Optimization of Peripheral Blood Mononuclear Cell Extraction from Small Volume of Blood Samples: Potential Implications for Children-Related Diseases
Abstract
:1. Introduction
2. Experimental Design
2.1. Materials
- Histopaque 1077 density gradient (Sigma Aldrich, St Louis, MO, USA; cat. n. 10771) orLymphocyte Separation Medium (Lonza, Walkersville, MD, USA; cat. n. 17-829E), or Lymphosep (Microgem, Naples, Italy; cat. n. L0560).
- Dulbecco’s Phosphate buffered saline D-PBS (Sigma-Aldrich, St. Louis, MO, USA; cat. n. D8662).
- Trypan blue (Sigma Aldrich, St Louis, MO, USA; cat. n. T8154).
- Ammonium-Chloride-Potassium (ACK) lysing buffer (Thermo Fisher, Waltham, MA, USA; cat. n. A1049201).
2.2. Equipment
- EDTA tubes (Fisher Scientific, Pittsburgh, PA, USA; model BD 367862).
- 2 mL and 5 mL sterile and disposable serological pipettes (BD Falcon Becton Dickinson Labware, Franklin Lakes, NJ, USA; cat. n. 357507 and n. 357543, respectively).
- 15 mL conical, sterile, polypropylene, centrifuge tubes (Fisher Scientific, Pittsburgh, PA, USA; model Corning 352196).
- Pipettes (Eppendorf Research, Milan, Italy; cat. n. 3123000020 (0.5–10 μL), n. 3123000047 (10–100 μL), n. 3123000063 (100–1000 μL), with relative sterile tips).
- Electronic pipette (Eppendorf Research, Milan, Italy; cat. n. 4430000018).
- Pasteur pipette.
- 1.5 mL tube (Eppendorf Research, Milan, Italy; cat. n. 0030125150).
- Burker cell counter (Sigma-Aldrich, St. Louis, MO, USA; cat. n. Z359629) and coverslip (Sigma-Aldrich, St. Louis, MO, USA; cat. n. Z375357).
- Centrifuge (Thermo Scientific, Cincinnati, OH, USA; model Heraeus Megafuge 1.0).
- Class II biological safety cabinet (Steril, Lecce, Italy; model. n. VBH 48 MP VBH).
- Inverted microscope (Leica Microsystems, Milan, Italy; model DMi1).
- Humidified 95% O2/5% CO2 water jacketed incubator, 37 °C (Thermo Scientific, Cincinnati, OH, USA; model Forma Series II) (optional).
- Flow Cytometer (Merck Millipore, Burlington, MA, USA; model Guava® easyCyte 5 HPL Benchtop Flow Cytometer).
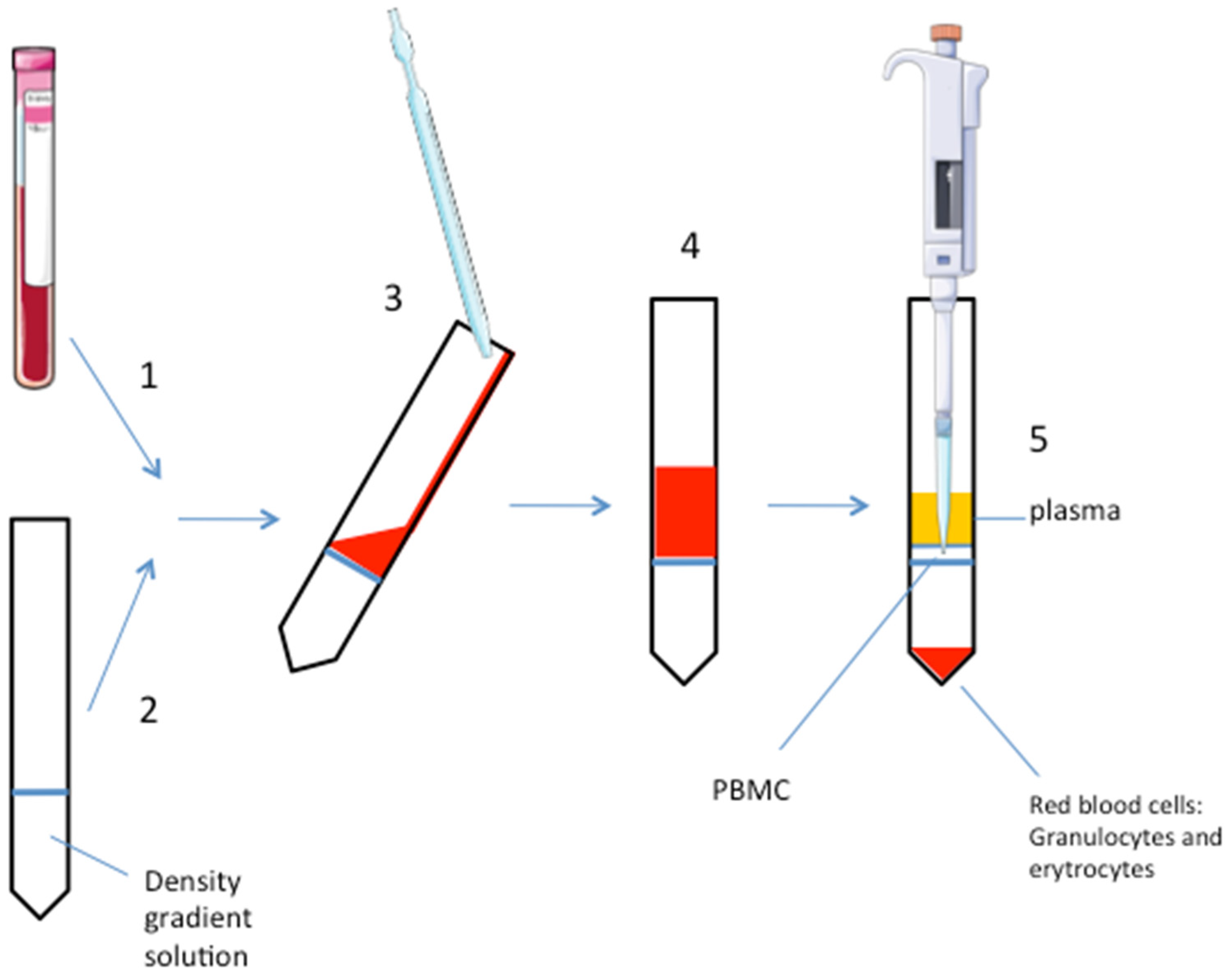
3. Procedure
- (1)
- Collect the blood sample in an EDTA-tube (volume: 2 mL).
- (2)
