Combined Fluorescence-Based in Vitro Assay for the Simultaneous Detection of Cell Viability and Alkaline Phosphatase Activity during Osteogenic Differentiation of Osteoblast Precursor Cells
Abstract
1. Introduction
2. Experimental Design
2.1. Materials
- 1.5x ALP Assay Buffer (75 mM Tris pH 9.3, 1.5 mM MgCl2, 0.15 mM ZnCl2 (all Carl Roth));
- ALP standard working solution (0.4 mU/µL) (shrimp alkaline phosphatase (Thermo Fisher));
- Phosphate-buffered saline (PBS) (Thermo Fisher);
- FDA (Thermo Fisher) working solution (2 µg/mL in PBS);
- 0.5% Triton-X-100 (Sigma-Aldrich, Steinheim, Germany) in PBS (Thermo Fisher) 4-Methylumbelliferyl Phosphate (4-MUP) (Thermo Fisher) working solution (100 µM in 1.5x ALP assay buffer);
- Distilled water for buffer preparation;
- MicroBCA kit (Thermo Fisher) (optional);
- Protein standard working solution (400 µg/mL) (optional, included in the MicroBCA kit).
2.2. Equipment
- 96-well cell culture plates transparent (Sarstedt, Nümbrecht, Germany) 96-well cell culture plates black or white (Kisker Biotech GmbH & Co. KG, Steinfurt, Germany)
- Fluorescence microplate reader (Wallac 1420 Victor 2, Perkin Elmer, Waltham, MA) Filter: 485/530 (ex/em) and 360/440 (ex/em)
2.3. Study Ethics
2.4. Primary Cells and Cell Lines
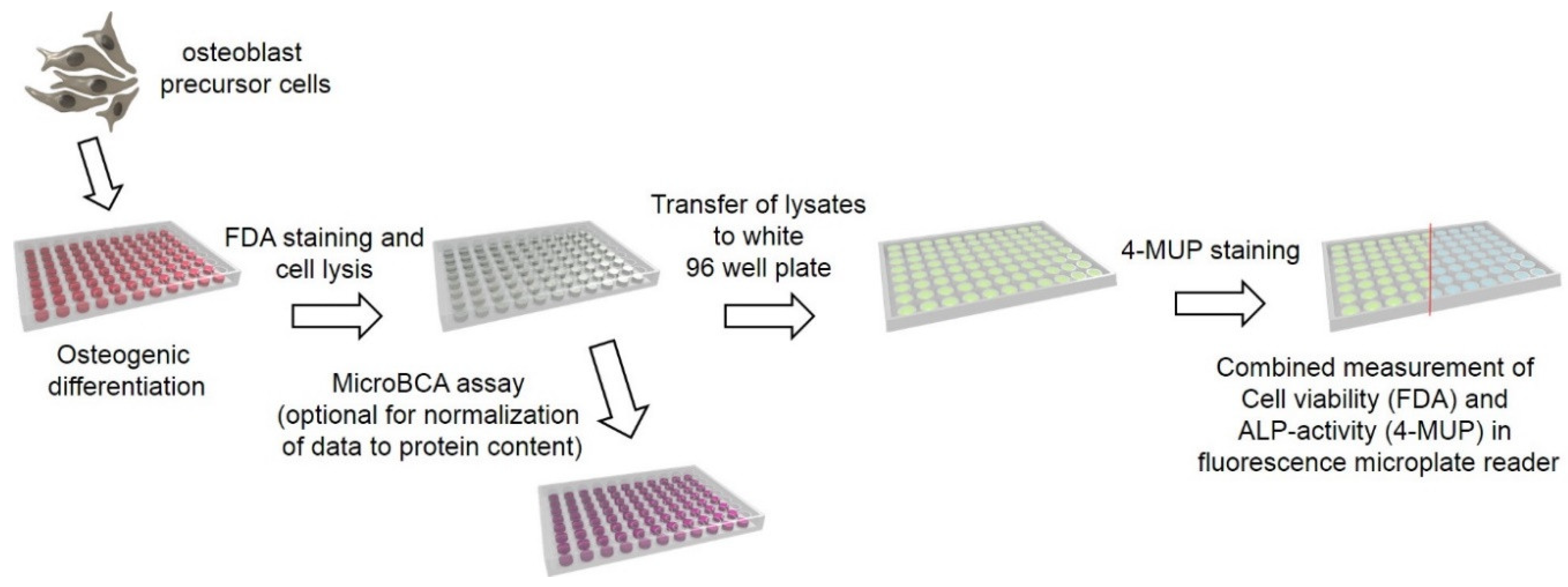
3. Procedure
- FDA staining (10 min)
- Culture cells in 96-well plates (5000 cells/well)
- Treat cells as desired
- Discard media from the 96-well cell culture plate
- Wash cells once with 100 µL PBS
- Add 100 µL of FDA working solution
- Incubate for 5 min at 37 °C
- Discard FDA working solution
- Wash cells once with 100 µL PBS
- Cell lysis (5 min)
- After the last wash, discard PBS and add 150 µL of 0.5% Triton-X-100 in PBS
- Incubate for 5 min at 37 °C
- Preparation of ALP and Protein standard (10 min)
- (1)
- ALP standard
- Add 50 µL of distilled H2O to wells A1 to H1 and A2 to H2 of a black or white 96-well cell culture plate
- Add 50 µL of ALP standard working solution to wells A1 and A2
- Transfer 50 µL of diluted ALP-standard from A1 to B1, from B1 to C1, from C1 to D1, from D1 to E1, from E1 to F1, from F1 to G1 and discard 50 µL from G1. The resulting standards range from 10 mU to 0.16 mU ALP
