Simple Summary
Antimicrobial resistance (AMR) is a concern that impacts both human and animal health. To understand AMR, a detailed analysis of 14 different studies was carried out. This study specifically focused on resistance to tetracycline, a common antibiotic, in E. coli bacteria present in cattle. The study found that 0.31 of the E. coli from beef cattle not treated with antibiotics were resistant to tetracycline. Surprisingly, when beef cattle were given the antibiotic through feed or injection, the resistance only rose slightly to 0.53 and 0.39, respectively. This challenges the common belief that using lots of antibiotics in livestock causes higher resistance. The results varied greatly across studies, likely due to different factors like cattle genetics, environment, management, and how antibiotics are given. Other factors, such as exposure to antibiotics in the environment, natural resistance mechanisms in E. coli, and the ability of bacteria to share resistance traits, could also play a part. The study was limited by inconsistent data and a lack of standardization across the studies. Hence, the results highlight the need for tailored research on AMR, to gain a comprehensive understanding of the influencing factors and to devise effective countermeasures.
Abstract
Antimicrobial resistance (AMR) is an emerging global concern, with the widespread use of antimicrobials in One Health contributing significantly to this phenomenon. Among various antimicrobials, tetracyclines are extensively used in the beef cattle industry, potentially contributing to the development of resistance in bacterial populations. This meta-analysis aimed to examine the association between tetracycline use in beef cattle and the development of tetracycline resistance in Escherichia coli isolates. A comprehensive search was conducted using multiple databases to gather relevant observational studies evaluating tetracycline use and tetracycline resistance in Escherichia coli isolates from beef cattle. The rate of tetracycline resistance from each study served as the effect measure and was pooled using a random-effects model, considering possible disparities among studies. The meta-analysis of 14 prospective longitudinal studies resulted in a 0.31 prevalence of tetracycline resistance in Escherichia coli in non-intervention (no exposure), contrasting numerically elevated resistance rates in the intervention (exposed) groups of 0.53 and 0.39 in those receiving tetracyclines via feed or systemically, respectively. Despite the observed numerical differences, no statistically significant differences existed between intervention and non-intervention groups, challenging the conventional belief that antimicrobial use in livestock inherently leads to increased AMR. The findings of this study underscore the need for additional research to fully understand the complex relationship between antimicrobial use and AMR development. A considerable degree of heterogeneity across studies, potentially driven by variations in study design and diverse presentation of results, indicates the intricate and complex nature of AMR development. Further research with standardized methodologies might help elucidate the relationship between tetracycline use and resistance in Escherichia coli isolated from beef cattle.
1. Introduction
The use of antimicrobials in animals plays a vital role in maintaining animal health and ensuring the safety and productivity of livestock industries. Antimicrobials are used mainly for the treatment and prevention of diseases [1]. The use of antimicrobials for prophylactic or preventative purposes is a common practice in various aspects of animal husbandry, especially in settings where the risk of infection is high. This is especially true in intensive farming environments such as beef feedlots where cattle are kept in close proximity to one another, creating conditions that can facilitate the spread of pathogens. Bovine Respiratory Disease Complex (BRDC) is one of the significant challenges for the beef industry, characterized by high morbidity and mortality rates, and high economic costs [2]. BRDC is a complex syndrome influenced by factors such as the health condition of the animal, environment, diet, transport, and immunity, posing a higher risk to cattle in feedlot systems [3]. Globally, the most common industry practice for preventing BRDC in high-risk cattle upon arrival at the feedlot is the metaphylactic use of antimicrobials [4,5,6]. The metaphylactic use of antimicrobials is employed to proactively control the potential development and spread of bovine respiratory disease (BRD) within a group of cattle, regardless of whether only a subset of animals exhibits respiratory disease symptoms [7,8,9]. To help prevent diseases, tetracycline is commonly used in food-producing animals and it can be administered to livestock using either as injections or mixed with feed [10]. The use of antimicrobials for the treatment or prevention of BRDC impacts the intended harmful bacteria but can also affect beneficial commensal bacteria residing within cattle.
About 70 to 90% of antimicrobials given to cattle are excreted intact or as metabolites in the urine and faeces [11]. It has been indicated that even small concentrations of antimicrobials have the potential to persist in the environment and contribute to the selection of AMR bacteria in the soil [12]. These bacteria can then spread through various pathways, including direct contact, consumption of contaminated food products, or environmental exposure [13]. Furthermore, historically in most developed countries, tetracyclines, have been utilized for purposes beyond their medicinal scope, acting as enhancers of feed efficiency and growth promoters in animals of agricultural importance [14]. Despite the advantages of antimicrobial use in food animals, there is increasing concern over the rise of antimicrobial-resistant bacteria, which potentially pose a hazard to animal and human health. The inappropriate use of antimicrobials creates a selective pressure that favours the survival and proliferation of resistant strains [15]. Nonetheless, it’s crucial to understand that the emergence of antimicrobial resistance is not solely attributable to inappropriate use, but rather it’s influenced by all kinds of usage, encompassing both appropriate and inappropriate applications [16]. This is because any exposure to antimicrobials can exert selective pressure on bacteria, leading them to develop resistance over time. Hence, it is of utmost importance to reduce the overall utilization of antimicrobials, while ensuring that any remaining use is strictly limited to situations where it is appropriate and necessary. Despite the recognized significance of this issue, there remains a knowledge gap regarding this risk. For instance, the magnitude of the risk and how it varies depending on the class of antimicrobial drug used, the duration of treatment, and the method of administration are not fully understood. This uncertainty highlights the need for further research in this area. To attain a comprehensive understanding of the existing situation, conducting a meta-analysis of studies carried out across diverse geographical locations stands out as one of the most effective approaches.