- Add the density gradient solution (i.e., polysucrose, ρ = 1.077 g/mL, or the highly branched copolymer of sucrose and epichlorohydin monomers: ficoll) to a clean 15 mL tube in a volume 1:1 to the blood sample. Critical step: do not dilute blood with D-PBS to avoid too much dilution of the small sample.
- (3)
- Carefully and slowly, add the blood sample on the gradient solution without mixing them to ensure a defined interphase separation. This step should be done by inclining the tube and adding the blood sample very slowly. Using the wall of the tube to help, pipet down the blood on the gradient solution.
- (4)
- Centrifuge at 1010× g for 30 min at room temperature. Do not use the rotor brake for avoiding phases mixing. When the rotor is totally stopped, carefully remove the tube, paying attention to not mix the phases.
- (5)
- After centrifugation, PBMC are stratified on the middle phase (cloudy white interface). At the top of the tube, there is the plasma, erytrocytes and granulocytes are stratified to the bottom. A well-defined interface is a marker of an optimal phase separation. Carefully aspirate the PBMC phase with a pipette and put it into a clean 15 mL tube. Another way is to aspirate and discard the upper plasma phase before pipetting the PBMC layer. Consider that the gradient solution could be toxic for the cells, so avoid too much time for this procedure step, if the experiment requires growing the PBMC in culture at the end of the extraction.
- (6)
- Add D-PBS in 1:1 in volume. Gently mix.
- (7)
- Centrifuge at 605× g for 10 min at room temperature with rotor brake on.
- (8)
- Remove and discard supernatant and add D-PBS, typically the volume ranges from 1 mL to 3 mL, depending on the pellet size; carefully re-suspend the pellet.
- (9)
- Centrifuge at 300× g for 10 min at room temperature with rotor brake on.
- (10)
- Remove and discard supernatant and re-suspend cells in the desired buffer (i.e., growing media, lysis buffer, or others).
- (11)
- Count cells with Trypan blue; or see next step (optional).
- (12)
- Acquire the sample on standard or multicolor flow cytometer and analyze it following a standard procedure (optional).