- Repeat the previous two steps for wells A2 to H2
- H1 and H2 represent the blanks
- (2)
- Protein standard (optional)
- Add 75 µL of distilled H2O to wells A1 to H1 and A2 to H2 of a transparent 96-well cell culture plate
- Add 75 µL of protein standard working solution to well A1 and A2
- Transfer 75 µL of diluted ALP-standard from A1 to B1, from B1 to C1, from C1 to D1, from D1 to E1, from E1 to F1, from F1 to G1 and discard 75 µL from G1. The resulting standards range from 200 µg/mL to 3.125 µg/mL BSA
- Repeat the previous two steps for wells A2 to H2
- H1 and H2 represent the blanks
- 4-MUP staining and fluorescence quantification (25 min)
- Mix cell lysates by pipetting up and down several times and transfer 50 µL of lysate into the empty wells of the white 96-well cell culture plate with the ALP standards
- ○
CRITICAL STEP: Mix and transfer gently to avoid foam formation
- Add 100 µL of 4-MUP working solution to all wells, including the standards and blanks
- Incubate for 20 min at 37 °C
- Quantify the fluorescence intensity of fluorescein at 485/535 (ex/em) with a microplate fluorescence reader
- Quantify the fluorescence intensity of 4’MU at 360/440 (ex/em) with a microplate fluorescence reader
- Total Protein measurement (optional) (2 h and 10 min)
- Transfer 75 µL of cell lysates into the empty wells of the prepared transparent 96-well cell culture plate containing the protein standard
- ○
CRITICAL STEP: Transfer gently to avoid foam formation
- Add 75 µL of Micro BCA working solution to all wells, including the standards and blanks
- Incubate for 2 h at 37 °C
- Cool the plate to room temperature (RT)
- Quantify the absorption in a microplate reader at ex 562 nm
4. Expected Results
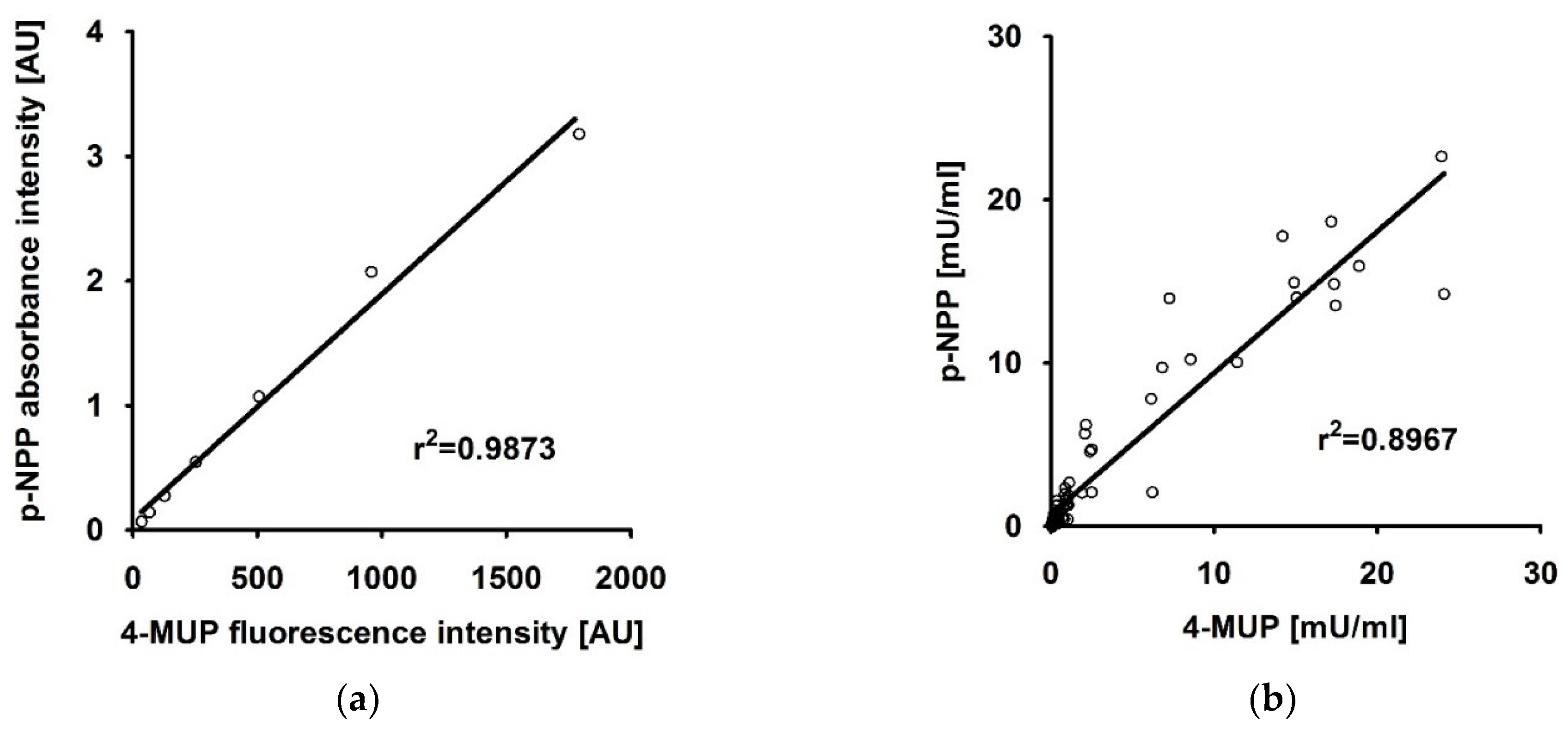
4.1. The Fluorescence-Based 4-MUP and Absorbance-Based p-NPP Assays Produce Comparable Results
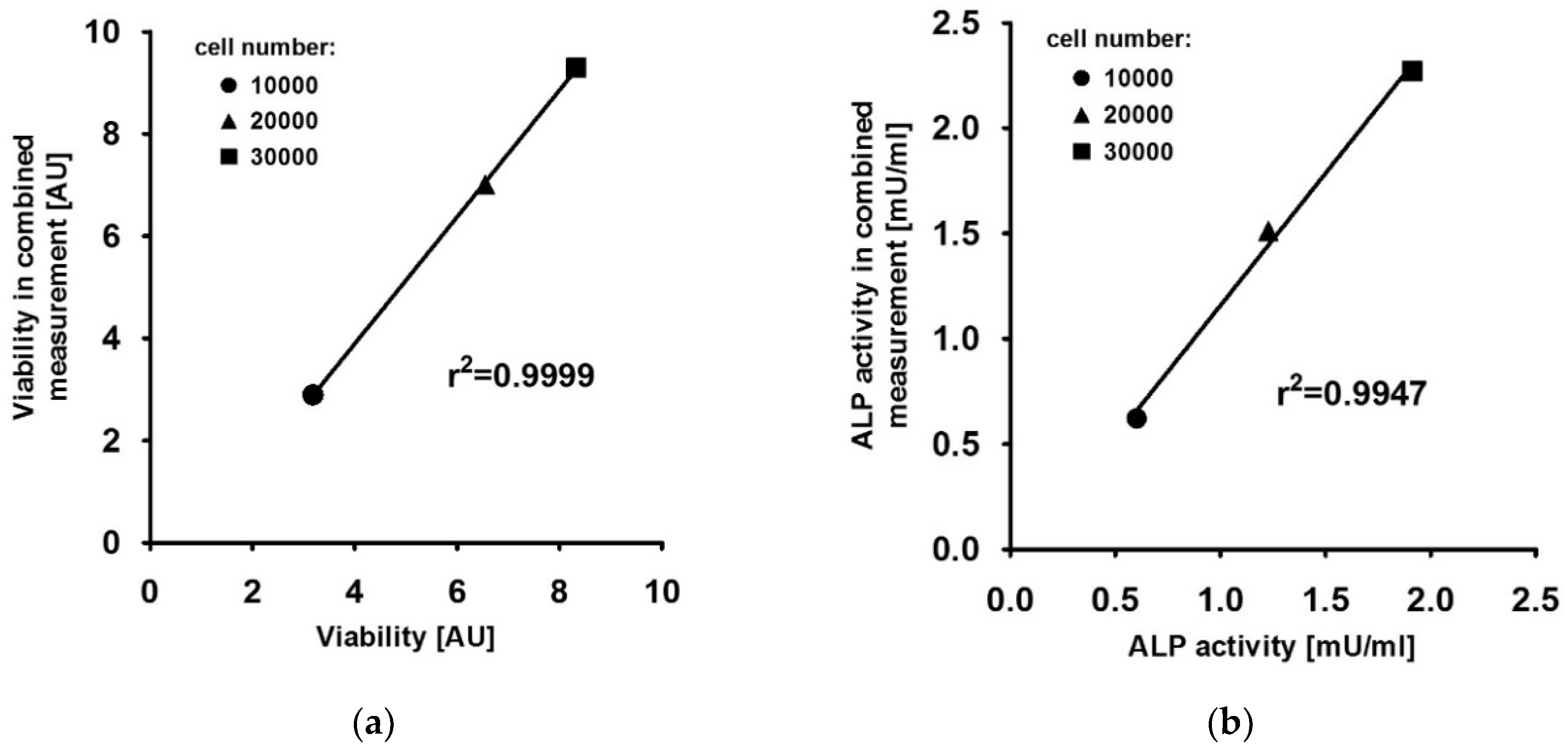
4.2. FDA Fluorescence Intensity Correlates With the Number of Viable Cells
4.3. FDA and 4-MUP Fluorescence Signals Do not Interfere in the Combined Approach
