ExoProK: A Practical Method for the Isolation of Small Extracellular Vesicles from Pleural Effusions
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients and Sample Collection
2.2. sEV Isolation
2.3. Transmission Electron Microscopy
2.4. Protein Quantification and Western Blot Analysis
2.5. Nanoparticle Tracking Analysis
2.6. RNA Isolation and Precipitation
2.7. cDNA Synthesis-qPCR
3. Results
3.1. sEV Isolation and Characterization of PE-Derived sEVs
3.2. Detection of sEV-Derived RNAs
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Light, R.W.; Macgregor, M.I.; Luchsinger, P.C.; Ball, W.C., Jr. Pleural effusions: The diagnostic separation of transudates and exudates. Ann. Intern. Med. 1972, 77, 507–513. [Google Scholar] [CrossRef] [PubMed]
- Daniil, Z.D.; Zintzaras, E.; Kiropoulos, T.; Papaioannou, A.I.; Koutsokera, A.; Kastanis, A.; Gourgoulianis, K.I. Discrimination of exudative pleural effusions based on multiple biological parameters. Eur. Respir. J. 2007, 30, 957–964. [Google Scholar] [CrossRef] [PubMed]
- Loveland, P.; Christie, M.; Hammerschlag, G.; Irving, L.; Steinfort, D. Diagnostic yield of pleural fluid cytology in malignant effusions: An Australian tertiary centre experience. Intern. Med. J. 2018, 48, 1318–1324. [Google Scholar] [CrossRef] [PubMed]
- Grosu, H.B.; Kazzaz, F.; Vakil, E.; Molina, S.; Ost, D. Sensitivity of Initial Thoracentesis for Malignant Pleural Effusion Stratified by Tumor Type in Patients with Strong Evidence of Metastatic Disease. Respiration 2018, 96, 363–369. [Google Scholar] [CrossRef]
- Mohan, A.; Agarwal, S.; Clauss, M.; Britt, N.S.; Dhillon, N.K. Extracellular vesicles: Novel communicators in lung diseases. Respir. Res. 2020, 21, 175. [Google Scholar] [CrossRef]
- Thery, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef]
- Witwer, K.W.; Thery, C. Extracellular vesicles or exosomes? On primacy, precision, and popularity influencing a choice of nomenclature. J. Extracell. Vesicles 2019, 8, 1648167. [Google Scholar] [CrossRef]
