Campylobacter Phage Isolation and Characterization: What We Have Learned So Far
Abstract
1. Introduction
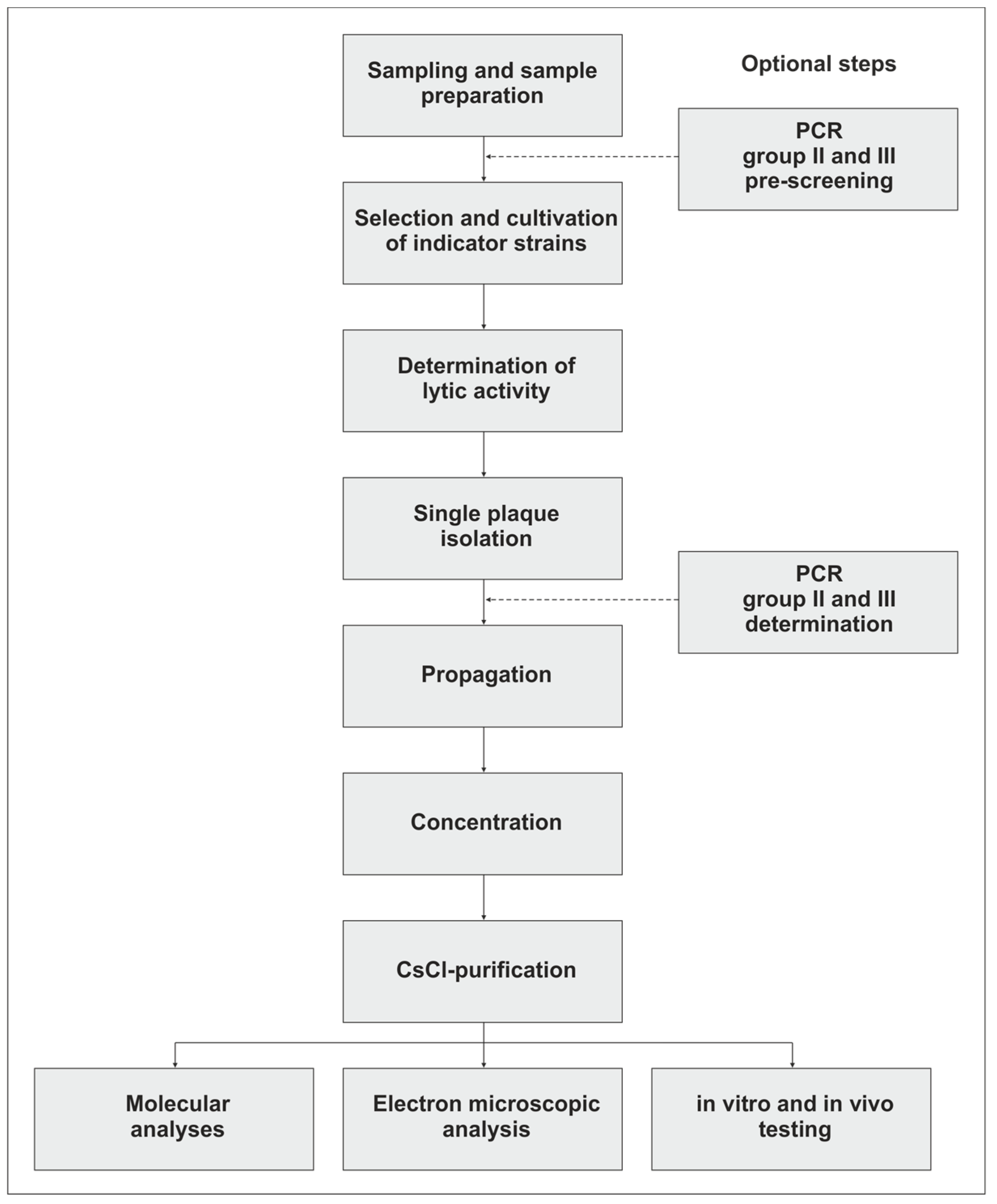
2. Isolation of Campylobacter Phages
3. Propagation, Concentration, and Purification of the Phages
4. Isolation and Analysis of Campylobacter Phage DNA
5. Studies Important for the Application of Campylobacter Phages
6. Conclusions
Author Contributions
Acknowledgments
Conflicts of Interest
References
- European Centre for Disease Prevention and Control (ECDC). Annual Epidemiological Report 2012: Annual Epidemiological Report Reporting on 2010 Surveillance Data and 2011 Epidemic Intelligence Data; ECDC: Solna Stad, Sweden, 2013. [Google Scholar]
- Moore, J.E.; Corcoran, D.; Dooley, J.S.; Fanning, S.; Lucey, B.; Matsuda, M.; McDowell, D.A.; Megraud, F.; Millar, B.C.; O’Mahony, R.; et al. Campylobacter. Vet. Res. 2005, 36, 351–382. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, S.J. The Consequences of Campylobacter Infection. Curr. Opin. Gastroenterol. 2017, 33, 14–20. [Google Scholar] [CrossRef] [PubMed]
- Shane, S.M. Campylobacter Infection of Commercial Poultry. Rev. Sci. Tech. 2000, 19, 376–395. [Google Scholar] [CrossRef] [PubMed]
- Blaser, M.J.; LaForce, F.M.; Wilson, N.A.; Wang, W.L. Reservoirs for Human Campylobacteriosis. J. Infect. Dis. 1980, 141, 665–669. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, T.; O’Brien, S.; Madsen, M. Campylobacters as Zoonotic Pathogens: A Food Production Perspective. Int. J. Food Microbiol. 2007, 117, 237–257. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.D.; Newell, D.G. Campylobacter in Poultry: Filling an Ecological Niche. Avian Dis. 2006, 50, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Rosenquist, H.; Nielsen, N.L.; Sommer, H.M.; Norrung, B.; Christensen, B.B. Quantitative Risk Assessment of Human Campylobacteriosis Associated with Thermophilic Campylobacter Species in Chickens. Int. J. Food Microbiol. 2003, 83, 87–103. [Google Scholar] [CrossRef]
- Havelaar, A.H.; Mangen, M.J.; de Koeijer, A.A.; Bogaardt, M.J.; Evers, E.G.; Jacobs-Reitsma, W.F.; van Pelt, W.; Wagenaar, J.A.; de Wit, G.A.; van der Zee, H.; et al. Effectiveness and Efficiency of Controlling Campylobacter on Broiler Chicken Meat. Risk Anal. 2007, 27, 831–844. [Google Scholar] [CrossRef]
- Klein, G.; Jansen, W.; Kittler, S.; Reich, F. Mitigation Strategies for Campylobacter spp. in Broiler at Pre-Harvest and Harvest Level. Berl. Münch. Tierärztl. Wochenschr. 2015, 128, 132–140. [Google Scholar]
- Umaraw, P.; Prajapati, A.; Verma, K.; Pathak, V.; Singh, V.P. Control of Campylobacter in Poultry Industry from Farm to Poultry Processing Unit: A Review. Crit. Rev. Food Sci. Nutr. 2017, 57, 659–665. [Google Scholar] [CrossRef]