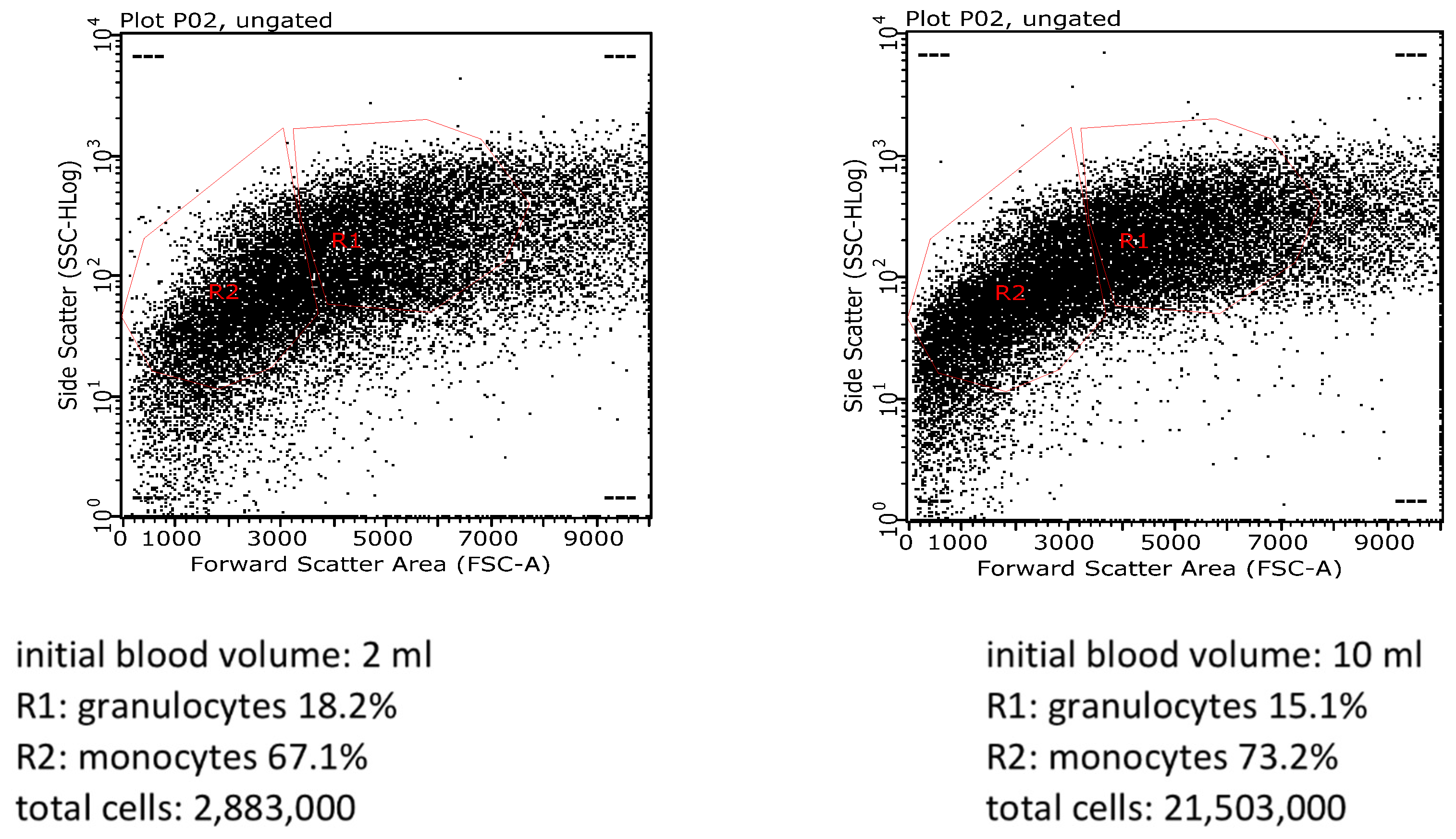
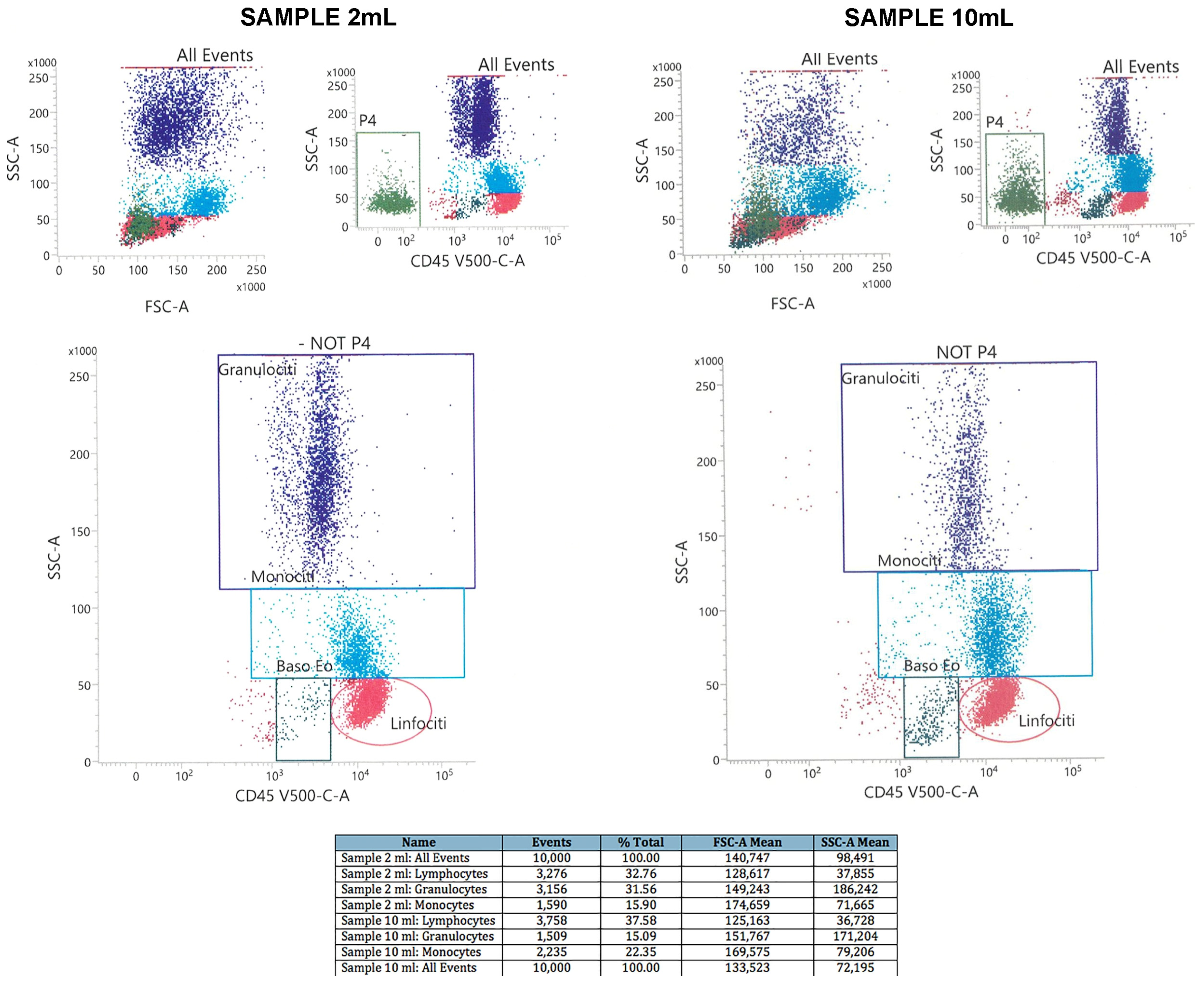
4. Expected Results
Troubleshooting
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kleiveland, C.R. Peripheral Blood Mononuclear Cells. In The Impact of Food Bioactives on Health; Verhoeckx, K., Cotter, P., López-Expósito, I., Kleiveland, C., Lea, T., Mackie, A., Eds.; Springer: Cham, Switzerland, 2015; pp. 161–167. [Google Scholar]
- Pourahmad, J.; Salimi, A. Isolated Human Peripheral Blood Mononuclear Cell (PBMC), a Cost Effective Tool for Predicting Immunosuppressive Effects of Drugs and Xenobiotics. Iran. J. Pharm. Res. 2015, 14, 979. [Google Scholar] [PubMed]
- Yang, D.N.; Wu, J.H.; Geng, L.; Cao, L.J.; Zhang, Q.J.; Luo, J.Q.; Kallen, A.; Hou, Z.H.; Qian, W.P.; Shi, Y.; et al. Efficacy of intrauterine perfusion of peripheral blood mononuclear cells (PBMC) for infertile women before embryo transfer: Meta-analysis. J. Obstet. Gynaecol. 2020, 40, 961–968. [Google Scholar] [CrossRef] [PubMed]