- Maacha, S.; Bhat, A.A.; Jimenez, L.; Raza, A.; Haris, M.; Uddin, S.; Grivel, J.C. Extracellular vesicles-mediated intercellular communication: Roles in the tumor microenvironment and anti-cancer drug resistance. Mol. Cancer 2019, 18, 55. [Google Scholar] [CrossRef]
- Silva, M.; Melo, S.A. Non-coding RNAs in Exosomes: New Players in Cancer Biology. Curr. Genom. 2015, 16, 295–303. [Google Scholar] [CrossRef]
- Rashed, M.H.; Bayraktar, E.; Helal, G.K.; Abd-Ellah, M.F.; Amero, P.; Chavez-Reyes, A.; Rodriguez-Aguayo, C. Exosomes: From Garbage Bins to Promising Therapeutic Targets. Int. J. Mol. Sci. 2017, 18, 538. [Google Scholar] [CrossRef]
- Mathieu, M.; Martin-Jaular, L.; Lavieu, G.; Thery, C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat. Cell Biol. 2019, 21, 9–17. [Google Scholar] [CrossRef]
- Yanez-Mo, M.; Siljander, P.R.; Andreu, Z.; Zavec, A.B.; Borras, F.E.; Buzas, E.I.; Buzas, K.; Casal, E.; Cappello, F.; Carvalho, J.; et al. Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles 2015, 4, 27066. [Google Scholar] [CrossRef]
- Malloci, M.; Perdomo, L.; Veerasamy, M.; Andriantsitohaina, R.; Simard, G.; Martinez, M.C. Extracellular Vesicles: Mechanisms in Human Health and Disease. Antioxid. Redox Signal. 2019, 30, 813–856. [Google Scholar] [CrossRef]
- Brennan, K.; Martin, K.; FitzGerald, S.P.; O’Sullivan, J.; Wu, Y.; Blanco, A.; Richardson, C.; Mc Gee, M.M. A comparison of methods for the isolation and separation of extracellular vesicles from protein and lipid particles in human serum. Sci. Rep. 2020, 10, 1039. [Google Scholar] [CrossRef]
- Patel, G.K.; Khan, M.A.; Zubair, H.; Srivastava, S.K.; Khushman, M.; Singh, S.; Singh, A.P. Comparative analysis of exosome isolation methods using culture supernatant for optimum yield, purity and downstream applications. Sci. Rep. 2019, 9, 5335. [Google Scholar] [CrossRef]
- Ding, M.; Wang, C.; Lu, X.; Zhang, C.; Zhou, Z.; Chen, X.; Zhang, C.Y.; Zen, K.; Zhang, C. Comparison of commercial exosome isolation kits for circulating exosomal microRNA profiling. Anal. Bioanal. Chem. 2018, 410, 3805–3814. [Google Scholar] [CrossRef]
