods and Protocols; Methods in Molecular Biology; Walker, J.M., Ed.; Humana Press: Totowa, NJ, USA, 2019; Volume 1864, ISBN 978-1-4939-8777-1. [Google Scholar]
- Christiaens, F.; Canher, B.; Lanssens, F.; Bisht, A.; Stael, S.; De Veylder, L.; Heyman, J. Pars Pro Toto: Every Single Cell Matters. Front. Plant Sci. 2021, 12, 656825. [Google Scholar] [CrossRef]
- Horsch, R.B.; Fry, J.E.; Hoffmann, N.L.; Wallroth, M.; Eichholtz, D.; Rogers, S.G.; Fraley, R.T. A Simple and General Method for Transferring Genes into Plants. Science 1985, 227, 1229–1231. [Google Scholar] [CrossRef] [PubMed]
- Heidari Japelaghi, R.; Haddad, R.; valizadeh, M.; Dorani Uliaie, E.; Jalali Javaran, M. High-Efficiency Improvement of Agrobacterium-Mediated Transformation of Tobacco (Nicotiana tabacum). J. Plant Mol. Breed. 2019, 6, 38–50. [Google Scholar] [CrossRef]
- Hellens, R.; Mullineaux, P.; Klee, H. Technical Focus:A guide to Agrobacterium binary Ti vectors. Trends Plant Sci. 2000, 5, 446–451. [Google Scholar] [CrossRef] [PubMed]
- Lee, L.-Y.; Gelvin, S.B. T-DNA Binary Vectors and Systems. Plant Physiol. 2008, 146, 325–332. [Google Scholar] [CrossRef]
- Wang, K. (Ed.) Agrobacterium Protocols, 2nd ed.; Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2006; ISBN 978-1-58829-536-1. [Google Scholar]
- Gartland, K.M.A.; Davey, M.R. Agrobacterium Protocols; Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 1995; Volume 44, ISBN 978-0-89603-302-3. [Google Scholar]
- Clemente, T. Nicotiana (Nicotiana tobaccum, Nicotiana benthamiana). In Agrobacterium Protocols; Humana Press: Totowa, NJ, USA, 2006; Volume 343, pp. 143–154. ISBN 978-1-59745-130-7. [Google Scholar]
- Vasil, V.; Hildebrandt, A.C. Differentiation of Tobacco Plants from Single, Isolated Cells in Microcultures. Science 1965, 150, 889–892. [Google Scholar] [CrossRef]
- Shen, K.; Xia, L.; Gao, X.; Li, C.; Sun, P.; Liu, Y.; Fan, H.; Li, X.; Han, L.; Lu, C.; et al. Tobacco as bioenergy and medical plant for biofuels and bioproduction. Heliyon 2024, 10, e33920. [Google Scholar] [CrossRef] [PubMed]
- Dasgupta, R.; Garcia, B.H.; Goodman, R.M. Systemic spread of an RNA insect virus in plants expressing plant viral movement protein genes. Proc. Natl. Acad. Sci. USA 2001, 98, 4910–4915. [Google Scholar] [CrossRef] [PubMed]
- Bally, J.; Nakasugi, K.; Jia, F.; Jung, H.; Ho, S.Y.W.; Wong, M.; Paul, C.M.; Naim, F.; Wood, C.C.; Crowhurst, R.N.; et al. The extremophile Nicotiana benthamiana has traded viral defence for early vigour. Nat. Plants 2015, 1, 15165. [Google Scholar] [CrossRef] [PubMed]
- Golubova, D.; Tansley, C.; Su, H.; Patron, N.J. Engineering Nicotiana benthamiana as a platform for natural product biosynthesis. Curr. Opin. Plant Biol. 2024, 81, 102611. [Google Scholar] [CrossRef]