Hence, the aim of this meta-analysis was to examine the association between tetracyclines use in beef cattle and the prevalence of tetracycline resistance in Escherichia coli (E. coli) isolates. This meta-analysis is specifically focused on tetracycline, a commonly used class of antimicrobials, and E. coli, a well-studied and prevalent bacterium. Due to the wide distribution and relatively well-documented interactions with antimicrobials, offering a robust basis for our study, E. coli was chosen as an ‘indicator organism’. We anticipate that our work will offer a better understanding of how the use of tetracycline over time affects the resistance of E. coli, providing valuable insights that could inform future strategies for combating antimicrobial resistance (AMR). Furthermore, this study could potentially shed light on how different antimicrobial classes and treatment durations contribute to AMR, and ultimately guides more responsible and effective usage of these antimicrobials.
2. Materials and Methods
The Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines were followed in developing and implementing the research questions, search, and screening protocols, and in reporting the results [17].
2.1. Search Strategy
A comprehensive literature search was conducted across databases including PubMed, Scopus, and Web of Science using the advanced search option of each of these databases. For each database, the search term: “(tetracycline) AND (cattle OR beef OR feedlot OR fattening)”, with slight modification to fit the specific database advanced search formatting requirements, was used to search in the title, abstract and keywords of an article. The search period concluded on 22 April 2023. In addition to the systematic search of articles in the three databases, to ensure literature saturation, manual searches of the reference lists of eligible and relevant articles were also carried out.
2.2. Inclusion and Exclusion Criteria
All articles downloaded from each database were imported into Endnote, exported to Covidence (https://app.covidence.org/, accessed on 22 April 2023) and screened in two steps: first, title and abstract, and second, full text. Both screening steps were based on the developed inclusion and exclusion criteria (Table 1). All articles uploaded on the Covidence website were screened by two authors and screening conflicts were resolved by another, senior author.

Table 1.
Inclusion and exclusion criteria.
Current literature on tetracycline resistance in feedlot cattle is limited, particularly for observational and case-control studies. Furthermore, inconsistencies in the presentation of results, despite the utilization of similar study designs, complicate the challenge of comparing and interpreting these studies. To mitigate these issues, our study incorporated observational data derived from feedlot cattle at both entry and exit points of feedlots. The data gathered at the point of feedlot exit was utilized as a control, facilitating comparison with other studies implementing case-control designs. These case-control studies typically employed interventions through the administration of various tetracycline analogues, delivered either via injection or incorporated into feed.
2.3. Data Extraction and Quality Assessment
Data extraction was conducted independently by two authors, using a pre-designed data extraction form. Extracted data included the first author’s name, publication year, study location, total sample size, number of tetracycline-resistant E. coli isolates, and other relevant study characteristics. The quality of eligible studies was independently assessed by two authors, YEM and GMW, using the Joanna Briggs Institute Qualitative Assessment and Review Instrument tool (JBI-QARI, available at https://jbi.global/critical-appraisal-tools, accessed 5 April 2023). Discrepancies were resolved through consensus or discussion with a third author, when necessary.
2.4. Outcomes and Variables
The primary outcome was the prevalence of tetracycline resistance in E. coli isolates derived from feedlot cattle. This was assessed based on observational data derived from both the entry and exit points of feedlots. Key variables included the type of tetracycline analogues used (tetracycline, oxytetracycline, chlortetracycline), the method of administration (injection or incorporated into feed), and the timing of sample collection (at entry or exit from feedlots).
2.5. Meta-Analysis
The meta-analysis was conducted using the ‘metafor’ package in R [18]. The proportion of tetracycline-resistant E. coli, in each study, was used as the effect measure for our binary outcomes. The proportion of tetracycline resistance was pooled using the random-effects model to account for potential heterogeneity among studies. Confidence in our effect measure was assessed using the random-effects model and confirmed by sensitivity analyses. Heterogeneity was assessed using the I^2 statistic and Q test. To explore potential sources of heterogeneity, Subgroup analyses were conducted based on predefined variables, such as intervention type and study design. Moreover, to assess the robustness of the findings, sensitivity analysis was conducted using the metafor package in R. The Graphic Display of Heterogeneity (GOSH) plots [19,20] were utilized to visually inspect the distribution of effect sizes and assess potential sources of heterogeneity. Additionally, an Influence analysis was conducted for each of the groups included in the study [21]. Given the nature of epidemiological data, which often exhibits noise and non-normal distributions, and due to its efficiency in handling these characteristics and its lack of reliance on assumptions of convex clusters or normal distributions, DBSCAN was selected for outlier detection [20]. Following the GOSH analysis, outliers identified by DBSCAN and those identified by Influence analysis were removed from the dataset. After removing these outliers, to ensure the integrity of the analysis, the meta-analysis was repeated. The potential presence of publication bias was assessed using Peter’s [22] and Egger’s [23] regression tests. In all analyses, a p-value less than 0.05 was considered to indicate statistical significance.