- Simonsen, J.B. What Are We Looking at? Extracellular Vesicles, Lipoproteins, or Both? Circ. Res. 2017, 121, 920–922. [Google Scholar] [CrossRef]
- Fessler, M.B. Next stop for HDL: The lung. Clin. Exp. Allergy 2012, 42, 340–342. [Google Scholar] [CrossRef]
- Sidhom, K.; Obi, P.O.; Saleem, A. A Review of Exosomal Isolation Methods: Is Size Exclusion Chromatography the Best Option? Int. J. Mol. Sci. 2020, 21, 6466. [Google Scholar] [CrossRef]
- Lee, J.S.; Hur, J.Y.; Kim, I.A.; Kim, H.J.; Choi, C.M.; Lee, J.C.; Kim, W.S.; Lee, K.Y. Liquid biopsy using the supernatant of a pleural effusion for EGFR genotyping in pulmonary adenocarcinoma patients: A comparison between cell-free DNA and extracellular vesicle-derived DNA. BMC Cancer 2018, 18, 1236. [Google Scholar] [CrossRef]
- Bard, M.P.; Hegmans, J.P.; Hemmes, A.; Luider, T.M.; Willemsen, R.; Severijnen, L.A.; van Meerbeeck, J.P.; Burgers, S.A.; Hoogsteden, H.C.; Lambrecht, B.N. Proteomic analysis of exosomes isolated from human malignant pleural effusions. Am. J. Respir. Cell Mol. Biol. 2004, 31, 114–121. [Google Scholar] [CrossRef]
- Qu, X.; Li, Q.; Yang, J.; Zhao, H.; Wang, F.; Zhang, F.; Zhang, S.; Zhang, H.; Wang, R.; Wang, Q.; et al. Double-Stranded DNA in Exosomes of Malignant Pleural Effusions as a Novel DNA Source for EGFR Mutation Detection in Lung Adenocarcinoma. Front. Oncol. 2019, 9, 931. [Google Scholar] [CrossRef]
- Hydbring, P.; De Petris, L.; Zhang, Y.; Branden, E.; Koyi, H.; Novak, M.; Kanter, L.; Haag, P.; Hurley, J.; Tadigotla, V.; et al. Exosomal RNA-profiling of pleural effusions identifies adenocarcinoma patients through elevated miR-200 and LCN2 expression. Lung Cancer 2018, 124, 45–52. [Google Scholar] [CrossRef]
- Park, J.O.; Choi, D.Y.; Choi, D.S.; Kim, H.J.; Kang, J.W.; Jung, J.H.; Lee, J.H.; Kim, J.; Freeman, M.R.; Lee, K.Y.; et al. Identification and characterization of proteins isolated from microvesicles derived from human lung cancer pleural effusions. Proteomics 2013, 13, 2125–2134. [Google Scholar] [CrossRef]