- Mehlhorn, D.G.; Wallmeroth, N.; Berendzen, K.W.; Grefen, C. 2in1 Vectors Improve in Planta BiFC and FRET Analyses. In The Plant Endoplasmic Reticulum; Hawes, C., Kriechbaumer, V., Eds.; Humana Press: Totowa, NJ, USA, 2018; Volume 1691, pp. 139–158. ISBN 978-1-4939-7388-0. [Google Scholar]
- Jones, J.B., Jr. Tomato Plant Culture in the Field, Greenhouse, and Home Garden, 1st ed.; CRC Press, Taylor & Francis Group: Boca Raton, FL, USA, 1998; ISBN 978-0-849-32025-5. [Google Scholar]
- Van Eck, J.; Keen, P.; Tjahjadi, M. Agrobacterium tumefaciens-Mediated Transformation of Tomato. In Transgenic Plants; Kumar, S., Barone, P., Smith, M., Eds.; Humana Press: Totowa, NJ, USA, 2019; Volume 1864, pp. 225–234. ISBN 978-1-4939-8777-1. [Google Scholar]
- Van Eck, J.; Kirk, D.D.; Walmsley, A.M. Tomato (Lycopersicum esculentum). In Agrobacterium Protocols; Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2006; Volume 343, pp. 459–474. ISBN 978-1-59745-130-7. [Google Scholar]
- Wang, Y.; Sun, C.; Ye, Z.; Li, C.; Huang, S.; Lin, T. The genomic route to tomato breeding: Past, present, and future. Plant Physiol. 2024, 195, 2500–2514. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; He, Q.; Wang, J.; Wang, B.; Zhao, J.; Huang, S.; Yang, T.; Tang, Y.; Yang, S.; Aisimutuola, P.; et al. Super-pangenome analyses highlight genomic diversity and structural variation across wild and cultivated tomato species. Nat. Genet. 2023, 55, 852–860. [Google Scholar] [CrossRef]
- Fossi, M.; Amundson, K.; Kuppu, S.; Britt, A.; Comai, L. Regeneration of Solanum tuberosum Plants from Protoplasts Induces Widespread Genome Instability. Plant Physiol. 2019, 180, 78–86. [Google Scholar] [CrossRef]
- Godec, T.; Beier, S.; Rodriguez-Granados, N.Y.; Sasidharan, R.; Abdelhakim, L.; Teige, M.; Usadel, B.; Gruden, K.; Petek, M. Haplotype-resolved genome assembly of the tetraploid potato cultivar Désirée. Sci. Data 2025, 12, 1044. [Google Scholar] [CrossRef] [PubMed]
- Amundson, K.R.; Henry, I.M.; Comai, L. The United States Potato Genebank Holding of cv. Desiree is a Somatic Mutant of cv. Urgenta. Am. J. Potato Res. 2023, 100, 27–38. [Google Scholar] [CrossRef]
- Albert, I.; Böhm, H.; Albert, M.; Feiler, C.E.; Imkampe, J.; Wallmeroth, N.; Brancato, C.; Raaymakers, T.M.; Oome, S.; Zhang, H.; et al. An RLP23–SOBIR1–BAK1 complex mediates NLP-triggered immunity. Nat. Plants 2015, 1, 15140. [Google Scholar] [CrossRef]
- Schenstnyi, K.; Strauß, A.; Dressel, A.; Morbitzer, R.; Wunderlich, M.; Andrade, A.G.; Phan, T.; Aguilera, P.D.L.A.; Brancato, C.; Berendzen, K.W.; et al. The tomato resistance gene Bs4 suppresses leaf watersoaking phenotypes induced by AVRHAH1, a transcription activator-like effector from tomato-pathogenic xanthomonads. New Phytol. 2022, 236, 1856–1870. [Google Scholar] [CrossRef]
