A Rapid and High Throughput MIC Determination Method to Screen Uranium Resistant Microorganisms
Abstract
:1. Introduction
2. Materials and Methods
2.1. Identification of Microorganisms
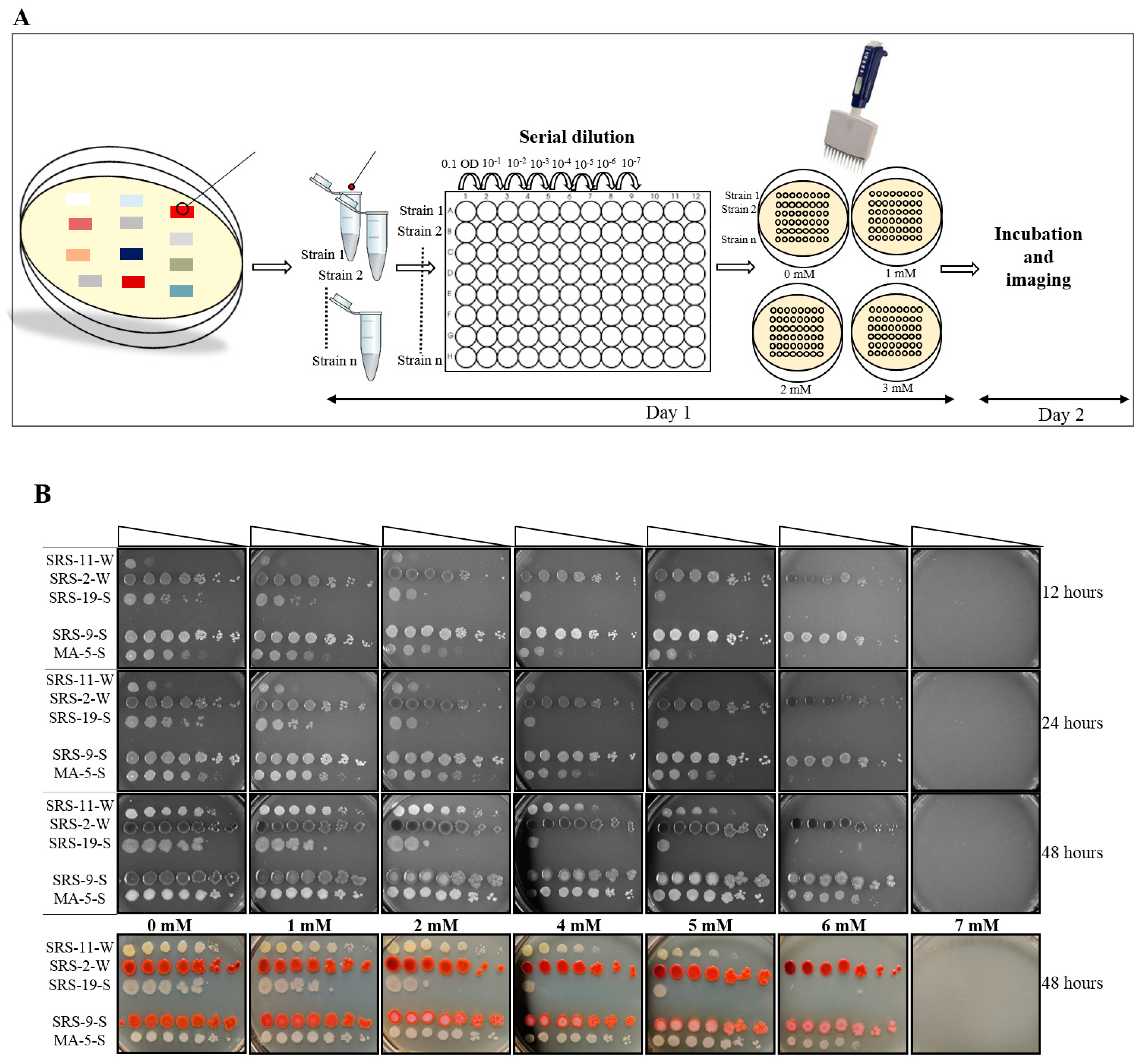
2.2. Plate Preparation
2.3. Plate MIC Method
2.3.1. Day 1
2.3.2. Day 2
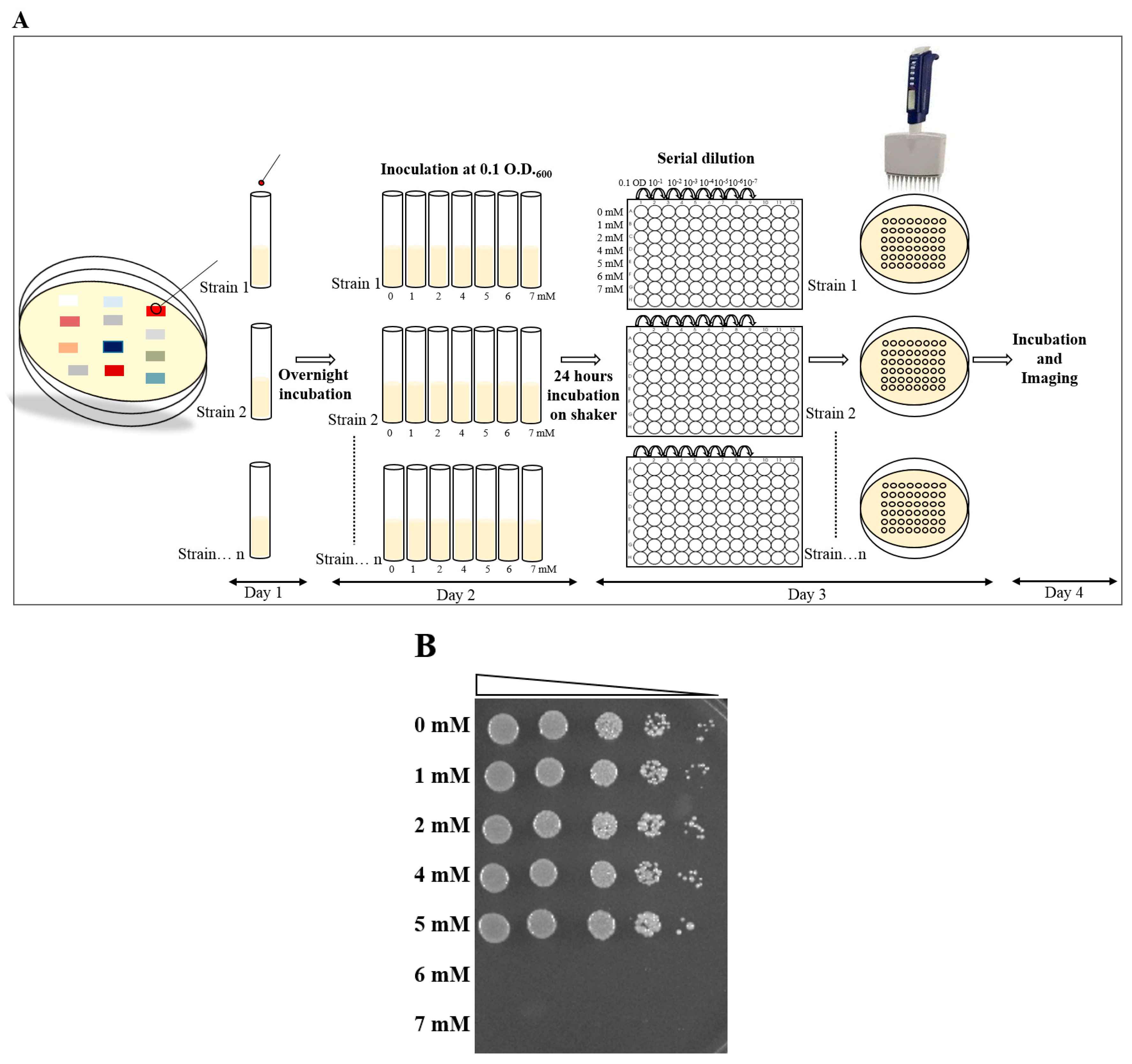
2.4. Broth Dilution Method
2.4.1. Day 1
2.4.2. Day 2
2.4.3. Day 3
2.4.4. Day 4
2.5. Spectrophotometric Count Method
2.5.1. Day 1
2.5.2. Day 2
2.5.3. Day 3
3. Results and Discussion
3.1. Screening of Bacterial Strains, Using the Plate MIC Method
3.2. MIC Determination, Using Broth Dilution Method, and Comparison to the Plate MIC Method
3.3. The MIC Determination, Using Spectrophotometric Count, and Comparison with the Plate MIC Method
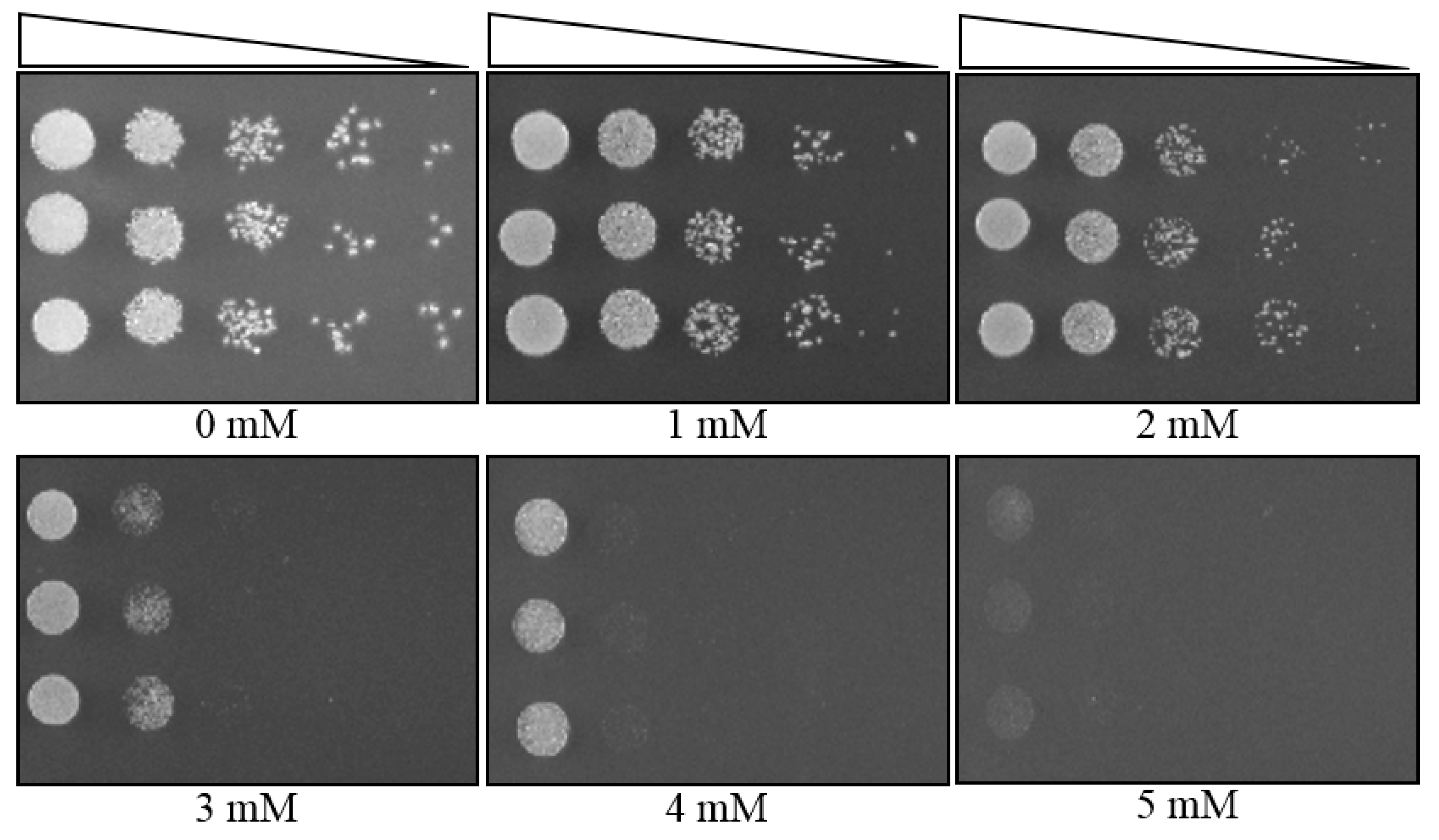
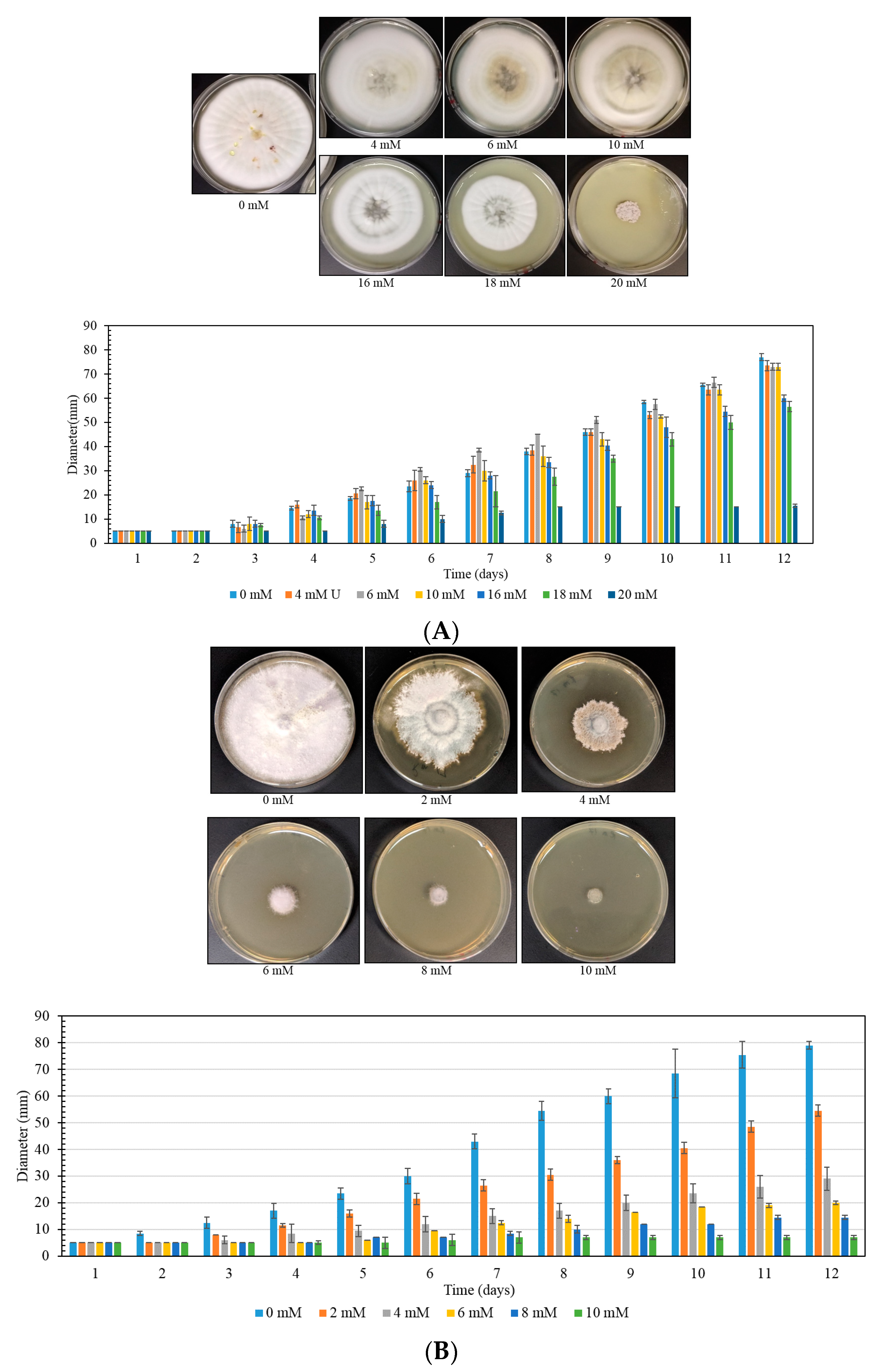
3.4. MIC Determination of a Yeast Strain, Using Plate MIC Method
3.5. MIC Determination of a Fungal Strain, Using Plate MIC Method
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Tchounwou, P.; Yedjou, C.G.; Patlolla, A.K.; Sutton, D.J. Heavy metal toxicity and the environment. Exp. Suppl. 2012, 101, 133–164. [Google Scholar] [PubMed] [Green Version]
