A Method of Well-Spread Pachytene Chromosome Preparations for Plant Species with Large Genomes Suitable for the Immunolocalization of Meiotic Proteins
Abstract
1. Introduction
2. Experimental Design
2.1. Materials and Equipment
2.1.1. Consumables
- Petri dish;
- Pasteur pipettes;
- Dissecting needle straight;
- Dissecting needle lancet-shaped;
- Pipette tips 1000 μL;
- Superfrost microscope slides (without adhesion);
- 24 × 24 mm coverslips;
- Scalpel;
- Filter paper;
- Box filled with ice;
- Liquid nitrogen.
2.1.2. Reagents
- Glacial acetic acid (CH3COOH);
- Sodium hydroxide (NaOH);
- Citric acid (Sigma-Aldrich Co., LLC, St. Louis, MO, USA, Cat. no.: C2404);
- Citric acid trisodium salt dihydrate (Panreac, Darmstadt, Germany, Cat. no.: 131655);
- Pectolyase from Aspergillus japonicus (Sigma-Aldrich Co., LLC, St. Louis, MO, USA, Cat. no.: P5936);
- Cellulase Onozuka R-10 (SERVA, Heidelberg, Germany, Cat. no.: 16419);
- Cytohelicase from Helix pomatia (Sigma-Aldrich Co., LLC, St. Louis, MO, USA, Cat. no.:C8274).
2.1.3. Equipment
- Water bath (Miulab, Hangzhou, Zhejiang, China; Cat. no.: SWT-100);
- Light Microscope Axio Imager M2 (Carl Zeiss AB, Stockholm, Sweden; Cat. no.: 490020-0004-000).
3. Procedure of Well-Spread Pachytene Chromosome Preparation
3.1. Anther Selection at the Middle-Late Pachytene Stage and Fixation
- Extract a single anther from the selected bud.
- Place the extracted anther in a 60% acetic acid solution and gently squash it on a microscope slide.
- Examine the squashed preparation under a light microscope to confirm the middle-late pachytene stage.
- If the stage is confirmed, collect the remaining anthers from the same bud into a tube containing Clark’s fixative.
- Leave the anthers in Clark’s fixative for 1 hour at room temperature.
- Anthers can be used immediately for chromosome preparations or stored in Clark’s fixative at −20 °C before use (no more than 5 months).
3.2. Enzymatic Digestion: Duration: 2:30 h
- Rinse the fixed anthers in water 3 times for 10 min each.
- Rinse the anthers with citrate buffer for 5 min.
- Transfer the anthers to the Petri dish with the enzyme mixture (50 μL of enzyme mixture per 10 anthers) for 120 min at 37 °C in a water bath.
- Stop the enzyme reaction by transferring the Petri dish with anthers to ice.
- The anthers should be submerged at the bottom of the enzyme drop using a dissecting needle. Try not to damage the integrity of the anthers.
- Conditions for proper enzyme treatment should be adapted for the species being analyzed (see our previous publication [32]).
3.3. Chromosome Squash Preparation—Duration: 10 min
- Carefully transfer one anther with a small drop of enzyme mixture to a slide using a lancet-shaped dissecting needle and crush the anther with a straight dissecting needle to obtain a fine cell suspension (Figure 1A,B).
- Add a drop of 45% acetic acid to the slide and gently mix. The cell suspension should become transparent (Figure 1C).
- Cover the cell suspension with a cover glass and gently tap on the entire area of the cover glass with a pipette tip.
- Cover the preparation with filter paper and press lightly to remove excess liquid, avoiding any horizontal movement.
- Freeze the slides in liquid nitrogen and remove the cover glass with a scalpel.
- Dry the slides in the air.
- The preparation can be used immediately for immunodetection.
- The anther disintegrates better in a small drop of liquid. Meanwhile, an insufficient amount of liquid can lead to rapid drying of the cell suspension on the slide. Be careful and attentive.
- Excessive pressure on the coverslip results in strong and uneven stretching of the pachytene chromosomes.
