A Rapid and Accurate Bioluminescence-Based Migration Assay Permitting Analysis of Tumor Cell/Stromal Cell Interactions
Abstract
1. Introduction
2. Experimental Design
2.1. Materials
2.1.1. Cell Lines
- Murine mammary gland carcinoma (4T1-luc2) (PerkinElmer, Waltham, MA, USA; Cat. No.: 124087)
- Human breast carcinoma MDA-MB-231-luc2 (MDA-luc2) (PerkinElmer; Waltham, MA, USA; Cat. No.: 124319)
- Human osteosarcoma (143B) (American Type Culture Collection; ATCC, Manassas, VA, USA; Cat. No.: CRL-8303)
- Human dermal fibroblast (NHDF) (Lonza, Basel, Switzerland; Cat. No.: CC-2511).
2.1.2. Reagents
- RPMI 1640 medium for 4T1-luc2 (Thermo Fisher Scientific, Waltham, MA, USA; Cat. No.: 21870-076)
- Minimum essential medium (MEM) for MDA-luc2 and 143B (Sigma-Aldrich, St. Louis, MO, USA; Cat. No.: M5650)
- Dulbecco’s modified eagle medium (DMEM) for NHDF (Sigma-Aldrich; Cat. No.: D5671)
- Fetal bovine serum (FBS) (GE Healthcare Bio-Sciences, Pittsburgh, PA, USA; Cat. No.: SH30071.03)
- 10X Trypsin-EDTA (Thermo Fisher Scientific; Cat. no.: 15400-054)
- Penicillin-streptomycin-neomycin (PSN) (Thermo Fisher Scientific; Cat. No.: 15640-055)
- Phosphate-buffered saline (PBS) (Sigma-Aldrich; Cat. no.: 806544)
- RediFect red-fluc-puromycin lentiviral particles (PerkinElmer; Waltham, MA, USA; Cat. no.: CLS960002)
- Puromycin (Invivogen, Pak Shek Kok, Hong Kong; Cat. No.: Ant-pr-1)
- Hexadimethrine bromide (polybrene) (Sigma-Aldrich; Cat. no.: TR-1003-G)
- D-luciferin (PerkinElmer; Cat. No.: 122796)
- Crystal violet solution (0.1% crystal violet in 2% ethanol, Sigma-Aldrich; Cat. No.: V5265)
- NuncTM EasYFlaskTM cell culture flask in 25 and 75 cm2 (Thermo Fisher Scientific; Cat. No.: 156367 and 156499)
- Costar® 24-well clear TC-treated multiple well plates (Corning, New York, NY, USA; Cat. No.: 3524)
- 6.5 mm Transwell® with 8.0 μm pore polycarbonate membrane insert (Corning; Cat. No.: 3422)
- Sterile cotton swabs
2.2. Equipment
- CO2 incubator at 37 °C with 95% humidity and 5% CO2 (Sanyo Electric, Osaka, Japan; Cat. No.: MCO-20AIC)
- IVIS® Spectrum in vivo imaging system (PerkinElmer; Cat. No.: IVISSPE)
- Inverted Bright-field/fluorescence microscopy (Olympus Life Science, Center Valley, PE, USA; Cat. No.: IX71)
2.3. Software
- Living Image® 4.4 software (PerkinElmer, Waltham, MA, USA; Cat. No.: 128113)
- Image-Pro® Plus (Media Cybernetics, Rockville, MD, USA)
- GraphPad Prism 6 (GraphPad Software, San Diego, CA, USA)
3. Procedure for the Co-Incubation of Bioluminescent and Non-Bioluminescent Cell Lines
3.1. Insertion of Cells into Transwells (Time for Completion: 20–30 min Not Including Incubation Period)
- Prepare separately luciferase-transduced and nonluciferase-transduced cells (1.25–2.5 × 105 cells/mL of each cell line) in media.
- Add 600 μL of media to the lower well of the 24-transwell plate.
- Add 100 μL of each of the cell suspension solution (luciferase-transduced cell and nonluciferase-transduced cell, total 200 μL) into the interior of the transwell.
- Gently insert the transwell into the lower well and incubate the plate for 6–24 h in a cell culture incubator.
3.2. Measurement of Migrated Cells
3.2.1. Initiation of IVIS® Spectrum and Living Image® software
- Start ‘Living Image® 4.4 software’
- Click ‘Initialize’ on the IVIS acquisition control panel to start up the system and raise the temperature (Figure 3 No. 1).
- Click ‘Imaging Wizard’ on the control panel (Figure 3 No. 2).
- Select ‘Bioluminescence’ and ‘Next’ in the imaging mode (Figure 3 No. 3–4).