- Jarup, L. Hazards of heavy metal contamination. Br. Med. Bull. 2003, 68, 167–182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jaishankar, M.; Tseten, T.; Anbalagan, N.; Mathew, B.B.; Beeregowda, K.N. Toxicity, mechanism and health effects of some heavy metals. Interdiscip. Toxicol. 2014, 7, 60–72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodriguez-Tudela, J.; Barchiesi, F.; Bille, J.; Chryssanthou, E.; Cuenca-Estrella, M.; Denning, D.; Donnelly, J.P.; Dupont, B.; Fegeler, W.; Moore, C.; et al. Method for the determination of minimum inhibitory concentration (MIC) by broth dilution of fermentative yeasts. Clin. Microbiol. Infect. 2003, 9, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Williams, K.H.; Bargar, J.R.; Lloyd, J.R.; Lovley, D. Bioremediation of uranium-contaminated groundwater: A systems approach to subsurface biogeochemistry. Curr. Opin. Biotechnol. 2013, 24, 489–497. [Google Scholar] [CrossRef]
- Bahafid, W.; Joutey, N.T.; Asri, M.; Sayel, H.; Tirry, N.; El Ghachtouli, N.; Sayel, N.T.H. Yeast Biomass: An Alternative for Bioremediation of Heavy Metals. Yeast Ind. Appl. 2017. [Google Scholar] [CrossRef] [Green Version]
- Ulhassan, Z.; Ali, S.; Rizwan, M.; Ibrahim, M.; Nafees, M.; Waseem, M. Role of Bioremediation Agents (Bacteria, Fungi, and Algae) in Alleviating Heavy Metal Toxicity. In Probiotics in Agroecosystem; Springer Science and Business Media LLC: Berlin/Heidelberg, Germany, 2017; pp. 517–537. [Google Scholar]