- Siniscalco, D.; Sapone, A.; Giordano, C.; Cirillo, A.; De Magistris, L.; Rossi, F.; Fasano, A.; Bradstreet, J.J.; Maione, S.; Antonucci, N. Cannabinoid Receptor Type 2, but not Type 1, is Up-Regulated in Peripheral Blood Mononuclear Cells of Children Affected by Autistic Disorders. J. Autism Dev. Disord. 2013, 43, 2686–2695. [Google Scholar] [CrossRef] [PubMed]
- Cuper, N.J.; Klaessens, J.; Jaspers, J.E.; De Roode, R.; Noordmans, H.J.; De Graaff, J.; Verdaasdonk, R.M. The use of near-infrared light for safe and effective visualization of subsurface blood vessels to facilitate blood withdrawal in children. Med. Eng. Phys. 2013, 35, 433–440. [Google Scholar] [CrossRef] [PubMed]
- Siniscalco, D.; Bradstreet, J.J.; Cirillo, A.; Antonucci, N. The in vitro GcMAF effects on endocannabinoid system transcriptionomics, receptor formation, and cell activity of autism-derived macrophages. J. Neuroinflamm. 2014, 11, 78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- HIV/AIDS Network Coordination: Cross-Network PBMC Processing Standard Operating Procedure. 2018. Available online: www.hanc.info (accessed on 20 February 2020).