3. Results
3.1. Study Selection
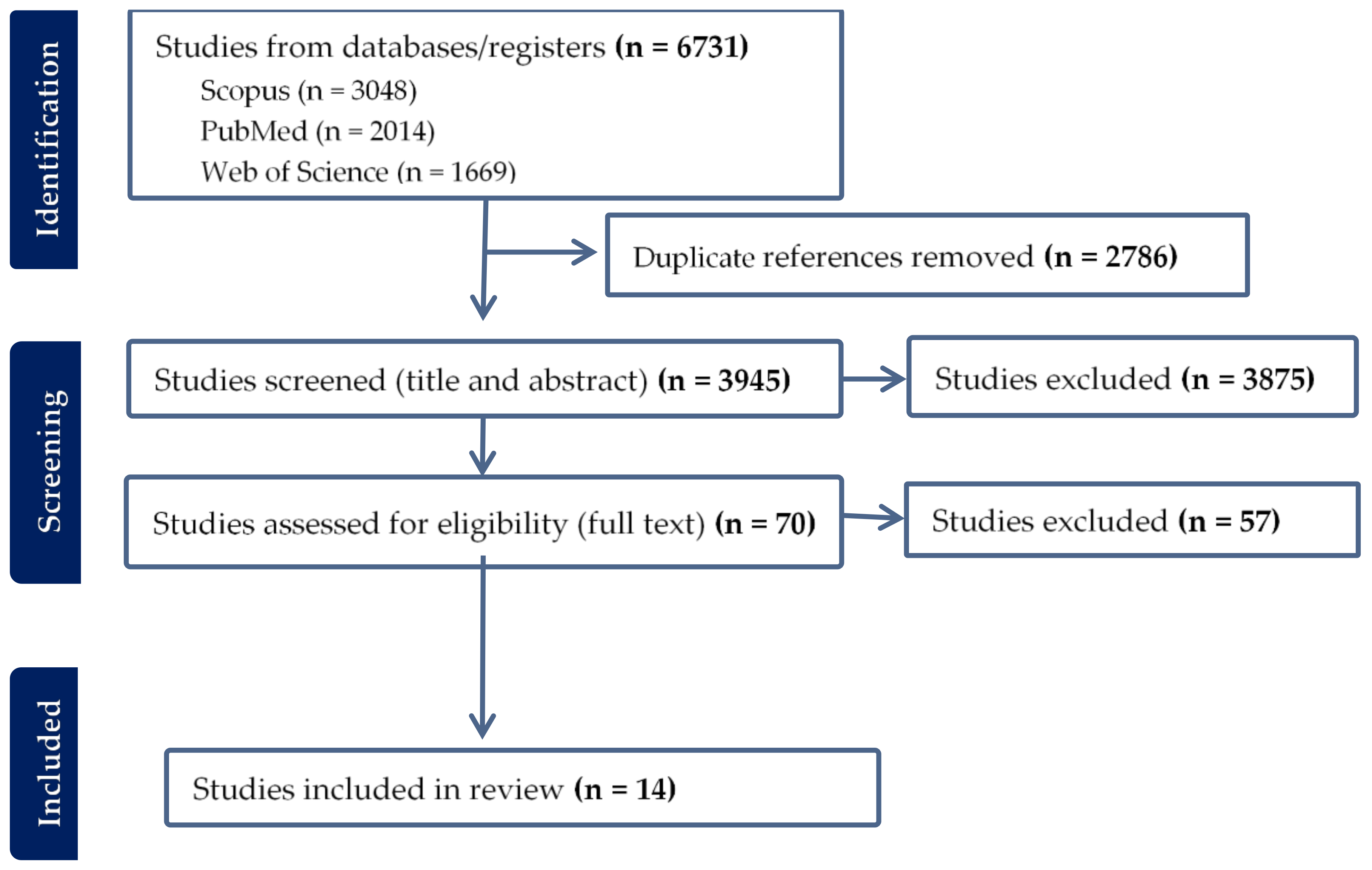
The systematic literature search yielded a total of 6731 articles, of which 1108 were found in all three databases (Figure 1 and Figure 2). After removing 2786 duplicates, 3945 articles were screened for relevance based on title and abstract. This resulted in the exclusion of 3875 articles, leaving 70 full-text articles for assessment. Upon conducting a full-text screening, studies that failed to meet the pre-established inclusion and exclusion criteria, or did not pass the quality appraisal tool, were excluded from the final analysis. The reasons for their exclusion were as follows: incorrect study design (n = 36), non-comparable results (n = 16), inappropriate study population (n = 2), full text not accessible (n = 1), language other than English (n = 1), and unsuitable sample type (n = 1). In addition to these, an article was manually retrieved from Google Scholar, thus elevating the total number of articles included in the meta-analysis to 14 (Figure 1).