- Igiri, B.E.; Okoduwa, S.I.R.; Idoko, G.O.; Akabuogu, E.P.; Adeyi, A.O.; Ejiogu, I.K. Toxicity and Bioremediation of Heavy Metals Contaminated Ecosystem from Tannery Wastewater: A Review. J. Toxicol. 2018, 2018, 2568038. [Google Scholar] [CrossRef]
- Agarwal, M.; Rathore, R.S.; Jagoe, C.; Chauhan, A. Multiple Lines of Evidences Reveal Mechanisms Underpinning Mercury Resistance and Volatilization by Stenotrophomonas sp. MA5 Isolated from the Savannah River Site (SRS), USA. Cells 2019, 8, 309. [Google Scholar] [CrossRef] [Green Version]
- Pathak, A.; Agarwal, M.; Rathore, R.S.; Chauhan, A. Gene Determinants for Mercury Bioremediation as Revealed by Draft Genome Sequence Analysis of Stenotrophomonas sp. Strain MA5. Microbiol. Resour. Announc. 2019, 8, e00130-19. [Google Scholar] [CrossRef] [Green Version]
- Machado, M.D.; Santos, M.S.; Gouveia, C.; Soares, E.; Soares, E. Removal of heavy metals using a brewer’s yeast strain of Saccharomyces cerevisiae: The flocculation as a separation process. Bioresour. Technol. 2008, 99, 2107–2115. [Google Scholar] [CrossRef]
- Czarny, J.; Staninska-Pięta, J.; Piotrowska-Cyplik, A.; Juzwa, W.; Wolniewicz, A.; Marecik, R.; Ławniczak, Ł.; Chrzanowski, Ł. Acinetobacter sp. as the key player in diesel oil degrading community exposed to PAHs and heavy metals. J. Hazard. Mater. 2020, 383, 121168. [Google Scholar] [CrossRef]