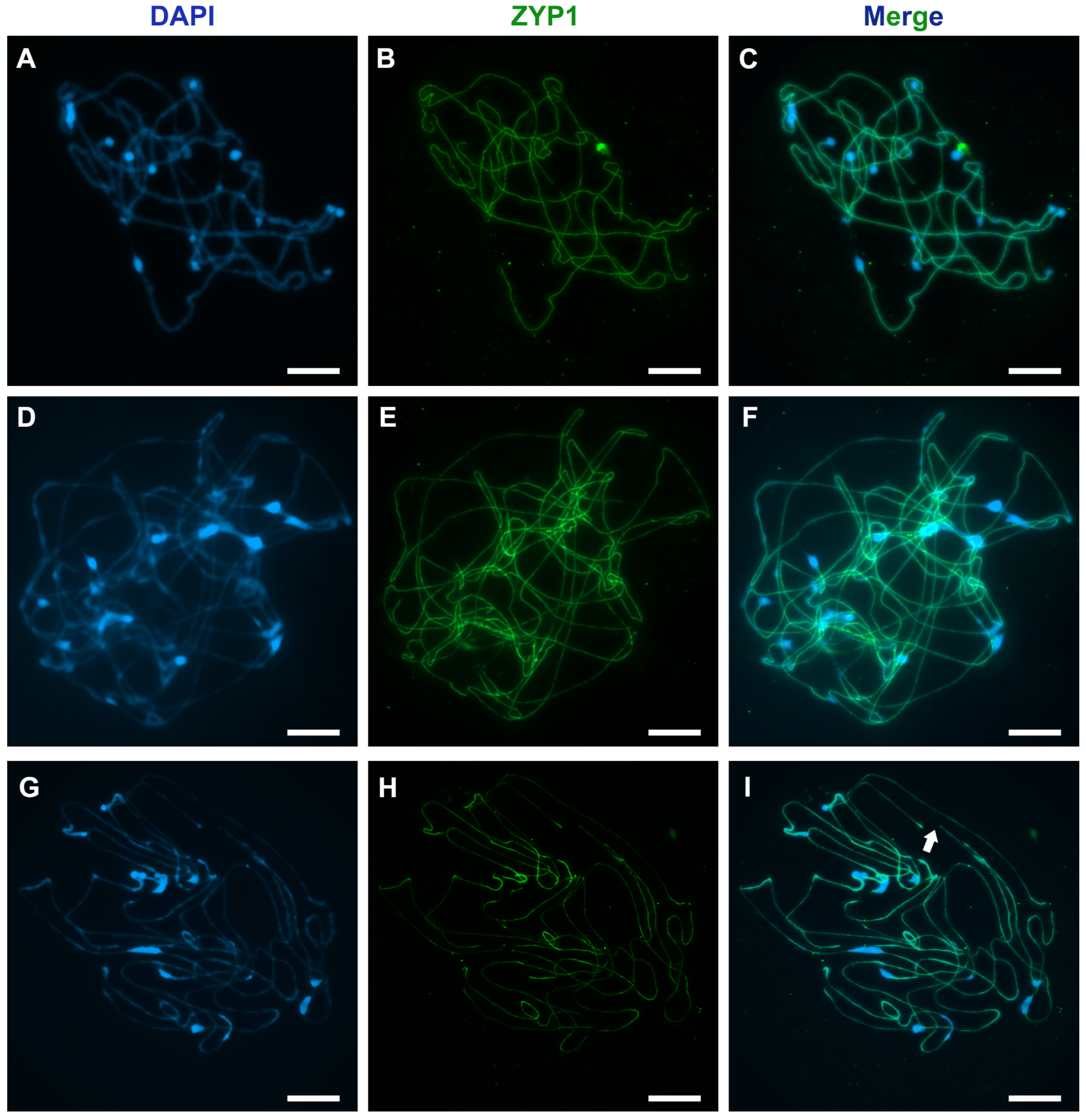
4. Expected Results
4.1. Troubleshooting
4.1.1. The Quality of the Chromosome Preparation Depends on the Cytoplasm Density
4.1.2. The Concentration of Acetic Acid Affects the Degree of Chromosome Stretching and Spreading
5. Reagent Setup
- Clark’s fixative: mix 3 volumes 96% ethanol and 1 volume glacial acetic acid. Use fresh solution.
- Citrate buffer: dissolve 0.558 g sodium citrate and 0.384 g citric acid in 100 mL distilled water. Adjust pH to 4.8 with NaOH (1 M). Sterilize and store at +4 °C.