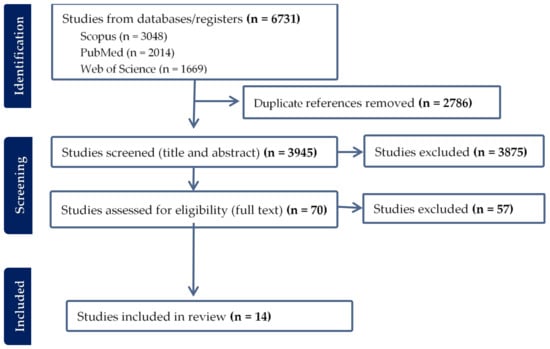
Figure 1.
Article collection and screening steps followed and a few included studies.
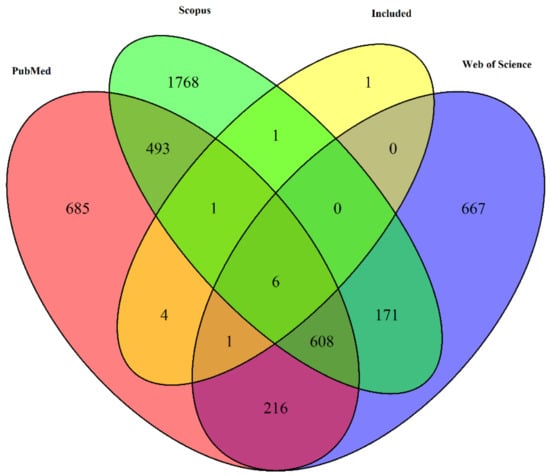
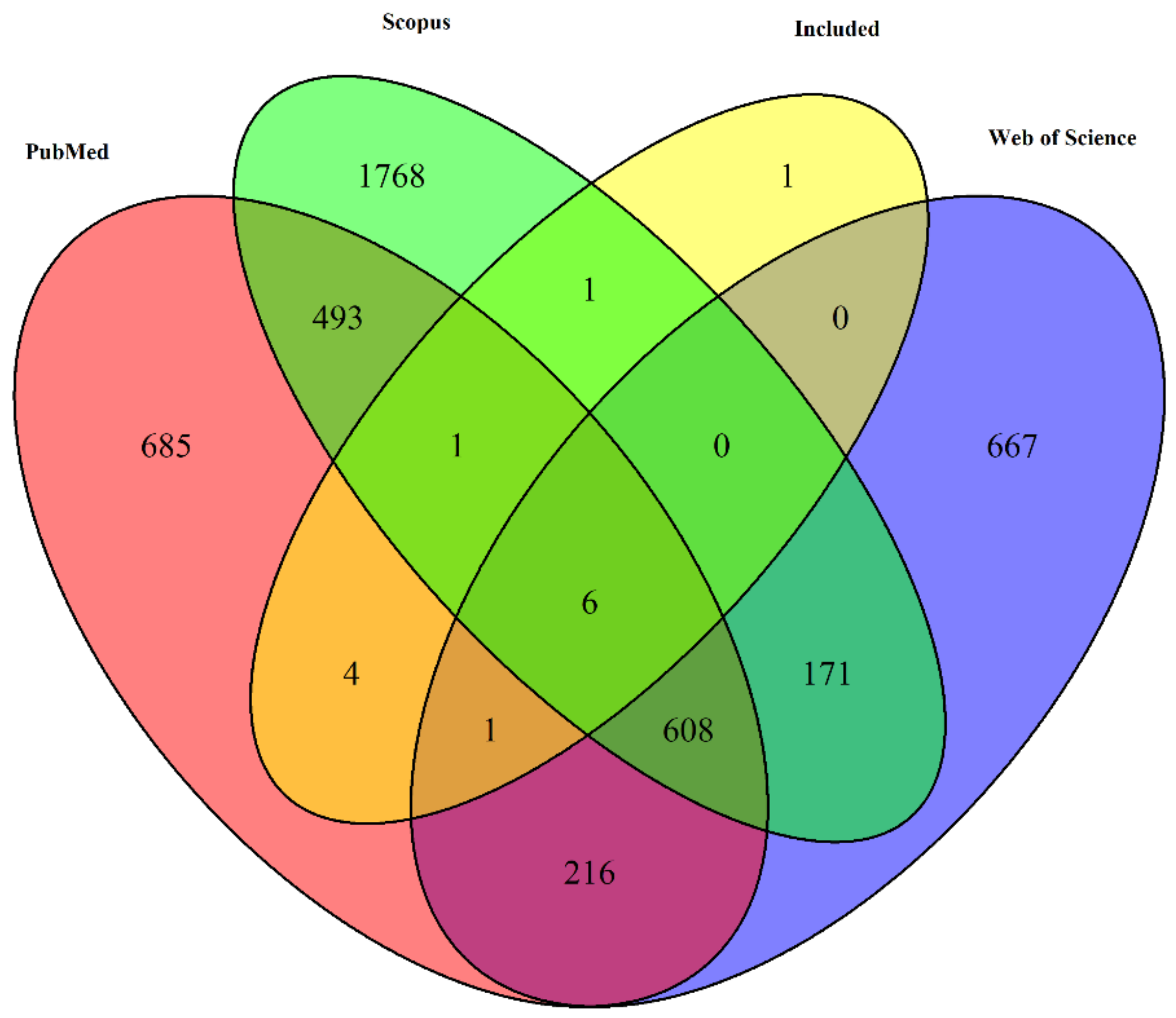
Figure 2.