5. Reagents Setup
- ALP Standard
- Stock solution: 1 U/µL Shrimp alkaline phosphatase (Thermo Fisher)
- Working solution: ALP stock solution diluted 1:2500 in distilled H2O (400 µU/µL)
- Protein Standard
- Stock solution: 2 mg/mL Albumin standard (Thermo Fisher)
- Working solution: Albumin standard stock solution diluted 1:5 in distilled H2O (400 µg/mL)
- 10x ALP Assay Buffer (storage at 4 °C)
- 500 mM TRIS pH 9.3 (Carl Roth, Karlsruhe, Germany)
- 10 mM MgCl2 (Carl Roth)
- 1 mM ZnCl2 (Carl Roth)
- FDA substrate solution
- Stock solution: 1 mg/mL FDA (Thermo Fisher) in acetone (Carl Roth) (storage at −20 °C)
- Working solution: FDA stock solution diluted 1:50 in PBS (20 µg/mL)
- 4-MUP substrate solution
- Stock solution: 5 mM 4-MUP (Thermo Fisher) (storage at 4 °C)
- Working solution: 4-MUP Stock solution diluted 1:50 in 1.5x ALP Assay Buffer (100 µM)
- Micro BCA substrate solution
- Micro BCA™ Protein Assay Kit: mix according to manufacturer’s instruction (Thermo Fisher)
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hench, L.L. The story of Bioglass. J. Mater. Sci. Mater. Med. 2006, 17, 967–978. [Google Scholar] [CrossRef] [PubMed]
- Birmingham, E.; Niebur, G.L.; McHugh, P.E.; Shaw, G.; Barry, F.P.; McNamara, L.M. Osteogenic differentiation of mesenchymal stem cells is regulated by osteocyte and osteoblast cells in a simplified bone niche. Eur Cell Mater. 2012, 23, 13–27. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, S.I.; Widholz, B.; Essers, C.; Becker, M.; Tulyaganov, D.U.; Moghaddam, A.; Gonzalo de Juan, I.; Westhauser, F. Superior biocompatibility and comparable osteoinductive properties: Sodium-reduced fluoride-containing bioactive glass belonging to the CaO-MgO-SiO2 system as a promising alternative to 45S5 bioactive glass. Bioact Mater. 2020, 5, 55–65. [Google Scholar] [CrossRef]
- Yao, X.Y.; Wang, Q.; Liu, Q.; Pang, M.; Du, X.M.; Zhao, B.; Li, Y.; Ruan, W.J. Ultrasensitive Assay of Alkaline Phosphatase Based on the Fluorescent Response Difference of the Metal-Organic Framework Sensor. ACS Omega 2020, 5, 712–717. [Google Scholar] [CrossRef]