- Meunier, M.; Guyard-Nicodeme, M.; Hirchaud, E.; Parra, A.; Chemaly, M.; Dory, D. Identification of Novel Vaccine Candidates against Campylobacter through Reverse Vaccinology. J. Immunol. Res. 2016, 2016, 5715790. [Google Scholar] [CrossRef] [PubMed]
- Meunier, M.; Guyard-Nicodeme, M.; Dory, D.; Chemaly, M. Control Strategies against Campylobacter at the Poultry Production Level: Biosecurity Measures, Feed Additives and Vaccination. J. Appl. Microbiol. 2016, 120, 1139–1173. [Google Scholar] [CrossRef] [PubMed]
- El-Shibiny, A.; Scott, A.; Timms, A.; Metawea, Y.; Connerton, P.; Connerton, I. Application of a Group II Campylobacter Bacteriophage to Reduce Strains of Campylobacter jejuni and Campylobacter coli Colonizing Broiler Chickens. J. Food Prot. 2009, 72, 733–740. [Google Scholar] [CrossRef] [PubMed]
- Hammerl, J.A.; Jäckel, C.; Alter, T.; Janzcyk, P.; Stingl, K.; Knüver, M.T.; Hertwig, S. Reduction of Campylobacter jejuni in Broiler Chicken by Successive Application of Group II and Group III Phages. PLoS ONE 2014, 9, e114785. [Google Scholar] [CrossRef]
- Orquera, S.; Gölz, G.; Hertwig, S.; Hammerl, J.A.; Sparborth, D.; Joldic, A.; Alter, T. Control of Campylobacter spp. And Yersinia enterocolitica by Virulent Bacteriophages. J. Mol. Genet. Med. 2012, 6, 273–278. [Google Scholar] [CrossRef] [PubMed]
- Gibson, J.R.; Fitzgerald, C.; Owen, R.J. Comparison of PFGE, Ribotyping and Phage-Typing in the Epidemiological Analysis of Campylobacter jejuni Serotype HS2 Infections. Epidemiol. Infect. 1995, 115, 215–225. [Google Scholar] [CrossRef] [PubMed]
- Grajewski, B.A.; Kusek, J.W.; Gelfand, H.M. Development of a Bacteriophage Typing System for Campylobacter jejuni and Campylobacter coli. J. Clin. Microbiol. 1985, 22, 13–18. [Google Scholar] [PubMed]
- Hopkins, K.L.; Desai, M.; Frost, J.A.; Stanley, J.; Logan, J.M. Fluorescent Amplified Fragment Length Polymorphism Genotyping of Campylobacter jejuni and Campylobacter coli Strains and Its Relationship with Host Specificity, Serotyping, and Phage Typing. J. Clin. Microbiol. 2004, 42, 229–235. [Google Scholar] [CrossRef]
- Khakhria, R.; Lior, H. Extended Phage-Typing Scheme for Campylobacter jejuni and Campylobacter coli. Epidemiol. Infect. 1992, 108, 403–414. [Google Scholar] [CrossRef]
- Sails, A.D.; Wareing, D.R.; Bolton, F.J.; Fox, A.J.; Curry, A. Characterisation of 16 Campylobacter jejuni and C. coli Typing Bacteriophages. J. Med. Microbiol. 1998, 47, 123–128. [Google Scholar] [CrossRef]