- Lin, J.; Wang, Y.; Zou, Y.Q.; Chen, X.; Huang, B.; Liu, J.; Xu, Y.M.; Li, J.; Zhang, J.; Yang, W.M.; et al. Differential miRNA expression in pleural effusions derived from extracellular vesicles of patients with lung cancer, pulmonary tuberculosis, or pneumonia. Tumor Biol. 2016, 37, 15835–15845. [Google Scholar] [CrossRef]
- Tamiya, H.; Mitani, A.; Saito, A.; Ishimori, T.; Saito, M.; Isago, H.; Jo, T.; Yamauchi, Y.; Tanaka, G.; Nagase, T. Exosomal MicroRNA Expression Profiling in Patients with Lung Adenocarcinoma-associated Malignant Pleural Effusion. Anticancer Res. 2018, 38, 6707–6714. [Google Scholar] [CrossRef]
- Lukic, A.; Wahlund, C.J.E.; Gomez, C.; Brodin, D.; Samuelsson, B.; Wheelock, C.E.; Gabrielsson, S.; Radmark, O. Exosomes and cells from lung cancer pleural exudates transform LTC4 to LTD4, promoting cell migration and survival via CysLT1. Cancer Lett. 2019, 444, 1–8. [Google Scholar] [CrossRef]
- Song, Z.; Cai, Z.; Yan, J.; Shao, Y.W.; Zhang, Y. Liquid biopsies using pleural effusion-derived exosomal DNA in advanced lung adenocarcinoma. Transl. Lung Cancer Res. 2019, 8, 392–400. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, Y.M.; Zou, Y.Q.; Lin, J.; Huang, B.; Liu, J.; Li, J.; Zhang, J.; Yang, W.M.; Min, Q.H.; et al. Identification of differential expressed PE exosomal miRNA in lung adenocarcinoma, tuberculosis, and other benign lesions. Medicine 2017, 96, e8361. [Google Scholar] [CrossRef]
- Ludwig, A.K.; De Miroschedji, K.; Doeppner, T.R.; Borger, V.; Ruesing, J.; Rebmann, V.; Durst, S.; Jansen, S.; Bremer, M.; Behrmann, E.; et al. Precipitation with polyethylene glycol followed by washing and pelleting by ultracentrifugation enriches extracellular vesicles from tissue culture supernatants in small and large scales. J. Extracell. Vesicles 2018, 7, 1528109. [Google Scholar] [CrossRef]
- Rider, M.A.; Hurwitz, S.N.; Meckes, D.G., Jr. ExtraPEG: A Polyethylene Glycol-Based Method for Enrichment of Extracellular Vesicles. Sci. Rep. 2016, 6, 23978. [Google Scholar] [CrossRef]