- Mohamed, R.M.; Abo-Amer, A.E. Isolation and characterization of heavy-metal resistant microbes from roadside soil and phylloplane. J. Basic Microbiol. 2012, 52, 53–65. [Google Scholar] [CrossRef] [PubMed]
- Yamina, B.; Tahar, B.; Laure, F.M. Isolation and screening of heavy metal resistant bacteria from wastewater: A study of heavy metal co-resistance and antibiotics resistance. Water Sci. Technol. 2012, 66, 2041–2048. [Google Scholar] [CrossRef] [PubMed]
- Alboghobeish, H.; Tahmourespour, A.; Doudi, M. The study of Nickel Resistant Bacteria (NiRB) isolated from wastewaters polluted with different industrial sources. J. Environ. Health Sci. Eng. 2014, 12, 44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agarwal, M.; Pathak, A.; Rathore, R.S.; Prakash, O.; Singh, R.; Jaswal, R.; Seaman, J.C.; Chauhan, A. Proteogenomic Analysis of Burkholderia Species Strains 25 and 46 Isolated from Uraniferous Soils Reveals Multiple Mechanisms to Cope with Uranium Stress. Cells 2018, 7, 269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andrews, J.M. Determination of minimum inhibitory concentrations. J. Antimicrob. Chemother. 2001, 48, 5–16. [Google Scholar] [CrossRef] [Green Version]