- Select ‘Open Filter’ and ‘Next’ in the imaging option (Figure 3 No. 5–6).
- Select ‘Well Plate’ in the imaging subject and ‘Next’ (Figure 3 No. 7–9).
3.2.2. Transwell Washing and Removal of Nonmigrated Cells (Time for Completion: 3–4 Min)
- Aspirate and discard the media containing nonmigrated cells from the inside of the transwell (Figure 2a).
- Wash the transwell twice in PBS (Figure 2b).
- Gently swab the interior of the transwell twice using water-soaked cotton swabs to remove nonmigrated cells attached to the interior of the transwell membrane (Figure 2c).
- Swab the interior of the transwell twice using dry cotton swabs to remove any residual moisture (Figure 2c).
3.2.3. Imaging the Transwells for Bioluminescence (Time for Completion: 1–2 min)
- Invert and place the transwell on the base of the IVIS® Spectrum imaging chamber (Figure 2d).
 CRITICAL STEP Make sure the transwell is placed within the imaging chamber as soon as possible after cleaning to prevent migrated cells from drying out.
CRITICAL STEP Make sure the transwell is placed within the imaging chamber as soon as possible after cleaning to prevent migrated cells from drying out.- ii.
- Gently pipette 40 μL D-luciferin solution upon the exterior membrane of the transwell (Figure 2e).
 CRITICAL STEP Ensure the transwell membrane is completely covered by the D-luciferin solution (Figure 2f and Figure A1a).
CRITICAL STEP Ensure the transwell membrane is completely covered by the D-luciferin solution (Figure 2f and Figure A1a).- iii.
- Click on “Acquire” in the control panel (Figure 3 No. 9).
3.3. Analyzing the Images to Quantify Migrated Cells (Time for Completion: 3–4 Min)
- Open the imaging file in the Living Image® 4.4 software and select ‘Radiance (Photons)’ in the units box (Figure 3 No. 10–11).