- Walker, S.E.; Lorsch, J. RNA purification—Precipitation methods. Methods Enzymol. 2013, 530, 337–343. [Google Scholar] [PubMed]
- Thery, C.; Amigorena, S.; Raposo, G.; Clayton, A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol. 2006, 30, 3.22.1–3.22.29. [Google Scholar] [CrossRef] [PubMed]
- Porcel, J.M.; Vives, M. Etiology and pleural fluid characteristics of large and massive effusions. Chest 2003, 124, 978–983. [Google Scholar] [CrossRef]
- Bica-Pop, C.; Cojocneanu-Petric, R.; Magdo, L.; Raduly, L.; Gulei, D.; Berindan-Neagoe, I. Overview upon miR-21 in lung cancer: Focus on NSCLC. Cell. Mol. Life Sci. 2018, 75, 3539–3551. [Google Scholar] [CrossRef]
- Dinh, T.K.; Fendler, W.; Chalubinska-Fendler, J.; Acharya, S.S.; O’Leary, C.; Deraska, P.V.; D’Andrea, A.D.; Chowdhury, D.; Kozono, D. Circulating miR-29a and miR-150 correlate with delivered dose during thoracic radiation therapy for non-small cell lung cancer. Radiat. Oncol. 2016, 11, 61. [Google Scholar] [CrossRef]
- Xue, X.; Wang, C.; Xue, Z.; Wen, J.; Han, J.; Ma, X.; Zang, X.; Deng, H.; Guo, R.; Asuquo, I.P.; et al. Exosomal miRNA profiling before and after surgery revealed potential diagnostic and prognostic markers for lung adenocarcinoma. Acta Biochim. Biophys. Sin. 2020, 52, 281–293. [Google Scholar] [CrossRef]
- Crossland, R.E.; Norden, J.; Bibby, L.A.; Davis, J.; Dickinson, A.M. Evaluation of optimal extracellular vesicle small RNA isolation and qRT-PCR normalisation for serum and urine. J. Immunol. Methods 2016, 429, 39–49. [Google Scholar] [CrossRef]
- Dixit, R.; Agarwal, K.C.; Gokhroo, A.; Patil, C.B.; Meena, M.; Shah, N.S.; Arora, P. Diagnosis and management options in malignant pleural effusions. Lung India 2017, 34, 160–166. [Google Scholar] [CrossRef]
- Antonangelo, L.; Sales, R.K.; Cora, A.P.; Acencio, M.M.; Teixeira, L.R.; Vargas, F.S. Pleural fluid tumour markers in malignant pleural effusion with inconclusive cytologic results. Curr. Oncol. 2015, 22, e336–e341. [Google Scholar] [CrossRef]