- Wiegand, I.; Hilpert, K.; Hancock, R.E. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 2008, 3, 163–175. [Google Scholar] [CrossRef]
- Stefánsson, A. Iron (III) hydrolysis and solubility at 25 degrees C. Environ. Sci. Technol. 2007, 41, 6117–6123. [Google Scholar] [CrossRef]
- Saxena, M.; Loza-Rosas, S.A.; Gaur, K.; Sharma, S.; Otero, S.C.P.; Tinoco, A.D. Exploring titanium(IV) chemical proximity to iron(III) to elucidate a function for Ti(IV) in the human body. Coord. Chem. Rev. 2018, 363, 109–125. [Google Scholar] [CrossRef]
- Crans, D.C.; Woll, K.A.; Prusinskas, K.; Johnson, M.D.; Norkus, E. Metal Speciation in Health and Medicine Represented by Iron and Vanadium. Inorg. Chem. 2013, 52, 12262–12275. [Google Scholar] [CrossRef]
- Kwon-Chung, K.J.; Polacheck, I.; Popkin, T.J. Melanin-lacking mutants of Cryptococcus neoformans and their virulence for mice. J. Bacteriol. 1982, 150, 1414–1421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jacobson, E.S. Pathogenic roles for fungal melanins. Clin. Microbiol. Rev. 2000, 13, 708–717. [Google Scholar] [CrossRef] [PubMed]
- Lau, G.W.; Ran, H.; Kong, F.; Hassett, D.J.; Mavrodi, D.V. Pseudomonas aeruginosa Pyocyanin Is Critical for Lung Infection in Mice. Infect. Immun. 