- Reible, B.; Schmidmaier, G.; Moghaddam, A.; Westhauser, F. Insulin-Like Growth Factor-1 as a Possible Alternative to Bone Morphogenetic Protein-7 to Induce Osteogenic Differentiation of Human Mesenchymal Stem Cells in Vitro. Int J. Mol. Sci. 2018, 19, 1674. [Google Scholar] [CrossRef]
- Westhauser, F.; Karadjian, M.; Essers, C.; Senger, A.S.; Hagmann, S.; Schmidmaier, G.; Moghaddam, A. Osteogenic differentiation of mesenchymal stem cells is enhanced in a 45S5-supplemented beta-TCP composite scaffold: An in-vitro comparison of Vitoss and Vitoss BA. PLoS One 2019, 14, e0212799. [Google Scholar] [CrossRef]
- Westhauser, F.; Widholz, B.; Nawaz, Q.; Tsitlakidis, S.; Hagmann, S.; Moghaddam, A.; Boccaccini, A.R. Favorable angiogenic properties of the borosilicate bioactive glass 0106-B1 result in enhanced in vivo osteoid formation compared to 45S5 Bioglass. J. Biomater. Sci. Polym. Ed. 2019, 7, 5161–5176. [Google Scholar] [CrossRef]
- Westhauser, F.; Hohenbild, F.; Arango-Ospina, M.; Schmitz, S.I.; Wilkesmann, S.; Hupa, L.; Moghaddam, A.; Boccaccini, A.R. Bioactive Glass (BG) ICIE16 Shows Promising Osteogenic Properties Compared to Crystallized 45S5-BG. Int J. Mol. Sci. 2020, 21, 1639. [Google Scholar] [CrossRef]
- Stefkova, K.; Prochazkova, J.; Pachernik, J. Alkaline phosphatase in stem cells. Stem Cells Int. 2015, 2015, 628368. [Google Scholar] [CrossRef]
- Karadjian, M.; Senger, A.S.; Essers, C.; Wilkesmann, S.; Heller, R.; Fellenberg, J.; Simon, R.; Westhauser, F. Human Platelet Lysate Can Replace Fetal Calf Serum as a Protein Source to Promote Expansion and Osteogenic Differentiation of Human Bone-Marrow-Derived Mesenchymal Stromal Cells. Cells 2020, 9, 918. [Google Scholar] [CrossRef]
- Rahaman, M.N.; Day, D.E.; Bal, B.S.; Fu, Q.; Jung, S.B.; Bonewald, L.F.; Tomsia, A.P. Bioactive glass in tissue engineering. Acta Biomater. 2011, 7, 2355–2373. [Google Scholar] [CrossRef] [PubMed]
- Karadjian, M.; Essers, C.; Tsitlakidis, S.; Reible, B.; Moghaddam, A.; Boccaccini, A.R.; Westhauser, F. Biological Properties of Calcium Phosphate Bioactive Glass Composite Bone Substitutes: Current Experimental Evidence. Int J. Mol. Sci. 2019, 20, 305. [Google Scholar] [CrossRef] [PubMed]