- Enzyme mixture: dissolve 0.003 g pectolyase, 0.003 g cellulase and 0.003 g cytohelicase in 100 μL citrate buffer.
- 45% and 60% acetic acid solution in water.
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| SC | Synaptonemal Complex |
| SE | Standard Error |
| SEM | Scanning Electron Microscopy |
| PMC | Pollen Mother Cell |
References
- Higgins, J.D.; Wright, K.M.; Bomblies, K.; Franklin, F.C.H. Cytological techniques to analyze meiosis in Arabidopsis arenosa for investigating adaptation to polyploidy. Front. Plant Sci. 2014, 4, 546. [Google Scholar] [CrossRef] [PubMed]
- Morgan, C.; Wegel, E. Cytological characterization of Arabidopsis arenosa polyploids by SIM. In Plant Meiosis: Methods and Protocols; Springer: Berlin/Heidelberg, Germany, 2020; pp. 37–46. [Google Scholar]
- Stack, S.M.; Shearer, L.A.; Lohmiller, L.D.; Anderson, L.K. Preparing maize synaptonemal complex spreads and sequential immunofluorescence and fluorescence in situ hybridization. In Plant Meiosis: Methods and Protocols; Springer: Berlin/Heidelberg, Germany, 2020; pp. 79–115. [Google Scholar]
- Anderson, L.K.; Lohmiller, L.D.; Tang, X.; Hammond, D.B.; Javernick, L.; Shearer, L.; Basu-Roy, S.; Martin, O.C.; Falque, M. Combined fluorescent and electron microscopic imaging unveils the specific properties of two classes of meiotic crossovers. Proc. Natl. Acad. Sci. USA 2014, 111, 13415–13420. [Google Scholar] [CrossRef] [PubMed]
- Higgins, J.D.; Perry, R.M.; Barakate, A.; Ramsay, L.; Waugh, R.; Halpin, C.; Armstrong, S.J.; Franklin, F.C.H. Spatiotemporal asymmetry of the meiotic program underlies the predominantly distal distribution of meiotic crossovers in barley. Plant Cell 2012, 24, 4096–4109. [Google Scholar] [CrossRef]
- Kudryavtseva, N.; Ermolaev, A.; Pivovarov, A.; Simanovsky, S.; Odintsov, S.; Khrustaleva, L. The Control of the Crossover Localization in Allium. Int. J. Mol. Sci. 2023, 24, 7066. [Google Scholar] [CrossRef]
- Desjardins, S.D.; Ogle, D.E.; Ayoub, M.A.; Heckmann, S.; Henderson, I.R.; Edwards, K.J.; Higgins, J.D. MutS homologue 4 and MutS homologue 5 maintain the obligate crossover in wheat despite stepwise gene loss following polyploidization. Plant Physiol. 2020, 183, 1545–1558. [Google Scholar] [CrossRef]
- Martinez-Garcia, M.; White, C.I.; Franklin, F.C.H.; Sanchez-Moran, E. The role of topoisomerase II in DNA repair and recombination in Arabidopsis thaliana. Int. J. Mol. Sci. 2021, 22, 13115. [Google Scholar] [CrossRef]