Venn diagram depicting the overlap of unique article titles retrieved from three different databases: PubMed, Web of Science, and Scopus, along with the articles that were included in the final selection.
From the 14 included studies, 4 were found exclusively in PubMed, 1 exclusively found in Scopus Seven of the 14 articles were found in all three databases (Figure 2).
3.2. Study Characteristics
The initial search protocol included both Enterococcus and Salmonella species. However, no eligible studies on these species were identified during the screening process. Consequently, our investigation was constrained to studies focusing solely on E. coli. In addition to analyzing minimum inhibitory concentration (MIC) data, we attempted to identify studies employing polymerase-chain reaction (PCR) or whole-genome sequencing (WGS) for the determination of tetracycline resistance. Despite our efforts, we were unable to locate studies employing these molecular methods for AMR determination that were comparable and could be included in the analysis.
Similarly, despite the search protocol encompassing articles published up until April 22, 2023, the studies ultimately included in our analysis were those published between 2005 and 2022. These studies originated from a diverse range of regions, namely Canada (n = 9), the United States (n = 2), Europe (n = 2), and Australia (n = 2). The sample sizes within these studies exhibited considerable variation, ranging from as few as 30 to as many as 3512 beef cattle per study. Cumulatively, our study included a total of 20,140 cattle.
3.3. Prevalence of Tetracycline Resistance in Escherichia coli Isolated from Beef Cattle without Intervention
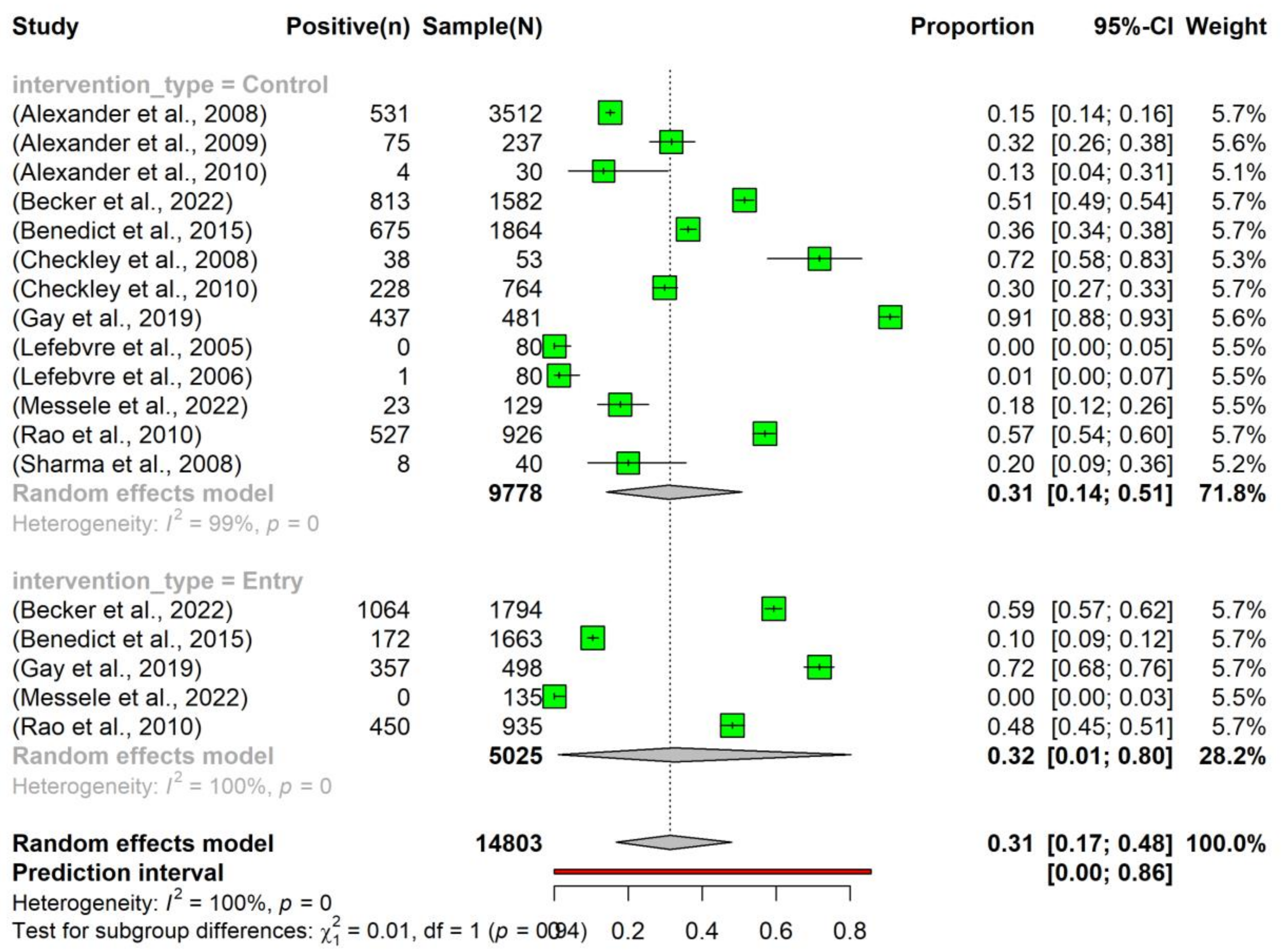