- Atterbury, R.J.; Connerton, P.L.; Dodd, C.E.; Rees, C.E.; Connerton, I.F. Application of Host-Specific Bacteriophages to the Surface of Chicken Skin Leads to a Reduction in Recovery of Campylobacter jejuni. Appl. Environ. Microbiol. 2003, 69, 6302–6306. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, C.M.; Gannon, B.W.; Halfhide, D.E.; Santos, S.B.; Hayes, C.M.; Roe, J.M.; Azeredo, J. The in vivo Efficacy of Two Administration Routes of a Phage Cocktail to Reduce Numbers of Campylobacter coli and Campylobacter jejuni in Chickens. BMC Microbiol. 2010, 10, 232. [Google Scholar] [CrossRef]
- Fischer, S.; Kittler, S.; Klein, G.; Glünder, G. Impact of a Single Phage and a Phage Cocktail Application in Broilers on Reduction of Campylobacter jejuni and Development of Resistance. PLoS ONE 2013, 8, e78543. [Google Scholar] [CrossRef] [PubMed]
- Goode, D.; Allen, V.M.; Barrow, P.A. Reduction of Experimental Salmonella and Campylobacter Contamination of Chicken Skin by Application of Lytic Bacteriophages. Appl. Environ. Microbiol. 2003, 69, 5032–5036. [Google Scholar] [CrossRef] [PubMed]
- Loc Carrillo, C.; Atterbury, R.J.; El-Shibiny, A.; Connerton, P.L.; Dillon, E.; Scott, A.; Connerton, I.F. Bacteriophage Therapy to Reduce Campylobacter jejuni Colonization of Broiler Chickens. Appl. Environ. Microbiol. 2005, 71, 6554–6563. [Google Scholar] [CrossRef] [PubMed]
- Wagenaar, J.A.; Van Bergen, M.A.; Müller, M.A.; Wassenaar, T.M.; Carlton, R.M. Phage Therapy Reduces Campylobacter jejuni Colonization in Broilers. Vet. Microbiol. 2005, 109, 275–283. [Google Scholar] [CrossRef] [PubMed]
- Kittler, S.; Fischer, S.; Abdulmawjood, A.; Glünder, G.; Klein, G. Effect of Bacteriophage Application on Campylobacter jejuni Loads in Commercial Broiler Flocks. Appl. Environ. Microbiol. 2013, 79, 7525–7533. [Google Scholar] [CrossRef]
- Javed, M.A.; Ackermann, H.W.; Azeredo, J.; Carvalho, C.M.; Connerton, I.; Evoy, S.; Hammerl, J.A.; Hertwig, S.; Lavigne, R.; Singh, A.; et al. A Suggested Classification for Two Groups of Campylobacter Myoviruses. Arch. Virol. 2014, 159, 181–190. [Google Scholar] [CrossRef]
- Bryner, J.H.; Ritchie, A.E.; Booth, G.D.; Foley, J.W. Lytic Activity of Vibrio Phages on Strains of Vibrio fetus Isolated from Man and Animals. Appl. Microbiol. 1973, 26, 404–409. [Google Scholar]
- Bryner, J.H.; Ritchie, A.E.; Foley, J.W.; Berman, D.T. Isolation and Characterization of a Bacteriophage for Vibrio fetus. J. Virol. 1970, 6, 94–99. [Google Scholar]
- Lis, L.; Connerton, I.F. The Minor Flagellin of Campylobacter jejuni (FlaB) Confers Defensive Properties against Bacteriophage Infection. Front. Microbiol. 2016, 7, 1908. [Google Scholar] [CrossRef] [PubMed]
- Hammerl, J.A.; Jäckel, C.; Hertwig, S. Genetics of Campylobacter Phages. Berl. Münch. Tierärztl. Wochenschr. 2015, 128, 148–154. (In German) [Google Scholar] [PubMed]