2004, 72, 4275–4278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, G.Y.; Essex, A.; Buchanan, J.T.; Datta, V.; Hoffman, H.M.; Bastian, J.F.; Fierer, J.; Nizet, V. Staphylococcus aureus golden pigment impairs neutrophil killing and promotes virulence through its antioxidant activity. J. Exp. Med. 2005, 202, 209–215. [Google Scholar] [CrossRef]
- Clauditz, A.; Resch, A.; Wieland, K.P.; Peschel, A.; Götz, F. Staphyloxanthin Plays a Role in the Fitness of Staphylococcus aureus and Its Ability To Cope with Oxidative Stress. Infect. Immun. 2006, 74, 4950–4953. [Google Scholar] [CrossRef] [Green Version]
- Nosanchuk, J.D.; Casadevall, A. Impact of Melanin on Microbial Virulence and Clinical Resistance to Antimicrobial Compounds. Antimicrob. Agents Chemother. 2006, 50, 3519–3528. [Google Scholar] [CrossRef] [Green Version]
- Liu, G.Y.; Nizet, V. Color me bad: microbial pigments as virulence factors. Trends Microbiol. 2009, 17, 406–413. [Google Scholar] [CrossRef] [Green Version]
- Durán, N.; Menck, C.F.M. Chromobacterium violaceum: A Review of Pharmacological and Industiral Perspectives. Crit. Rev. Microbiol. 2001, 27, 201–222. [Google Scholar] [CrossRef]
- Pérez-Tomás, R.; Montaner, B.; Llagostera, E.; Soto-Cerrato, V. The prodigiosins, proapoptotic drugs with anticancer properties. Biochem. Pharmacol. 2003, 66, 1447–1452. [Google Scholar] [CrossRef]
- Williamson, N.R.; Fineran, P.C.; Leeper, F.J.; Salmond, G.P.C. The biosynthesis and regulation of bacterial prodiginines. Nat. Rev. Genet. 2006, 4, 887–899. [Google Scholar] [CrossRef]