The overall pooled prevalence of tetracycline resistance in E. coli isolated from beef cattle without intervention was 0.31 (95% Confidence Interval (CI): 0.17–0.48). Notably, there was no significant difference in the prevalence between the ‘Entry’ (0.32, 95% CI: 0.01–0.80) and ‘Control’ (0.31, 95% CI: 0.14–0.51) subgroups (Figure 3).
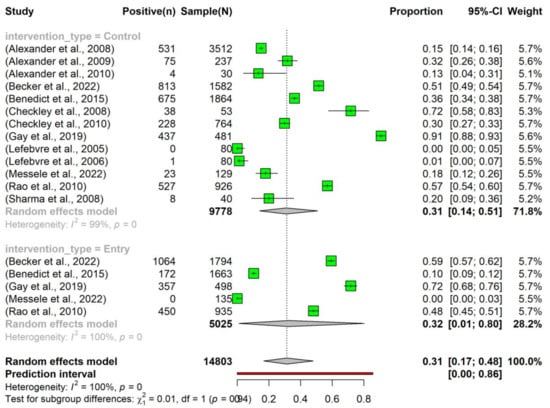
Figure 3.
Prevalence of tetracycline resistance in Escherichia coli isolates obtained from beef cattle without antimicrobial intervention. Samples were collected upon the entry of cattle into the feedlots (‘Entry’) and at the time of their exit (‘Control’). NOTE: Some of the studies classified under the ‘Control’ subgroup were part of case-control studies (n = 7) [24,25,26,27,28,29,30], while others originated from cohort observational studies (n = 6) [31,32,33,34,35,36].
3.4. Prevalence of Tetracycline Resistance in Escherichia coli Isolated from Beef Cattle after Intervention
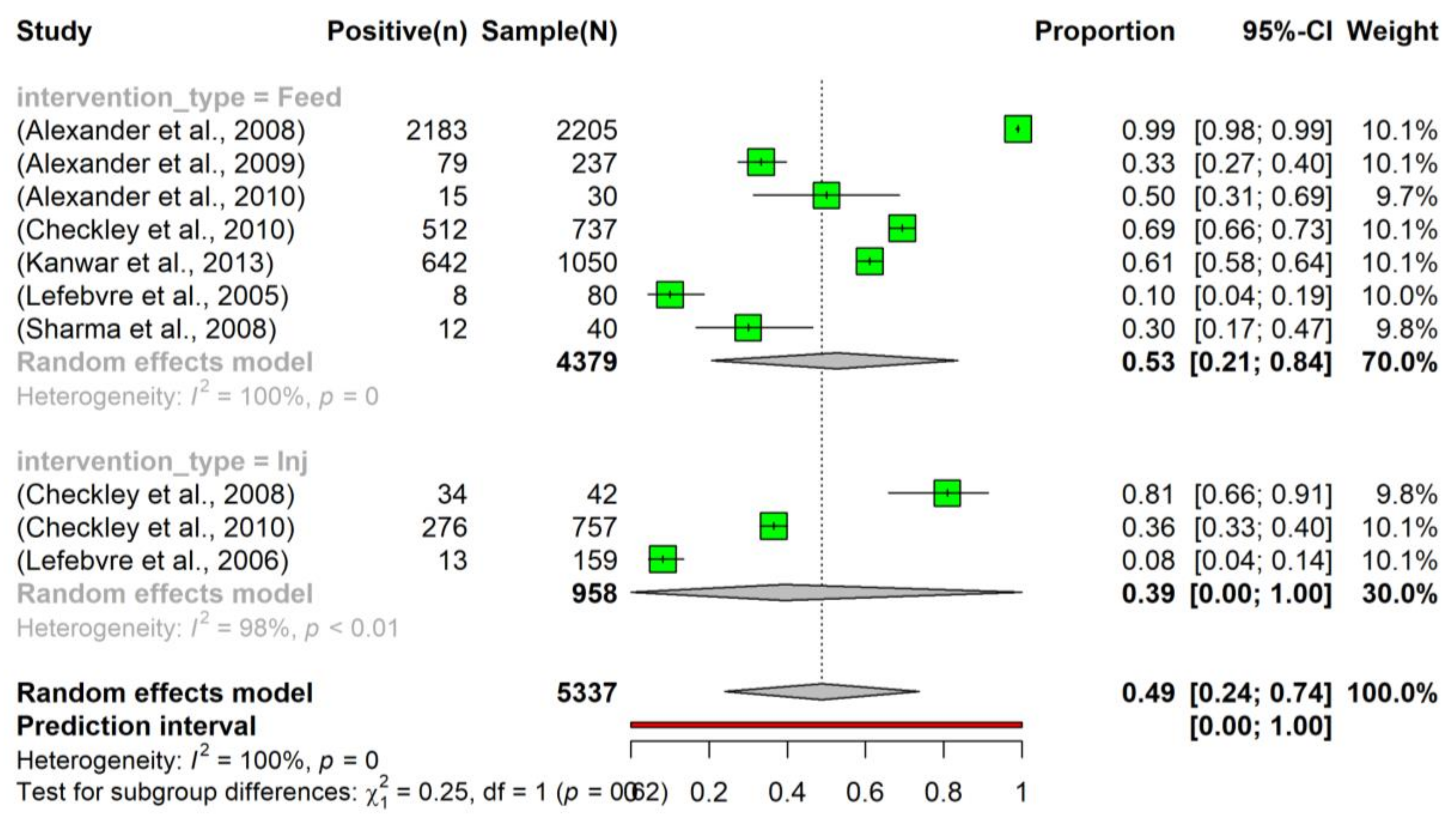
The pooled prevalence of tetracycline resistance in E. coli isolated from beef cattle that had been administered tetracycline in feed was slightly higher (0.53, 95% CI: 0.21–0.84) compared to those that had received tetracycline via injection (0.39, 95% CI: 0.00–1.00). Moreover, considering all interventions, the overall pooled prevalence of tetracycline resistance (0.49, 95% CI: 0.24–0.74) was higher than the prevalence observed in the control group (0.31, 95% CI: 0.01–0.80) (Figure 4). Despite the observed slight increase, the intervention did not have a significant effect on altering the prevalence of tetracycline resistance in E. coli.
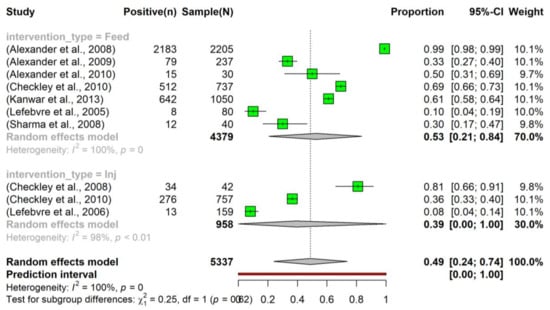
Figure 4.