- Hammerl, J.A.; Jäckel, C.; Reetz, J.; Beck, S.; Alter, T.; Lurz, R.; Barretto, C.; Brüssow, H.; Hertwig, S. Campylobacter jejuni Group III Phage CP81 Contains Many T4-Like Genes without Belonging to the T4-Type Phage Group: Implications for the Evolution of T4 Phages. J. Virol. 2011, 85, 8597–8605. [Google Scholar] [CrossRef] [PubMed]
- Hansen, V.M.; Rosenquist, H.; Baggesen, D.L.; Brown, S.; Christensen, B.B. Characterization of Campylobacter Phages Including Analysis of Host Range by Selected Campylobacter Penner Serotypes. BMC Microbiol. 2007, 7, 90. [Google Scholar] [CrossRef] [PubMed]
- Loc Carrillo, C.M.; Connerton, P.L.; Pearson, T.; Connerton, I.F. Free-Range Layer Chickens as a Source of Campylobacter Bacteriophage. Antonie Van Leeuwenhoek 2007, 92, 275–284. [Google Scholar] [CrossRef] [PubMed]
- Timms, A.R.; Cambray-Young, J.; Scott, A.E.; Petty, N.K.; Connerton, P.L.; Clarke, L.; Seeger, K.; Quail, M.; Cummings, N.; Maskell, D.J.; et al. Evidence for a Lineage of Virulent Bacteriophages That Target Campylobacter. BMC Genom. 2010, 11, 214. [Google Scholar] [CrossRef] [PubMed]
- Hammerl, J.A.; Jäckel, C.; Reetz, J.; Hertwig, S. The Complete Genome Sequence of Bacteriophage CP21 Reveals Modular Shuffling in Campylobacter Group II Phages. J. Virol. 2012, 86, 8896. [Google Scholar] [CrossRef]
- Timms, A.R.; Al Khandari, S.; Wilson, R.; Rowsell, J.; Connerton, I.F. Campylobacter Phage CPX, Complete Genome. 2011; unpublished. [Google Scholar]
- Brathwaite, K.J.; Siringan, P.; Connerton, P.L.; Connerton, I.F. Host Adaption to the Bacteriophage Carrier State of Campylobacter jejuni. Res. Microbiol. 2015, 166, 504–515. [Google Scholar] [CrossRef]
- Kropinski, A.M.; Arutyunov, D.; Foss, M.; Cunningham, A.; Ding, W.; Singh, A.; Pavlov, A.R.; Henry, M.; Evoy, S.; Kelly, J.; et al. Genome and Proteome of Campylobacter jejuni Bacteriophage NCTC 12673. Appl. Environ. Microbiol. 2011, 77, 8265–8271. [Google Scholar] [CrossRef]
- Janez, N.; Peterka, M.; Accetto, T. Complete Genome Sequences of Group III Campylobacter Bacteriophages PC5 and PC14. Genome Announc. 2016, 4, e01030-16. [Google Scholar] [CrossRef]
- O’Sullivan, L.; Lucid, A.; Neve, H.; Franz, C.; Bolton, D.; McAuliffe, O.; Paul Ross, R.; Coffey, A. Comparative Genomics of CP8viruses with Special Reference to Campylobacter Phage vb_Cjem_Los1, Isolated from a Slaughterhouse in Ireland. Arch. Virol. 2018, 163, 2139–2154. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, C.M.; Kropinski, A.M.; Lingohr, E.J.; Santos, S.B.; King, J.; Azeredo, J. The Genome and Proteome of a Campylobacter coli Bacteriophage vb_Ccom-Ibb_35 Reveal Unusual Features. Virol. J. 2012, 9, 35. [Google Scholar] [CrossRef] [PubMed]