- Armstrong, S.J.; Caryl, A.P.; Jones, G.H.; Franklin, F.C.H. Asy1, a protein required for meiotic chromosome synapsis, localizes to axis-associated chromatin in Arabidopsis and Brassica. J. Cell Sci. 2002, 115, 3645–3655. [Google Scholar] [CrossRef]
- Armstrong, S.; Osman, K. Immunolocalization of meiotic proteins in Arabidopsis thaliana: Method 2. In Plant Meiosis: Methods and Protocols; Springer: Berlin/Heidelberg, Germany, 2013; pp. 103–107. [Google Scholar]
- Higgins, J.D. Analyzing meiosis in barley. In Plant Meiosis: Methods and Protocols; Springer: Berlin/Heidelberg, Germany, 2013; pp. 135–144. [Google Scholar]
- Yang, C.; Sofroni, K.; Hamamura, Y.; Hu, B.; Elbasi, H.T.; Balboni, M.; Chu, L.; Stang, D.; Heese, M.; Schnittger, A. ZYP1-mediated recruitment of PCH2 to the synaptonemal complex remodels the chromosome axis leading to crossover restriction. Nucleic Acids Res. 2022, 50, 12924–12937. [Google Scholar] [CrossRef]
- Zhu, L.; Dluzewska, J.; Fernández-Jiménez, N.; Ranjan, R.; Pelé, A.; Dziegielewski, W.; Szymanska-Lejman, M.; Hus, K.; Górna, J.; Pradillo, M.; et al. The kinase ATR controls meiotic crossover distribution at the genome scale in Arabidopsis. Plant Cell 2025, 37, koae292. [Google Scholar] [CrossRef]
- Steckenborn, S.; Cuacos, M.; Ayoub, M.; Feng, C.; Schubert, V.; Hoffie, I.; Hensel, G.; Kumlehn, J.; Heckmann, S. The meiotic topoisomerase VI B subunit (MTOPVIB) is essential for meiotic DNA double-strand break formation in barley (Hordeum vulgare L.). Plant Reprod. 2023, 36, 1–15. [Google Scholar] [CrossRef]
- Parra-Nunez, P.; Fernández-Jiménez, N.; Pachon-Penalba, M.; Sanchez-Moran, E.; Pradillo, M.; Santos, J.L. Synthetically induced Arabidopsis thaliana autotetraploids provide insights into the analysis of meiotic mutants with altered crossover frequency. New Phytol. 2024, 241, 197–208. [Google Scholar] [CrossRef] [PubMed]
- Son, N.; Kim, H.; Kim, J.; Park, J.; Byun, D.; Park, S.J.; Kim, H.; Park, Y.M.; Bourguet, P.; Berger, F.; et al. The histone variant H2A. W restricts heterochromatic crossovers in Arabidopsis. Proc. Natl. Acad. Sci. USA 2025, 122, e2413698122. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.; Cai, B.; Hamamura, Y.; Schnittger, A.; Yang, C. SCFRMF-dependent degradation of the nuclear lamina releases the somatic chromatin mobility restriction for meiotic recombination. Sci. Adv. 2025, 11, eadr4567. [Google Scholar] [CrossRef]
- Pochon, G.; Henry, I.M.; Yang, C.; Lory, N.; Fernández-Jiménez, N.; Böwer, F.; Hu, B.; Carstens, L.; Tsai, H.T.; Pradillo, M.; et al. The Arabidopsis Hop1 homolog ASY1 mediates cross-over assurance and interference. PNAS Nexus 2023, 2, pgac302. [Google Scholar] [CrossRef]