- Lotvall, J.; Hill, A.F.; Hochberg, F.; Buzas, E.I.; Di Vizio, D.; Gardiner, C.; Gho, Y.S.; Kurochkin, I.V.; Mathivanan, S.; Quesenberry, P.; et al. Minimal experimental requirements for definition of extracellular vesicles and their functions: A position statement from the International Society for Extracellular Vesicles. J. Extracell. Vesicles 2014, 3, 26913. [Google Scholar] [CrossRef]
- Gatta, A.T.; Carlton, J.G. The ESCRT-machinery: Closing holes and expanding roles. Curr. Opin. Cell Biol. 2019, 59, 121–132. [Google Scholar] [CrossRef]
- Cheng, Y.; Zeng, Q.; Han, Q.; Xia, W. Effect of pH, temperature and freezing-thawing on quantity changes and cellular uptake of exosomes. Protein Cell 2019, 10, 295–299. [Google Scholar] [CrossRef]
- Betzel, C.; Bellemann, M.; Pal, G.P.; Bajorath, J.; Saenger, W.; Wilson, K.S. X-ray and model-building studies on the specificity of the active site of proteinase K. Proteins 1988, 4, 157–164. [Google Scholar] [CrossRef]
- Betzel, C.; Singh, T.P.; Visanji, M.; Peters, K.; Fittkau, S.; Saenger, W.; Wilson, K.S. Structure of the complex of proteinase K with a substrate analogue hexapeptide inhibitor at 2.2-A resolution. J. Biol. Chem. 1993, 268, 15854–15858. [Google Scholar] [CrossRef]
- Chubb, S.P.; Williams, R.A. Biochemical Analysis of Pleural Fluid and Ascites. Clin. Biochem. Rev. 2018, 39, 39–50. [Google Scholar]
- Gao, X.; Yuan, S.; Jayaraman, S.; Gursky, O. Differential stability of high-density lipoprotein subclasses: Effects of particle size and protein composition. J. Mol. Biol. 2009, 387, 628–638. [Google Scholar] [CrossRef][Green Version]
- Gursky, O. Structural stability and functional remodeling of high-density lipoproteins. FEBS Lett. 2015, 589 Pt A, 2627–2639. [Google Scholar] [CrossRef]
- Mendez, A.J.; Oram, J.F. Limited proteolysis of high density lipoprotein abolishes its interaction with cell-surface binding sites that promote cholesterol efflux. Biochim. Biophys. Acta 1997, 1346, 285–299. [Google Scholar] [CrossRef]