- Adams, M.J.; Lefkowitz, E.J.; King, A.M.; Harrach, B.; Harrison, R.L.; Knowles, N.J.; Kropinski, A.M.; Krupovic, M.; Kuhn, J.H.; Mushegian, A.R.; et al. Ratification Vote on Taxonomic Proposals to the International Committee on Taxonomy of Viruses (2016). Arch. Virol. 2016, 161, 2921–2949. [Google Scholar] [CrossRef] [PubMed]
- Jäckel, C.; Hammerl, J.A.; Reetz, J.; Kropinski, A.M.; Hertwig, S. Campylobacter Group II Phage CP21 Is the Prototype of a New Subgroup Revealing a Distinct Modular Genome Organization and Host Specificity. BMC Genom. 2015, 16, 629. [Google Scholar]
- Gencay, Y.E.; Birk, T.; Sorensen, M.C.; Brondsted, L. Methods for Isolation, Purification, and Propagation of Bacteriophages of Campylobacter jejuni. Methods Mol. Biol. 2017, 1512, 19–28. [Google Scholar]
- Sorensen, M.C.; Gencay, Y.E.; Brondsted, L. Methods for Initial Characterization of Campylobacter jejuni Bacteriophages. Methods Mol. Biol. 2017, 1512, 91–105. [Google Scholar]
- Janez, N.; Loc-Carrillo, C. Use of Phages to Control Campylobacter spp. J. Microbiol. Methods 2013, 95, 68–75. [Google Scholar] [CrossRef]
- Owens, J.; Barton, M.D.; Heuzenroeder, M.W. The Isolation and Characterization of Campylobacter jejuni Bacteriophages from Free Range and Indoor Poultry. Vet. Microbiol. 2013, 162, 144–150. [Google Scholar] [CrossRef]
- Furuta, M.; Nasu, T.; Umeki, K.; Hoang Minh, D.; Honjoh, K.I.; Miyamoto, T. Characterization and Application of Lytic Bacteriophages against Campylobacter jejuni Isolated from Poultry in Japan. Biocontrol Sci. 2017, 22, 213–221. [Google Scholar] [CrossRef]
- Firlieyanti, A.S.; Connerton, P.L.; Connerton, I.F. Campylobacters and Their Bacteriophages from Chicken Liver: The Prospect for Phage Biocontrol. Int. J. Food Microbiol. 2016, 237, 121–127. [Google Scholar] [CrossRef]
- El-Shibiny, A.; Connerton, P.L.; Connerton, I.F. Enumeration and Diversity of Campylobacters and Bacteriophages Isolated During the Rearing Cycles of Free-Range and Organic Chickens. Appl. Environ. Microbiol. 2005, 71, 1259–1266. [Google Scholar] [CrossRef]
- Jäckel, C.; Hammerl, J.A.; Rau, J.; Hertwig, S. A Multiplex Real-Time PCR for the Detection and Differentiation of Campylobacter Phages. PLoS ONE 2017, 12, e0190240. [Google Scholar] [CrossRef]
- Atterbury, R.J.; Connerton, P.L.; Dodd, C.E.; Rees, C.E.; Connerton, I.F. Isolation and Characterization of Campylobacter Bacteriophages from Retail Poultry. Appl. Environ. Microbiol. 2003, 69, 4511–4518. [Google Scholar] [CrossRef]
- Janez, N.; Kokosin, A.; Zaletel, E.; Vranac, T.; Kovac, J.; Vuckovic, D.; Smole Mozina, S.; Curin Serbec, V.; Zhang, Q.; Accetto, T.; et al. Identification and Characterisation of New Campylobacter Group III Phages of Animal Origin. FEMS Microbiol. Lett. 2014, 359, 64–71. [Google Scholar] [CrossRef] [PubMed]