- Lhuissier, F.G.; Offenberg, H.H.; Wittich, P.E.; Vischer, N.O.; Heyting, C. The mismatch repair protein MLH1 marks a subset of strongly interfering crossovers in tomato. Plant Cell 2007, 19, 862–876. [Google Scholar] [CrossRef]
- Liu, X.; Cao, Y.; Du, G.; Zhang, C.; Xu, M.; Cheng, Z.; Shen, Y.; Yu, H. OsRAD51 plays a vital role in promoting homologous recombination in rice meiosis. Int. J. Mol. Sci. 2022, 23, 9906. [Google Scholar] [CrossRef]
- Hesse, S.; Zelkowski, M.; Mikhailova, E.I.; Keijzer, C.J.; Houben, A.; Schubert, V. Ultrastructure and dynamics of synaptonemal complex components during meiotic pairing and synapsis of standard (A) and accessory (B) rye chromosomes. Front. Plant Sci. 2019, 10, 773. [Google Scholar] [CrossRef]
- Hoffman, E.A.; Frey, B.L.; Smith, L.M.; Auble, D.T. Formaldehyde crosslinking: A tool for the study of chromatin complexes. J. Biol. Chem. 2015, 290, 26404–26411. [Google Scholar] [CrossRef]
- Wang, K.; Tang, D.; Wang, M.; Lu, J.; Yu, H.; Liu, J.; Qian, B.; Gong, Z.; Wang, X.; Chen, J.; et al. MER3 is required for normal meiotic crossover formation, but not for presynaptic alignment in rice. J. Cell Sci. 2009, 122, 2055–2063. [Google Scholar] [CrossRef]
- Castellani, M.; Zhang, M.; Thangavel, G.; Mata-Sucre, Y.; Lux, T.; Campoy, J.A.; Marek, M.; Huettel, B.; Sun, H.; Mayer, K.F.; et al. Meiotic recombination dynamics in plants with repeat-based holocentromeres shed light on the primary drivers of crossover patterning. Nat. Plants 2024, 10, 423–438. [Google Scholar] [CrossRef]
- Kalfusová, R.; Herklotz, V.; Kumke, K.; Houben, A.; Kovařík, A.; Ritz, C.M.; Lunerová, J. Epigenetic histone H3 phosphorylation marks discriminate between univalent-and bivalent-forming chromosomes during canina asymmetrical meiosis. Ann. Bot. 2024, 133, 435–446. [Google Scholar] [CrossRef] [PubMed]
- Bhat, A.H.; Hussein, S. Fixation and different types of fixatives: Their role and functions: A review. Int. J. Clin. Diagn. Pathol. 2021, 4, 113–119. [Google Scholar] [CrossRef]
- Chelysheva, L.; Grandont, L.; Vrielynck, N.; Le Guin, S.; Mercier, R.; Grelon, M. An easy protocol for studying chromatin and recombination protein dynamics during Arabidopsis thaliana meiosis: Immunodetection of cohesins, histones and MLH1. Cytogenet. Genome Res. 2010, 129, 143–153. [Google Scholar] [CrossRef]
- Albini, S.; Jones, G. Synaptonemal complex spreading in Allium cepa and Allium fistulosum. II. Pachytene observations: The SC karyotype and the correspondence of late recombination nodules and chiasmata. Genome 1988, 30, 399–410. [Google Scholar] [CrossRef]