- Liu, Q.; Yu, Z.; Yuan, S.; Xie, W.; Li, C.; Hu, Z.; Xiang, Y.; Wu, N.; Wu, L.; Bai, L.; et al. Circulating exosomal microRNAs as prognostic biomarkers for non-small-cell lung cancer. Oncotarget 2017, 8, 13048–13058. [Google Scholar] [CrossRef]
- Shi, J. Considering Exosomal miR-21 as a Biomarker for Cancer. J. Clin. Med. 2016, 5, 42. [Google Scholar] [CrossRef] [PubMed]
- Fabbri, M.; Paone, A.; Calore, F.; Galli, R.; Gaudio, E.; Santhanam, R.; Lovat, F.; Fadda, P.; Mao, C.; Nuovo, G.J.; et al. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc. Natl. Acad. Sci. USA 2012, 109, E2110–E2116. [Google Scholar] [CrossRef] [PubMed]
- Alipoor, S.D.; Tabarsi, P.; Varahram, M.; Movassaghi, M.; Dizaji, M.K.; Folkerts, G.; Garssen, J.; Adcock, I.M.; Mortaz, E. Serum Exosomal miRNAs Are Associated with Active Pulmonary Tuberculosis. Dis. Markers 2019, 2019, 1907426. [Google Scholar] [CrossRef] [PubMed]

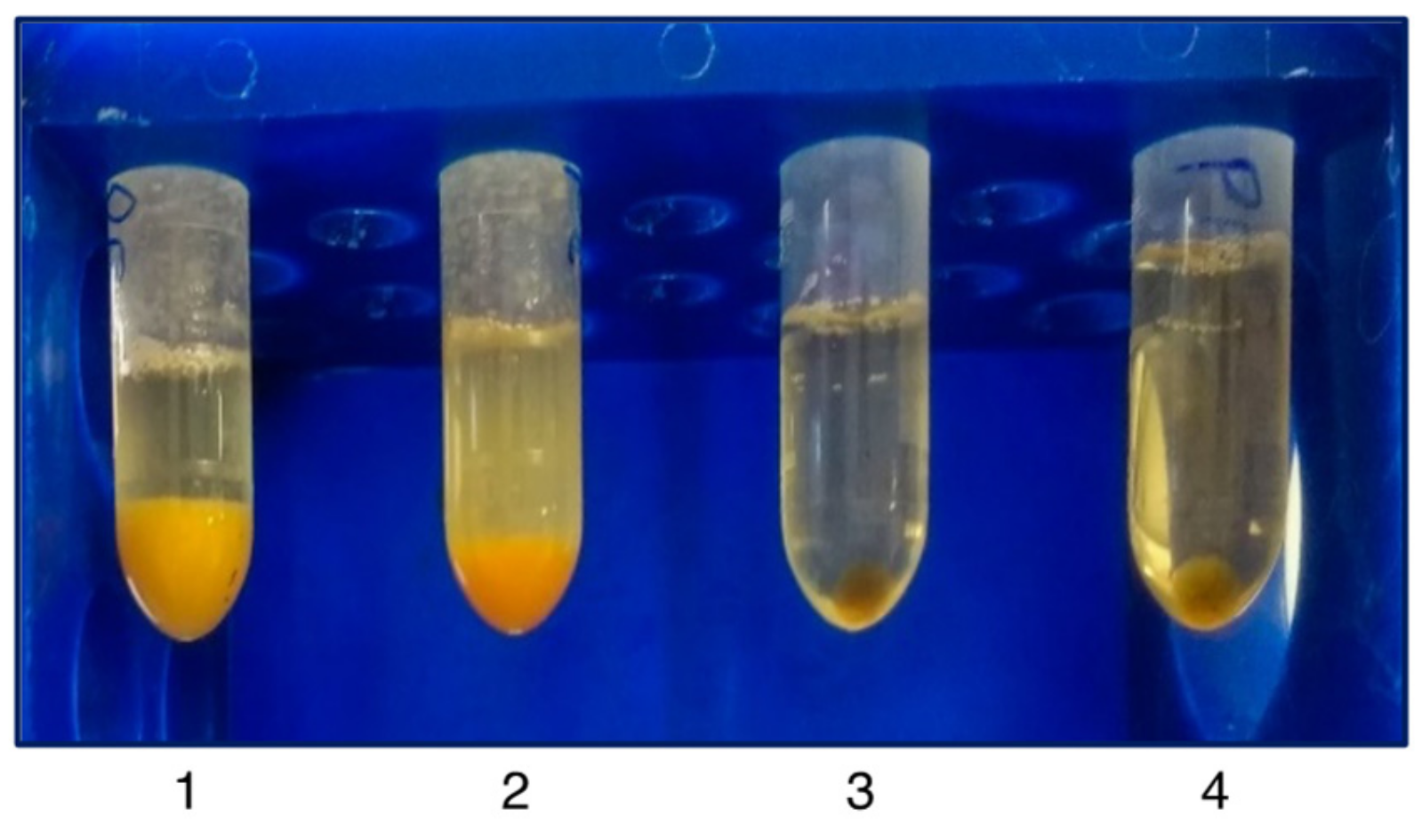
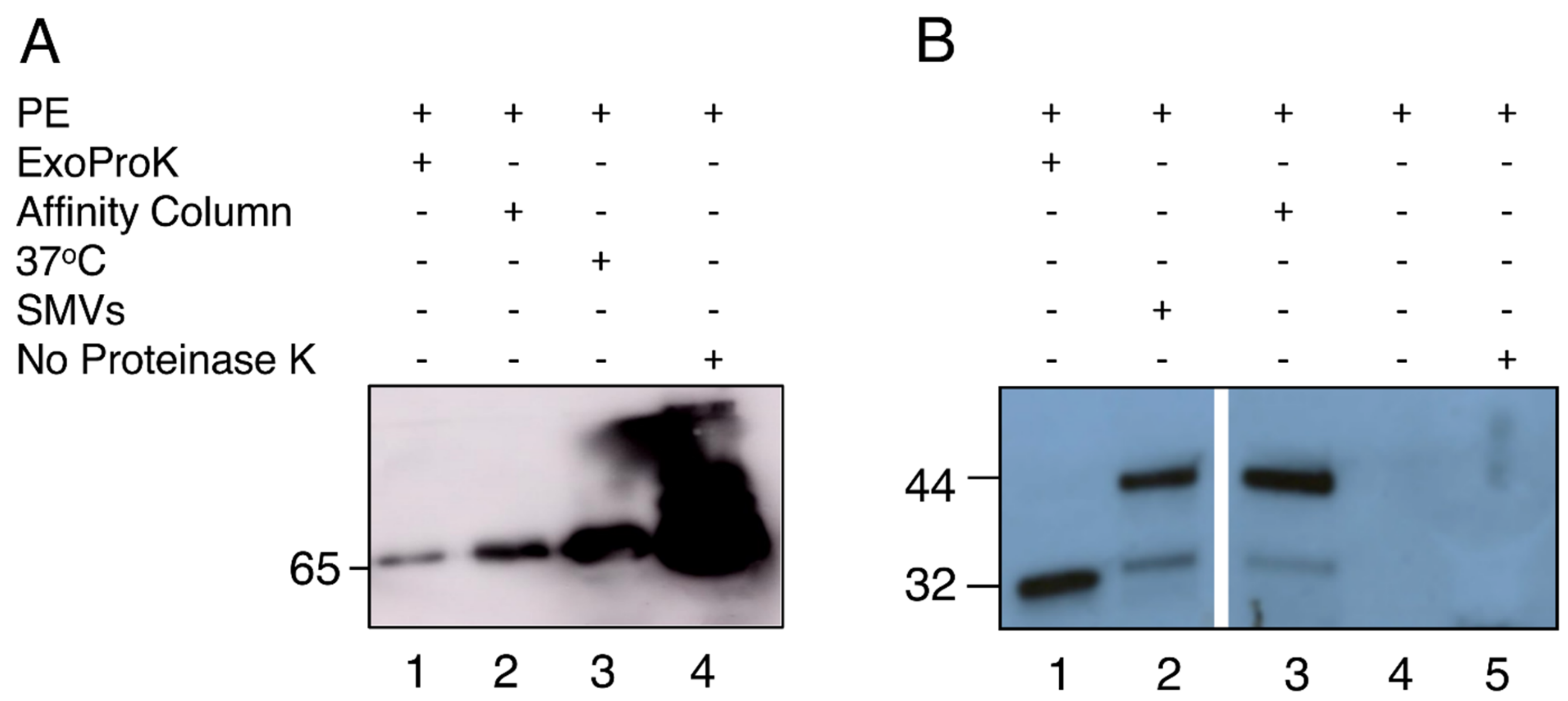

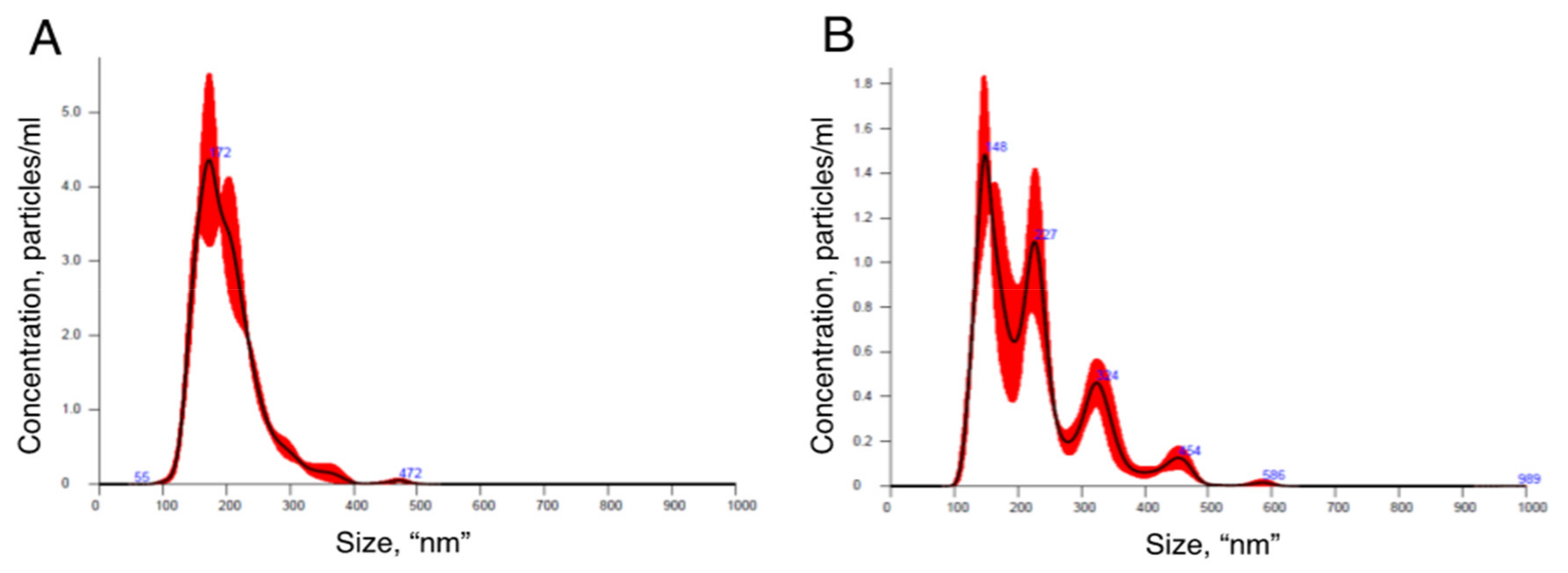
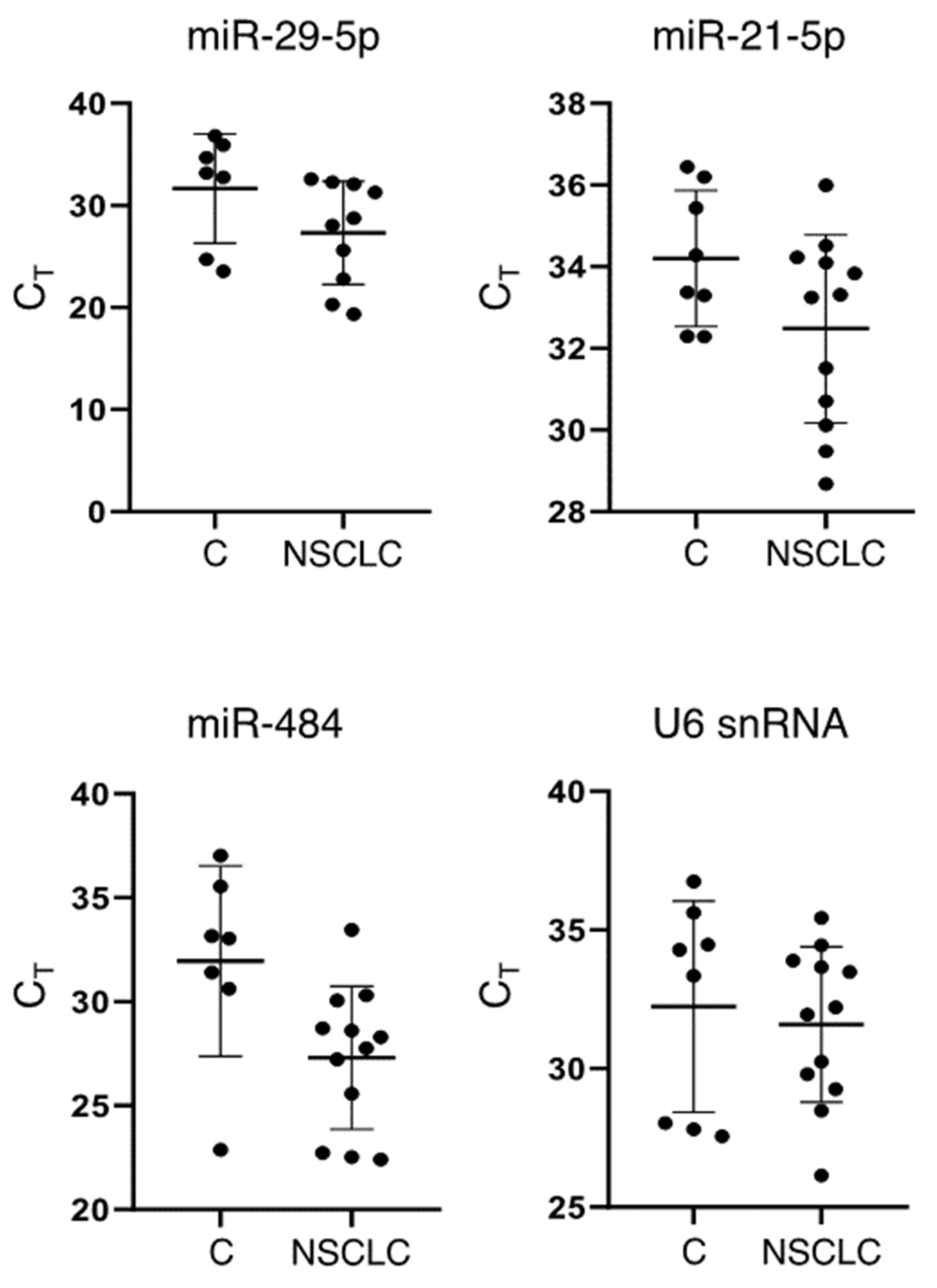
| Vesicle Isolation Technique | Downstream Analysis | Project | Publication |
|---|---|---|---|
| Differential Ultracentrifugation | DNA—qPCR | Detect EGFR mutations in sEV enriched DNA | [20] |
| Sucrose gradient Ultracentrifugation | Protein Mass-spec | Protein biomarker discovery | [21] |
| Polymer precipitation | DNA—qPCR | Validate diagnostic value EGFR mutations | [22] |
| Affinity column | RNA—qPCR | RNA biomarker discovery | [23] |
| Sucrose gradient Ultracentrifugation | Protein Mass-spec | Protein biomarker discovery | [24] |
| Differential Ultracentrifugation | RNA sequencing | RNA biomarker discovery | [25] |
| Polymer precipitation | RNA—qPCR | RNA biomarker validation | [26] |
| Sucrose gradient Ultracentrifugation | Cell migration—proliferation | Determine the effect of PE sEV on cultured cells | [27] |
| Differential Ultracentrifugation | DNA sequencing | Detect common oncogene mutations in sEV-enriched DNA | [28] |