Prevalence of tetracycline resistance in Escherichia coli isolates sourced from beef cattle subjected to antimicrobial intervention. The tetracycline was administered sub-therapeutically via either feed (indicated as ‘Feed’; n = 7) [24,25,26,28,29,30,37] or an injection (denoted as ‘Inj’; n = 3) [27,28,34]. To examine the dynamics of tetracycline resistance following an intervention, samples were collected at various time points post-administration.
3.5. Heterogeneity and Subgroup Analysis
Substantial heterogeneity was observed among the included studies in both the intervention and non-intervention groups (I2 = 100%, p < 0.01). A subgroup analysis was carried out to investigate potential sources of this heterogeneity. Within the intervention group, there was no significant difference (p > 0.05) between the sub-therapeutic administration of tetracycline through injection or feed supplementation. Similarly, there was no statistically significant difference (p > 0.05) between the subgroups within the nonintervention group. After removing outliers identified by sensitivity analysis, there was no significant difference (p > 0.05) between the intervention and nonintervention groups.
3.6. Sensitivity and Publication Bias Analysis
Results from the GOSH and Influence analyses were instrumental in identifying potential outliers, assessing the influence of individual studies on the overall effect estimate, and evaluating the stability of the pooled results. For the group without intervention, both GOSH and Influence analysis identified articles ‘Alexander et al., 2008′ [26] and ‘Benedict et al., 2015′ [32] as outliers. Following exclusion from the analysis, the overall pooled prevalence increased to 0.34 (95% CI: 0.18–0.53), with an I2 value of 99%, a reduction from the original estimates (Supplementary Figure S1).