- Böyum, A. Isolation of mononuclear cells and granulocytes from human blood. Isolation of monuclear cells by one centrifugation, and of granulocytes by combining centrifugation and sedimentation at 1 g. Scand. J. Clin. Lab. Investig. Suppl. 1968, 97, 77–89. [Google Scholar]
- Taghizadeh, N.; Heard, G.; Davidson, A.; Williams, K.; Story, D. The experiences of children with autism spectrum disorder, their caregivers and health care providers during day procedure: A mixed methods study. Pediatr. Anesth. 2019, 29, 927–937. [Google Scholar] [CrossRef] [PubMed]
- Olson, W.C.; Smolkin, M.E.; Farris, E.M.; Fink, R.J.; Czarkowski, A.R.; Fink, J.H.; Chianese-Bullock, K.A.; Slingluff, C.L., Jr. Shipping blood to a central laboratory in multicenter clinical trials: Effect of ambient temperature on specimen temperature, and effects of temperature on mononuclear cell yield, viability and immunologic function. J. Transl. Med. 2011, 9, 26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palmirotta, R.; De Marchis, M.L.; Ludovici, G.; Leone, B.; Savonarola, A.; Ialongo, C.; Spila, A.; De Angelis, F.; Ferroni, P.; Della-Morte, D.; et al. Impact of preanalytical handling and timing for peripheral blood mononuclear cells isolation and RNA studies: The experience of the Interinstitutional Multidisciplinary BioBank (BioBIM). Int. J. Biol. Markers 2012, 27, e90–e98. [Google Scholar] [CrossRef] [PubMed]
- Corkum, C.P.; Ings, D.P.; Burgess, C.; Karwowska, S.; Kroll, W.; Michalak, T.I. Immune cell subsets and their gene expression profiles from human PBMC isolated by Vacutainer Cell Preparation Tube (CPT™) and standard density gradient. BMC Immunol. 2015, 16, 48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petriz, J.; Bradford, J.A.; Ward, M.D. No lyse no wash flow cytometry for maximizing minimal sample preparation. Methods 2018, 134–135, 149–163. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Patrone, D.; Alessio, N.; Antonucci, N.; Brigida, A.L.; Peluso, G.; Galderisi, U.; Siniscalco, D. Optimization of Peripheral Blood Mononuclear Cell Extraction from Small Volume of Blood Samples: Potential Implications for Children-Related Diseases. Methods Protoc. 2022, 5, 20. https://doi.org/10.3390/mps5020020
Patrone D, Alessio N, Antonucci N, Brigida AL, Peluso G, Galderisi U, Siniscalco D. Optimization of Peripheral Blood Mononuclear Cell Extraction from Small Volume of Blood Samples: Potential Implications for Children-Related Diseases. Methods and Protocols. 2022; 5(2):20. https://doi.org/10.3390/mps5020020
Chicago/Turabian StylePatrone, Deanira, Nicola Alessio, Nicola Antonucci, Anna Lisa Brigida, Gianfranco Peluso, Umberto Galderisi, and Dario Siniscalco. 2022. "Optimization of Peripheral Blood Mononuclear Cell Extraction from Small Volume of Blood Samples: Potential Implications for Children-Related Diseases" Methods and Protocols 5, no. 2: 20. https://doi.org/10.3390/mps5020020
APA StylePatrone, D., Alessio, N., Antonucci, N., Brigida, A. L., Peluso, G., Galderisi, U., & Siniscalco, D. (2022). Optimization of Peripheral Blood Mononuclear Cell Extraction from Small Volume of Blood Samples: Potential Implications for Children-Related Diseases. Methods and Protocols, 5(2), 20. https://doi.org/10.3390/mps5020020