- Wittmann, J.; Brancato, C.; Berendzen, K.W.; Dreiseikelmann, B. Development of a tomato plant resistant to Clavibacter michiganensis using the endolysin gene of bacteriophage CMP1 as a transgene. Plant Pathol. 2016, 65, 496–502. [Google Scholar] [CrossRef]
- McCormick, S.; Niedermeyer, J.; Fry, J.; Barnason, A.; Horsch, R.; Fraley, R. Leaf disc transformation of cultivated tomato (L. esculentum) using Agrobacterium tumefaciens. Plant Cell Rep. 1986, 5, 81–84. [Google Scholar] [CrossRef] [PubMed]
- Curtis, I.S.; Davey, M.R.; Power, J.B. Leaf disk transformation. Methods Mol. Biol. 1995, 44, 59–70. [Google Scholar] [CrossRef] [PubMed]
- Sheerman, S.; Bevan, M.W. A rapid transformation method for Solanum tuberosum using binary Agrobacterium tumefaciens vectors. Plant Cell Rep. 1988, 7, 13–16. [Google Scholar] [CrossRef]
- De Block, M. Genotype-independent leaf disc transformation of potato (Solanum tuberosum) using Agrobacterium tumefaciens. Theor. Appl. Genet. 1988, 76, 767–774. [Google Scholar] [CrossRef] [PubMed]
- Larebeke, N.V.; Engler, G.; Holsters, M.; Den Elsacker, S.V.; Zaenen, I.; Schilperoort, R.A.; Schell, J. Large plasmid in Agrobacterium tumefaciens essential for crown gall-inducing ability. Nature 1974, 252, 169–170. [Google Scholar] [CrossRef]
- Koncz, C.; Schell, J. The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Molec. Gen. Genet. 1986, 204, 383–396. [Google Scholar] [CrossRef]
- Hellens, R.P.; Edwards, E.A.; Leyland, N.R.; Bean, S.; Mullineaux, P.M. pGreen: A versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 2000, 42, 819–832. [Google Scholar] [CrossRef]
- Lampropoulos, A.; Sutikovic, Z.; Wenzl, C.; Maegele, I.; Lohmann, J.U.; Forner, J. GreenGate—A Novel, Versatile, and Efficient Cloning System for Plant Transgenesis. PLoS ONE 2013, 8, e83043. [Google Scholar] [CrossRef] [PubMed]
- Deblaere, R.; Bytebier, B.; De Greve, H.; Deboeck, F.; Schell, J.; Van Montagu, M.; Leemans, J. Efficient octopine Ti plasmid-derived vectors for Agrobacterium-mediated gene transfer to plants. Nucleic Acids Res. 1985, 13, 4777–4788. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A. Agrobacterium-Mediated Transformation of Potato Genotypes. In Agrobacterium Protocols; Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 1995; Volume 44, pp. 121–128. ISBN 978-0-89603-302-3. [Google Scholar]
- Vinterhalter, D. Protocols for Agrobacterium mediated transformation of potato. Fruit Veg. Cereal Sci. Biotechnol. 2008, 2, 1–15. Available online: http://www.globalsciencebooks.info/Online/GSBOnline/OnlineFVCSB_2_SI1.html (accessed on 16 July 2025).