- Hoppe, A.; Guldal, N.S.; Boccaccini, A.R. A review of the biological response to ionic dissolution products from bioactive glasses and glass-ceramics. Biomaterials 2011, 32, 2757–2774. [Google Scholar] [CrossRef] [PubMed]
- Daguano, J.; Milesi, M.T.B.; Rodas, A.C.D.; Weber, A.F.; Sarkis, J.E.S.; Hortellani, M.A.; Zanotto, E.D. In vitro biocompatibility of new bioactive lithia-silica glass-ceramics. Mater. Sci. Eng. C 2019, 94, 117–125. [Google Scholar] [CrossRef]
- Kargozar, S.; Lotfibakhshaeish, N.; Ebrahimi-Barough, S.; Nazari, B.; Hill, R.G. Stimulation of Osteogenic Differentiation of Induced Pluripotent Stem Cells (iPSCs) Using Bioactive Glasses: An in vitro Study. Front. Bioeng Biotechnol. 2019, 7, 355. [Google Scholar] [CrossRef]
- Talukdar, Y.; Rashkow, J.; Lalwani, G.; Kanakia, S.; Sitharaman, B. The effects of graphene nanostructures on mesenchymal stem cells. Biomaterials 2014, 35, 4863–4877. [Google Scholar] [CrossRef]
- Bejarano, J.; Detsch, R.; Boccaccini, A.R.; Palza, H. PDLLA scaffolds with Cu- and Zn-doped bioactive glasses having multifunctional properties for bone regeneration. J. Biomed. Mater. Res. A 2017, 105, 746–756. [Google Scholar] [CrossRef]
- Gade, T.P.; Motley, M.W.; Beattie, B.J.; Bhakta, R.; Boskey, A.L.; Koutcher, J.A.; Mayer-Kuckuk, P. Imaging of alkaline phosphatase activity in bone tissue. PLoS One 2011, 6, e22608. [Google Scholar] [CrossRef][Green Version]
- Jiajia, L.; Shinghung, M.; Jiacheng, Z.; Jialing, W.; Dilin, X.; Shengquan, H.; Zaijun, Z.; Qinwen, W.; Yifan, H.; Wei, C. Assessment of Neuronal Viability Using Fluorescein Diacetate-Propidium Iodide Double Staining in Cerebellar Granule Neuron Culture. J. Vis. Exp. 2017, 123, e55442. [Google Scholar] [CrossRef]
- Reible, B.; Schmidmaier, G.; Prokscha, M.; Moghaddam, A.; Westhauser, F. Continuous stimulation with differentiation factors is necessary to enhance osteogenic differentiation of human mesenchymal stem cells in-vitro. Growth Factors 2017, 35, 179–188. [Google Scholar] [CrossRef]