- Lima-E-Silva, A.; De Carvalho, M.A.R.; De Souza, S.A.L.; Dias, P.M.T.; Filho, R.G.D.S.; Saramago, C.S.D.M.; Bento, C.A.D.M.; Hofer, E. Heavy metal tolerance (Cr, Ag and Hg) in bacteria isolated from sewage. Braz. J. Microbiol. 2012, 43, 1620–1631. [Google Scholar] [CrossRef] [Green Version]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Agarwal, M.; Rathore, R.S.; Chauhan, A. A Rapid and High Throughput MIC Determination Method to Screen Uranium Resistant Microorganisms. Methods Protoc. 2020, 3, 21. https://doi.org/10.3390/mps3010021
Agarwal M, Rathore RS, Chauhan A. A Rapid and High Throughput MIC Determination Method to Screen Uranium Resistant Microorganisms. Methods and Protocols. 2020; 3(1):21. https://doi.org/10.3390/mps3010021
Chicago/Turabian StyleAgarwal, Meenakshi, Rajesh Singh Rathore, and Ashvini Chauhan. 2020. "A Rapid and High Throughput MIC Determination Method to Screen Uranium Resistant Microorganisms" Methods and Protocols 3, no. 1: 21. https://doi.org/10.3390/mps3010021
APA StyleAgarwal, M., Rathore, R. S., & Chauhan, A. (2020). A Rapid and High Throughput MIC Determination Method to Screen Uranium Resistant Microorganisms. Methods and Protocols, 3(1), 21. https://doi.org/10.3390/mps3010021