| Differential Ultracentrifugation | RNA sequencing | RNA biomarker discovery—validation | [29] |
| NSCLC Patients n = 12 | Control Subjects (non-NSCLC-Related) n = 8 | |
|---|---|---|
| Age, years (median) | 72 | 65 |
| Male/Female | 8/4 | 5/3 |
| Exudates | 12 | 8 |
| Smoking status | ||
| Smoker/Ex-smoker | 11 | 6 |
| Non-smoker | 1 | 2 |
| Control Subjects’ Characteristics | ||
| Tuberculosis | - | 1 |
| Post-operative | - | 1 |
| Kidney disease | - | 1 |
| Inflammation | - | 2 |
| Heart failure | - | 1 |
| Rheumatoid arthritis | - | 1 |
| Connective tissue disease | - | 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Antonopoulos, D.; Tsilioni, I.; Tsiara, S.; Moustaka, E.; Ladias, S.; Perlepe, G.; Theoharides, T.C.; Gourgoulianis, K.I.; Balatsos, N.A.A. ExoProK: A Practical Method for the Isolation of Small Extracellular Vesicles from Pleural Effusions. Methods Protoc. 2021, 4, 31. https://doi.org/10.3390/mps4020031
Antonopoulos D, Tsilioni I, Tsiara S, Moustaka E, Ladias S, Perlepe G, Theoharides TC, Gourgoulianis KI, Balatsos NAA. ExoProK: A Practical Method for the Isolation of Small Extracellular Vesicles from Pleural Effusions. Methods and Protocols. 2021; 4(2):31. https://doi.org/10.3390/mps4020031
Chicago/Turabian StyleAntonopoulos, Dionysios, Irene Tsilioni, Sophia Tsiara, Eirini Moustaka, Spyridon Ladias, Garyfallia Perlepe, Theoharis C. Theoharides, Konstantinos I. Gourgoulianis, and Nikolaos A. A. Balatsos. 2021. "ExoProK: A Practical Method for the Isolation of Small Extracellular Vesicles from Pleural Effusions" Methods and Protocols 4, no. 2: 31. https://doi.org/10.3390/mps4020031
APA StyleAntonopoulos, D., Tsilioni, I., Tsiara, S., Moustaka, E., Ladias, S., Perlepe, G., Theoharides, T. C., Gourgoulianis, K. I., & Balatsos, N. A. A. (2021). ExoProK: A Practical Method for the Isolation of Small Extracellular Vesicles from Pleural Effusions. Methods and Protocols, 4(2), 31. https://doi.org/10.3390/mps4020031