- Niedbała, G.; Niazian, M.; Sabbatini, P. Modeling Agrobacterium-Mediated Gene Transformation of Tobacco (Nicotiana tabacum)—A Model Plant for Gene Transformation Studies. Front. Plant Sci. 2021, 12, 695110. [Google Scholar] [CrossRef]
- De Saeger, J.; Park, J.; Chung, H.S.; Hernalsteens, J.-P.; Van Lijsebettens, M.; Inzé, D.; Van Montagu, M.; Depuydt, S. Agrobacterium strains and strain improvement: Present and outlook. Biotechnol. Adv. 2021, 53, 107677. [Google Scholar] [CrossRef] [PubMed]
- Sivankalyani, V.; Takumi, S.; Thangasamy, S.; Ashakiran, K.; Girija, S. Punctured-hypocotyl method for high-efficient transformation and adventitious shoot regeneration of tomato. Sci. Hortic. 2014, 165, 357–364. [Google Scholar] [CrossRef]
- Zhao, X.; Dong, Z.; Xie, X.; Zhang, Z.; Wu, Y.; Zhang, Q.; Sun, W.; Du, X.; Cao, X.; Hu, Q.A. Optimization of Leaf-disk Transformation and High Frequency Regeneration of Nicotianan benthamiana. Plant Gene and Trait. 2020, 11, 1–10. [Google Scholar] [CrossRef]
- Rajewski, A.C.; Elkins, K.B.; Henry, A.; Van Eck, J.; Litt, A. In vitro plant regeneration and Agrobacterium tumefaciens –mediated transformation of Datura stramonium (Solanaceae). Appl. Plant Sci. 2019, 7, e01220. [Google Scholar] [CrossRef]
- Ebinuma, H.; Sugita, K.; Matsunaga, E.; Yamakado, M. Selection of marker-free transgenic plants using the isopentenyl transferase gene. Proc. Natl. Acad. Sci. USA 1997, 94, 2117–2121. [Google Scholar] [CrossRef] [PubMed]
- Kiernan, J.M.; Goldberg, K.-B.; Young, M.J.; Schoelz, J.E.; Shepherd, R.J. Transformation and regeneration of Nicotiana edwardsonii. Plant Sci. 1989, 64, 67–78. [Google Scholar] [CrossRef]
- Shams, S.; Naeem, B.; Ma, L.; Li, R.; Zhang, Z.; Cao, Y.; Yu, H.; Feng, X.; Qiu, Y.; Wu, H.; et al. Developing an Optimized Protocol for Regeneration and Transformation in Pepper. Genes 2024, 15, 1018. [Google Scholar] [CrossRef] [PubMed]
- Yamoune, A.; Zdarska, M.; Depaepe, T.; Rudolfova, A.; Skalak, J.; Berendzen, K.W.; Mira-Rodado, V.; Fitz, M.; Pekarova, B.; Nicolas Mala, K.L.; et al. Cytokinins regulate spatially specific ethylene production to control root growth in Arabidopsis. Plant Commun. 2024, 5, 101013. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.S.; Chae, S.C.; Soh, W.Y.; Suh, M.C.; Hong, C.B. Agrobacterium Mediated Transformation of Nicotiana glauca, a Woody Plant. Mol. Cells 1993, 3, 345–347. [Google Scholar] [CrossRef]
- Komari, T.; Takakura, Y.; Ueki, J.; Kato, N.; Ishida, Y.; Hiei, Y. Binary Vectors and Super-Binary Vectors. In Agrobacterium Protocols; Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2006; Volume 343, pp. 15–42. ISBN 978-1-59745-130-7. [Google Scholar]
- Qi, Y. (Ed.) Plant Genome Editing with CRISPR Systems: Methods and Protocols; Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2019; Volume 1917, ISBN 978-1-4939-8990-4. [Google Scholar]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brancato, C.; Heusch, N.; Berendzen, K.W. Compendium of Agrobacterium-Mediated Tissue Culture Transformation Methods of Various Solanaceae Species. Methods Protoc. 2025, 8, 107. https://doi.org/10.3390/mps8050107
Brancato C, Heusch N, Berendzen KW. Compendium of Agrobacterium-Mediated Tissue Culture Transformation Methods of Various Solanaceae Species. Methods and Protocols. 2025; 8(5):107. https://doi.org/10.3390/mps8050107
Chicago/Turabian StyleBrancato, Caterina, Najmeh Heusch, and Kenneth Wayne Berendzen. 2025. "Compendium of Agrobacterium-Mediated Tissue Culture Transformation Methods of Various Solanaceae Species" Methods and Protocols 8, no. 5: 107. https://doi.org/10.3390/mps8050107
APA StyleBrancato, C., Heusch, N., & Berendzen, K. W. (2025). Compendium of Agrobacterium-Mediated Tissue Culture Transformation Methods of Various Solanaceae Species. Methods and Protocols, 8(5), 107. https://doi.org/10.3390/mps8050107