- Gencay, Y.E.; Sorensen, M.C.H.; Wenzel, C.Q.; Szymanski, C.M.; Brondsted, L. Phase Variable Expression of a Single Phage Receptor in Campylobacter jejuni NCTC 12662 Influences Sensitivity toward Several Diverse CPS-Dependent Phages. Front. Microbiol. 2018, 9, 82. [Google Scholar] [CrossRef] [PubMed]
- Sorensen, M.C.; Gencay, Y.E.; Birk, T.; Baldvinsson, S.B.; Jäckel, C.; Hammerl, J.A.; Vegge, C.S.; Neve, H.; Brondsted, L. Primary Isolation Strain Determines Both Phage Type and Receptors Recognised by Campylobacter jejuni Bacteriophages. PLoS ONE 2015, 10, e0116287. [Google Scholar] [CrossRef] [PubMed]
- Sorensen, M.C.; van Alphen, L.B.; Harboe, A.; Li, J.; Christensen, B.B.; Szymanski, C.M.; Brondsted, L. Bacteriophage F336 Recognizes the Capsular Phosphoramidate Modification of Campylobacter jejuni NCTC 11168. J. Bacteriol. 2011, 193, 6742–6749. [Google Scholar] [CrossRef]
- Baldvinsson, S.B.; Sorensen, M.C.; Vegge, C.S.; Clokie, M.R.; Brondsted, L. Campylobacter jejuni Motility Is Required for Infection of the Flagellotropic Bacteriophage F341. Appl. Environ. Microbiol. 2014, 80, 7096–7106. [Google Scholar] [CrossRef] [PubMed]
- Ng, L.K.; Stiles, M.E.; Taylor, D.E. Comparison of Basal Media for Culturing Campylobacter jejuni and Campylobacter coli. J. Clin. Microbiol. 1985, 21, 226–230. [Google Scholar]
- Frost, J.A.; Kramer, J.M.; Gillanders, S.A. Phage Typing of Campylobacter jejuni and Campylobacter coli and Its Use as an Adjunct to Serotyping. Epidemiol. Infect. 1999, 123, 47–55. [Google Scholar] [CrossRef]
- Sambrook, J.; Russell, D. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2001. [Google Scholar]
- Fischer, S.; Kittler, S.; Klein, G.; Glünder, G. Microplate-Test for the Rapid Determination of Bacteriophage-Susceptibility of Campylobacter Isolates-Development and Validation. PLoS ONE 2013, 8, e53899. [Google Scholar] [CrossRef] [PubMed]
- Rabinovitch, A.; Aviram, I.; Zaritsky, A. Bacterial Debris-an Ecological Mechanism for Coexistence of Bacteria and Their Viruses. J. Theor. Biol. 2003, 224, 377–383. [Google Scholar] [CrossRef]
- Ackermann, H.W. Basic Phage Electron Microscopy. Methods Mol. Biol. 2009, 501, 113–126. [Google Scholar] [PubMed]
| Phage Group | CP220virus (Group II) | CP8virus (Group III) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Phage | CP220 | CPt10 | CP21 | IBB_35 | CP81 | NCTC 12673 | CPX | CP8 | CP30A | PC5 | PC14 | vB_CjeM_Los1 |
| Source | Chicken | Environ- ment | Water, organic farm | Poultry ceca | Chicken skin | Poultry excreta | Retail chicken | Chicken ceca | Poultry excreta | Chicken ceca | Chicken ceca | Poultry excreta |