- Click and open ‘ROI Tools’ in the tool palette (Figure 3 No. 12).
- Click the
button next to ‘Measurement ROIs’ and select ‘Auto All’ on the drop-down list (Figure 3 No. 12).
 CRITICAL STEP The software automatically draws measurement regions of interest (ROIs) on all images. The ROI label shows the total intensity within the ROI and the threshold %.
CRITICAL STEP The software automatically draws measurement regions of interest (ROIs) on all images. The ROI label shows the total intensity within the ROI and the threshold %.- iv.
- Use the threshold % slider and click ‘Measure ROIs’ button in the ROI tools to show the ROI measurements table (Figure 3 No. 12–13).
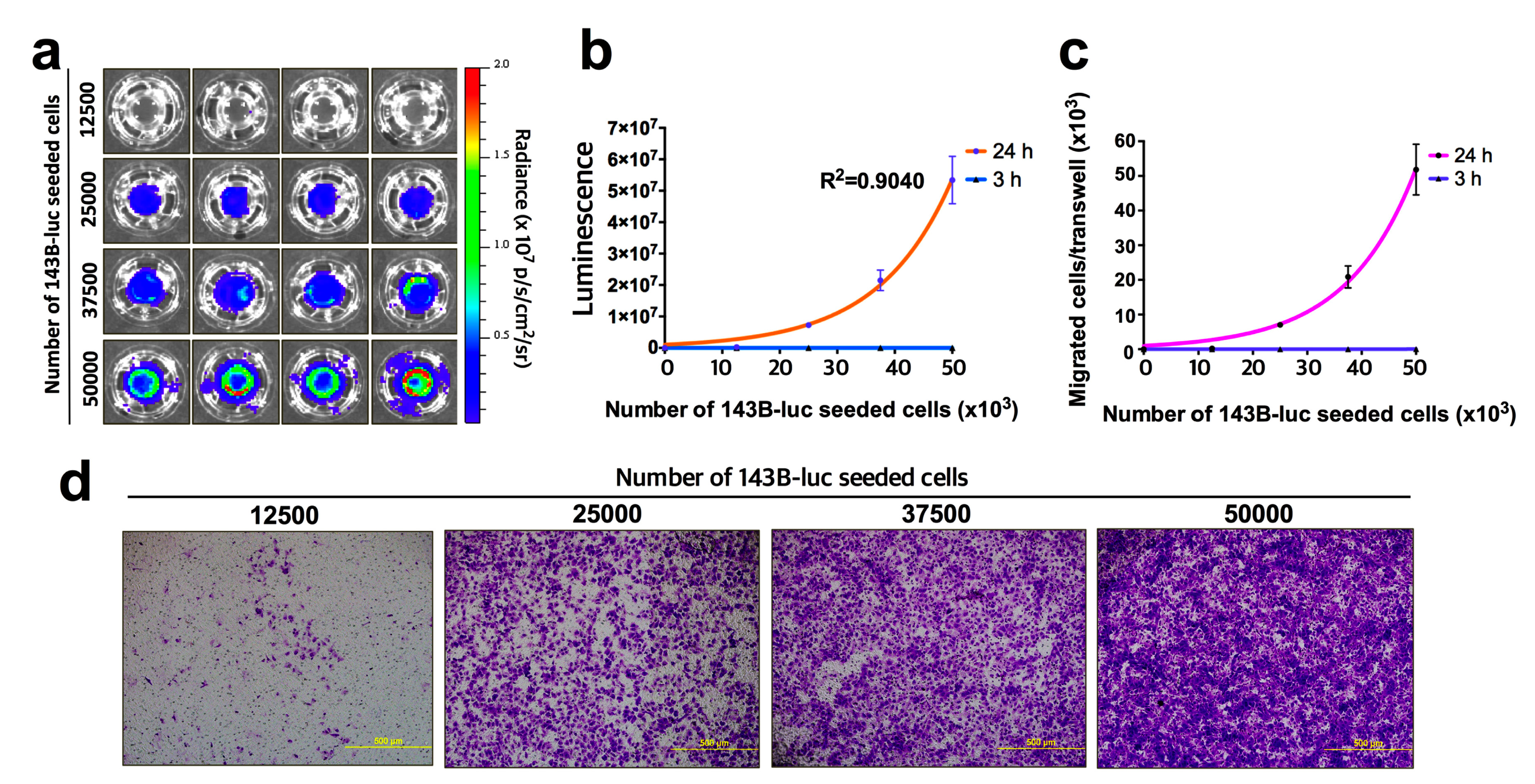
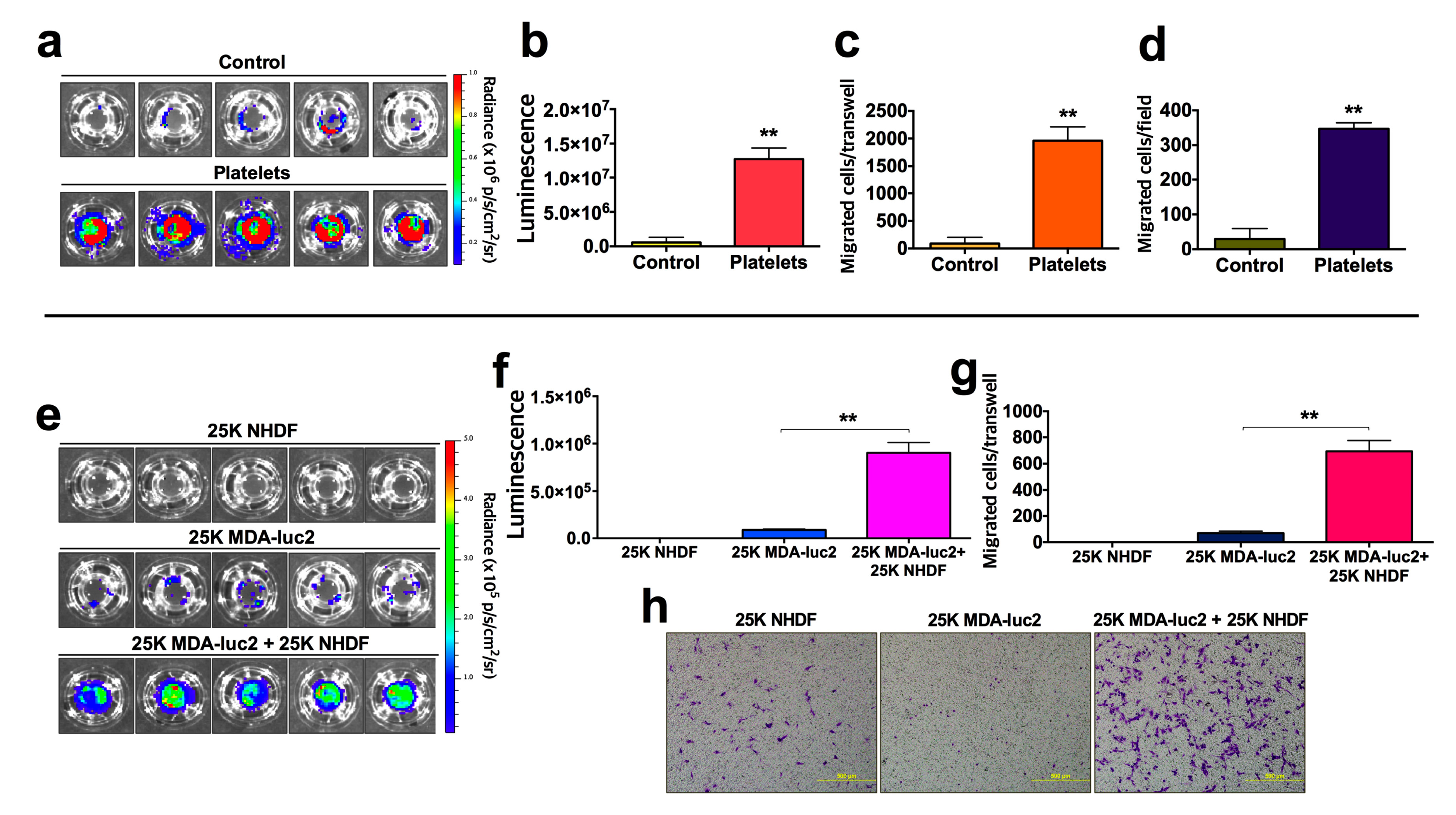
4. Expected Results
5. Reagents Setup
5.1. Establishment of Luciferase-Transduced 143B Cells
5.2. D-Luciferin Solution
5.3. Crystal Violet Staining
Author Contributions
Funding
Conflicts of Interest
Appendix A


| Processing Steps | Bioluminescence | Cell Count (Classical) | Colorimetry or Fluorometry |
|---|---|---|---|
| Wash | ✔ | ✔ | ✔ |
| Transwell Cleaning | ✔ | ✔ | ✔ |
| Cell Staining | ✕ | ✔ | ✔ |
| Cell Detachment | ✕ | ✕ | ✔ |
| Cell Lysis | ✕ | ✕ | ✔ |
| Cell Count | ✕ | ✔ | ✕ |
| Quantification Steps | |||
| Data Collection and Analysis | Low light imaging system and software | Light microscope with image software | Plate reader and software |
| Detection Reagent | Firefly luciferase with D-luciferin | Crystal violot Hematoxylin Toluidin blue | Crystal violot or Calcein AM |
| Assay Time | Less than 10 min | Approximately 1 h | Approximately 1 h |
| Problem | Possible reason | Solution |
|---|---|---|
| Absence of a bioluminescent signal |
|
|
| D-luciferin solution leaks down inside of transwell (Figure A1) |