- Widholz, B.; Tsitlakidis, S.; Reible, B.; Moghaddam, A.; Westhauser, F. Pooling of Patient-Derived Mesenchymal Stromal Cells Reduces Inter-Individual Confounder-Associated Variation without Negative Impact on Cell Viability, Proliferation and Osteogenic Differentiation. Cells 2019, 8, 633. [Google Scholar] [CrossRef] [PubMed]
- Radin, S.; Reilly, G.; Bhargave, G.; Leboy, P.S.; Ducheyne, P. Osteogenic effects of bioactive glass on bone marrow stromal cells. J. Biomed. Mater. Res. Part A 2005, 73, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Wilkesmann, S.; Fellenberg, J.; Nawaz, Q.; Reible, B.; Moghaddam, A.; Boccaccini, A.R.; Westhauser, F. Primary osteoblasts, osteoblast precursor cells or osteoblast-like cell lines: Which human cell types are (most) suitable for characterizing 45S5-bioactive glass? J. Biomed. Mater. Res. Part A 2020, 108, 663–674. [Google Scholar] [CrossRef] [PubMed]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wilkesmann, S.; Westhauser, F.; Fellenberg, J. Combined Fluorescence-Based in Vitro Assay for the Simultaneous Detection of Cell Viability and Alkaline Phosphatase Activity during Osteogenic Differentiation of Osteoblast Precursor Cells. Methods Protoc. 2020, 3, 30. https://doi.org/10.3390/mps3020030
Wilkesmann S, Westhauser F, Fellenberg J. Combined Fluorescence-Based in Vitro Assay for the Simultaneous Detection of Cell Viability and Alkaline Phosphatase Activity during Osteogenic Differentiation of Osteoblast Precursor Cells. Methods and Protocols. 2020; 3(2):30. https://doi.org/10.3390/mps3020030
Chicago/Turabian StyleWilkesmann, Sebastian, Fabian Westhauser, and Joerg Fellenberg. 2020. "Combined Fluorescence-Based in Vitro Assay for the Simultaneous Detection of Cell Viability and Alkaline Phosphatase Activity during Osteogenic Differentiation of Osteoblast Precursor Cells" Methods and Protocols 3, no. 2: 30. https://doi.org/10.3390/mps3020030
APA StyleWilkesmann, S., Westhauser, F., & Fellenberg, J. (2020). Combined Fluorescence-Based in Vitro Assay for the Simultaneous Detection of Cell Viability and Alkaline Phosphatase Activity during Osteogenic Differentiation of Osteoblast Precursor Cells. Methods and Protocols, 3(2), 30. https://doi.org/10.3390/mps3020030