- Sherman, J.D.; Stack, S.M. Two-dimensional spreads of synaptonemal complexes from solanaceous plants. VI. High-resolution recombination nodule map for tomato (Lycopersicon esculentum). Genetics 1995, 141, 683–708. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Kirov, I.; Khrustaleva, L.; Van Laere, K.; Soloviev, A.; Meeus, S.; Romanov, D.; Fesenko, I. DRAWID: User-friendly java software for chromosome measurements and idiogram drawing. Comp. Cytogenet. 2017, 11, 747. [Google Scholar] [CrossRef]
- Kirov, I.; Divashuk, M.; Van Laere, K.; Soloviev, A.; Khrustaleva, L. An easy “SteamDrop” method for high quality plant chromosome preparation. Mol. Cytogenet. 2014, 7, 1–10. [Google Scholar] [CrossRef]
- Rahman, M.A.; Sultana, N.; Ayman, U.; Bhakta, S.; Afrose, M.; Afrin, M.; Haque, Z. Alcoholic fixation over formalin fixation: A new, safer option for morphologic and molecular analysis of tissues. Saudi J. Biol. Sci. 2022, 29, 175–182. [Google Scholar] [CrossRef]

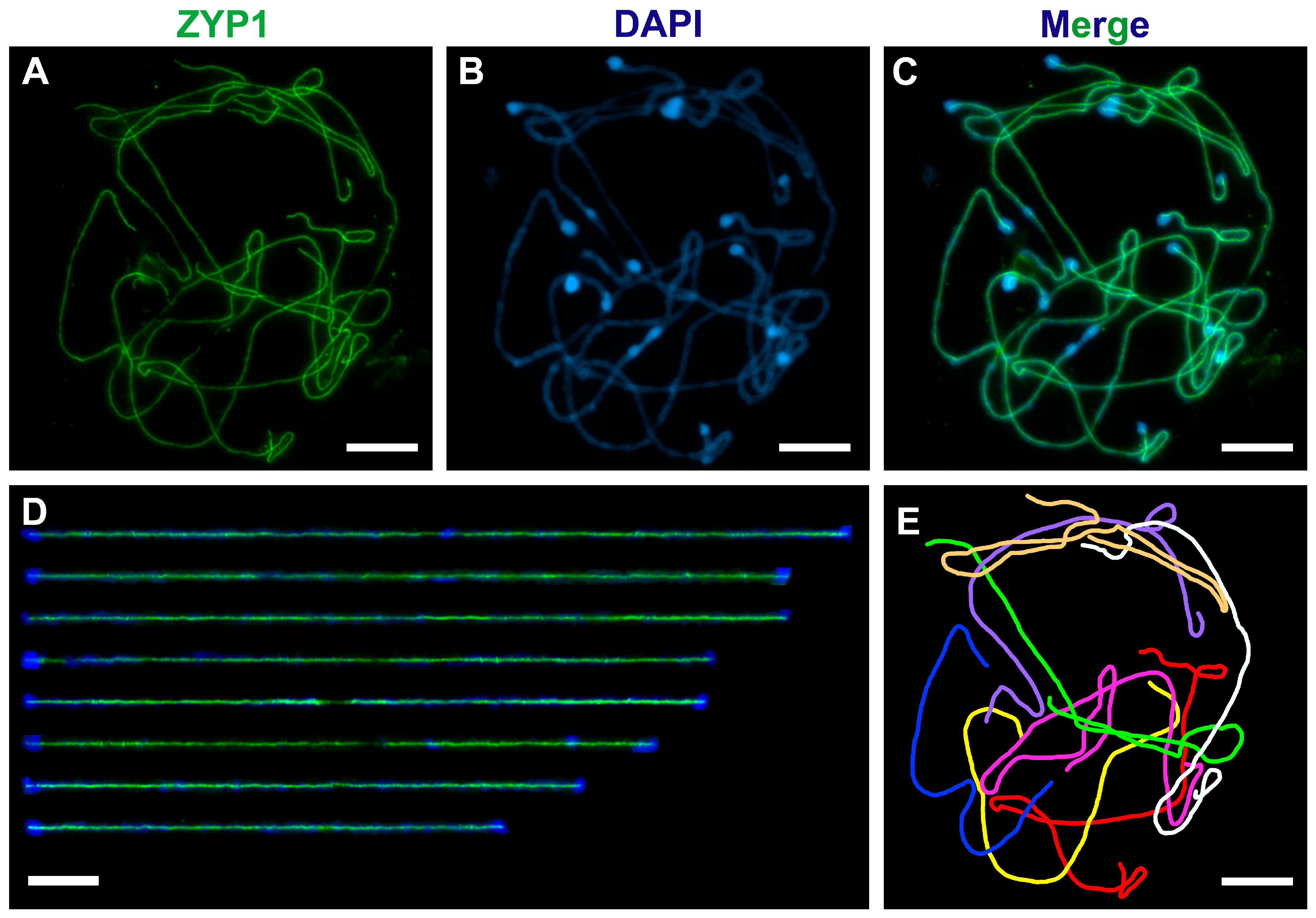

| Chromosome | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | Total |
|---|---|---|---|---|---|---|---|---|---|
| Mean SC length ± SE * (μm) | 121.93 ± 6.99 | 113.44 ± 6.31 | 108.22 ± 5.46 | 97.63 ± 4.94 | 86.72 ± 3.85 | 81.18 ± 3.94 | 77.64 ± 3.60 | 67.56 ± 3.08 | 754.32 |
| Relative SC length | 0.16 | 0.15 | 0.14 | 0.13 | 0.11 | 0.11 | 0.10 | 0.09 | 1.00 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kudryavtseva, N.; Ermolaev, A.; Khrustaleva, L. A Method of Well-Spread Pachytene Chromosome Preparations for Plant Species with Large Genomes Suitable for the Immunolocalization of Meiotic Proteins. Methods Protoc. 2025, 8, 54. https://doi.org/10.3390/mps8030054
Kudryavtseva N, Ermolaev A, Khrustaleva L. A Method of Well-Spread Pachytene Chromosome Preparations for Plant Species with Large Genomes Suitable for the Immunolocalization of Meiotic Proteins. Methods and Protocols. 2025; 8(3):54. https://doi.org/10.3390/mps8030054
Chicago/Turabian StyleKudryavtseva, Natalya, Aleksey Ermolaev, and Ludmila Khrustaleva. 2025. "A Method of Well-Spread Pachytene Chromosome Preparations for Plant Species with Large Genomes Suitable for the Immunolocalization of Meiotic Proteins" Methods and Protocols 8, no. 3: 54. https://doi.org/10.3390/mps8030054
APA StyleKudryavtseva, N., Ermolaev, A., & Khrustaleva, L. (2025). A Method of Well-Spread Pachytene Chromosome Preparations for Plant Species with Large Genomes Suitable for the Immunolocalization of Meiotic Proteins. Methods and Protocols, 8(3), 54. https://doi.org/10.3390/mps8030054