|
|
References
- Boyden, S. The Chemotactic Effect Of Mixtures Of Antibody And Antigen On Polymorphonuclear Leucocytes. J. Exp. Med. 1962, 115, 453–466. [Google Scholar] [CrossRef] [PubMed]
- Kramer, N.; Walzl, A.; Unger, C.; Rosner, M.; Krupitza, G.; Hengstschläger, M.; Dolznig, H. In vitro cell migration and invasion assays. Mutat. Res. Mutat. Res. 2013, 752, 10–24. [Google Scholar] [CrossRef] [PubMed]
- Friedl, P.; Wolf, K. Plasticity of cell migration: a multiscale tuning model. J. Exp. Med. 2010, 207. [Google Scholar] [CrossRef]
- Shinde, R.; Perkins, J.; Contag, C.H. Luciferin Derivatives for Enhanced in Vitro and in Vivo Bioluminescence Assays†. Biochem. 2006, 45, 11103–11112. [Google Scholar] [CrossRef] [PubMed]



© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yoon, J.; Parish, C.R.; Coupland, L.A. A Rapid and Accurate Bioluminescence-Based Migration Assay Permitting Analysis of Tumor Cell/Stromal Cell Interactions. Methods Protoc. 2020, 3, 10. https://doi.org/10.3390/mps3010010
Yoon J, Parish CR, Coupland LA. A Rapid and Accurate Bioluminescence-Based Migration Assay Permitting Analysis of Tumor Cell/Stromal Cell Interactions. Methods and Protocols. 2020; 3(1):10. https://doi.org/10.3390/mps3010010
Chicago/Turabian StyleYoon, Jinsoo, Christopher R. Parish, and Lucy A. Coupland. 2020. "A Rapid and Accurate Bioluminescence-Based Migration Assay Permitting Analysis of Tumor Cell/Stromal Cell Interactions" Methods and Protocols 3, no. 1: 10. https://doi.org/10.3390/mps3010010
APA StyleYoon, J., Parish, C. R., & Coupland, L. A. (2020). A Rapid and Accurate Bioluminescence-Based Migration Assay Permitting Analysis of Tumor Cell/Stromal Cell Interactions. Methods and Protocols, 3(1), 10. https://doi.org/10.3390/mps3010010