| Year of isolation | 2003 | 1989 | 2011 | n.a. | 2008/2009 | Before 1985 | n.a. | n.a. | n.a. | 2011/2012 | 2011/2012 | n.a. |
| Country | United Kingdom | United Kingdom | Germany | Portugal | Germany | USA | United Kingdom | United Kingdom | United Kingdom | Slovenia | Slovenia | Ireland |
| Family | Myo- viridae | Myo- viridae | Myo- viridae | Myo- viridae | Myo- viridae | Myo- viridae | Myo- viridae | Myo- viridae | Myo- viridae | Myo-viridae | Myo- viridae | Myo- viridae |
| Host range | C. jejuni, C. coli | C. jejuni, C. coli | C. jejuni, C. coli | C. jejuni, C. coli | C. jejuni | C. jejuni | C. jejuni | C. jejuni | C. jejuni | C. jejuni | C. jejuni | C. jejuni |
| Restriction | Refractory | Refractory | Refractory | Refractory | Refractory | Refractory | Refractory | Refractory | Refractory | Refractory | Refractory | n.a. |
| Sequencing/ platform | Shotgun seq. DNA libraries | 454 FLX pyroseq. and PCR/ Sanger | 454 FLX pyroseq., and PCR/ Sanger | 454 FLX pyroseq. | 454 FLX pyroseq. | Fidelity System | 454 FLX pyroseq. | n.a. | 454 FLX pyroseq. | 454 FLX pyroseq. | 454 FLX pyroseq. | Illumina |
| Genome size (bp) | 177,493 | 175,720 | 182,833 | 172,065 | 132,454 | 135,041 | 132,662 | 132,667 | 133,572 | 131,095 | 134,927 | 134,073 |
| Complex repeat regions | + | + | + | n.a. | - | - | - | - | - | - | - | - |
| GC content (%) | 27.4 | 27.3 | 27.2 | 27.4 | 26.1 | 26.2 | 26.0 | 26.0 | 26.1 | 26.1 | 26.2 | 26.2 |
| PFGE size (kb) | ~197 | n.a. | ~209 | ~204 | ~145 | ~170 | n.a. | ~140 | n. a. | ~150 | ~150 | n.a. |
| Accession no. | FN667788 | FN667789 | NC019507 | HM246720-4 | FR823450 | NC015464 | NC016562.1 | KF148616 | NC018861 | KX229736 | KX236333 | KX879627 |
| Reference | [37] | [37] | [38,46] | [44] | [34] | [41] | n.a. | [43] | [40] | [42] | [42] | [43] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jäckel, C.; Hammerl, J.A.; Hertwig, S. Campylobacter Phage Isolation and Characterization: What We Have Learned So Far. Methods Protoc. 2019, 2, 18. https://doi.org/10.3390/mps2010018
Jäckel C, Hammerl JA, Hertwig S. Campylobacter Phage Isolation and Characterization: What We Have Learned So Far. Methods and Protocols. 2019; 2(1):18. https://doi.org/10.3390/mps2010018
Chicago/Turabian StyleJäckel, Claudia, Jens Andre Hammerl, and Stefan Hertwig. 2019. "Campylobacter Phage Isolation and Characterization: What We Have Learned So Far" Methods and Protocols 2, no. 1: 18. https://doi.org/10.3390/mps2010018
APA StyleJäckel, C., Hammerl, J. A., & Hertwig, S. (2019). Campylobacter Phage Isolation and Characterization: What We Have Learned So Far. Methods and Protocols, 2(1), 18. https://doi.org/10.3390/mps2010018