Similarly, in the intervention group, articles ‘Alexander et al., 2008′ [26] and ‘Lefebvre et al., 2006′ [34] were identified as outliers by both GOSH and Influence analysis. Removal of these two studies resulted in a revised overall pooled prevalence of 0.46 (95% CI: 0.26–0.67) and an I2 value of 98%, again higher than the original estimates (Supplementary Figure S2).
A regression test for funnel plot asymmetry was conducted separately for the intervention and without intervention groups to assess potential publication bias within each. For the intervention group, the test was not statistically significant (p = 0.8029), suggesting no evidence of publication bias. The limit estimate as the standard error approached zero was 0.2079, with a 95% confidence interval of −0.6119 to 1.0276. This wide confidence interval, encompassing both negative and positive values, indicated uncertainty about the true effect size when the standard error was nearly zero. Conversely, in the non-intervention group, the test for funnel plot asymmetry similarly found no statistically significant evidence of publication bias (p = 0.9733). The limit estimate for this group was −0.2476 with a 95% confidence interval of −0.7334 to 0.2382. Once again, this wide confidence interval suggested uncertainty about the true effect size in studies with a low standard error. Taken together, these results suggested that, for both groups, the studies included in our meta-analysis were not significantly influenced by publication bias.
4. Discussion
The focus of this meta-analysis was to provide a comprehensive assessment of tetracycline resistance prevalence in E. coli populations isolated from beef cattle across a range of geographically diverse studies. The meta-analysis included 14 studies on the prevalence of tetracycline resistance of E. coli isolated from beef cattle. The overall pooled prevalence of resistance in non-intervention group E. coli was 0.31. In contrast, a higher prevalence was observed in intervention groups, where tetracycline was administered via feed (0.53) or injection (0.39). However, the increased prevalence of resistance in the intervention groups was not significantly different compared to the non-intervention groups. Substantial heterogeneity was observed among the included studies, yet the sources of this heterogeneity remained elusive as no significant differences were found upon subgroup analysis.
In addition to the scarcity of observational cohort and case-control studies on tetracycline resistance prevalence in E. coli populations isolated from beef cattle, this meta-analysis faced another challenge: variability in the reporting of results across studies. Inconsistencies in the presentation of results among available studies, even among those employing similar study designs, added to the difficulty of extracting, comparing, and interpreting these studies, resulting in the inclusion of only 14 articles in the current study. This may have decreased the total number of studies potentially suitable for inclusion in the meta-analysis, highlighting the need for standardized data reporting methods in AMR research. Standardization in reporting in the future will facilitate synergy in systematic reviews and meta-analyses studies.
In the current study, the prevalence of tetracycline resistance in E. coli isolated from non-intervention beef cattle was 0.31. Interestingly, the introduction of tetracycline as an intervention—either via feed or injection—elicited a moderate elevation in resistance prevalence to 0.53 in the group receiving tetracycline through feed and a modest elevation to 0.39 in those administered systemically. This increase in tetracycline resistance among intervention subgroups may be due to the selective pressure exerted by the antimicrobial, which favors the survival and propagation of resistant strains [38,39]. It is possible that the relatively higher level of resistance observed in the feed subgroup resulted from prolonged, gradual antimicrobial exposure, and subtherapeutic levels, thereby fostering an increase in resistance. However, additional factors, such as dosage, administration frequency, and biological heterogeneity among cattle, may contribute to this finding.
A subtle trend toward a higher prevalence of tetracycline resistance in intervention groups, particularly distinguished by the route of administration was observed. While we noted a trend towards a higher prevalence of tetracycline resistance in the intervention groups, this was not statistically significant. Therefore, it is important to interpret our results with caution. The absence of statistical significance may not necessarily negate the existence of an effect. Rather, it reveals the complexities and inherent difficulties associated with such types of analyses, such as the diversity in study designs, variation in sample sizes, wide confidence intervals, and high heterogeneity across our dataset, which may have potentially obscured a true effect if it exists. Interestingly, the consistent prevalence of tetracycline resistance in non-intervention groups, including both “entry” and “control” groups, points towards the influence of other factors apart from mere tetracycline exposure. This high baseline resistance could also have potentially impeded our ability to discern a statistically significant difference when compared with the intervention group. The lack of significant differences between the intervention and non-intervention groups contradicts the notion that the widespread use of antimicrobials in animals results in the development of AMR. Likewise, earlier studies reported no significant variations in the occurrence of AMR in isolates obtained from animals, regardless of whether they were exposed to antimicrobials or not [40,41,42]. Environmental exposure to tetracycline residues [43], innate resistance mechanisms in certain E. coli strains [44], and the ability of these bacteria to share resistance genes [45] may be major contributors to the higher prevalence of resistant E. coli populations in non-intervention groups. In addition, in the presence of multi-resistance genetic elements, the use of other antimicrobials may have inadvertently promoted tetracycline resistance [46]. Recent evidence also highlights the potential for bacteria to develop resistance when exposed to non-antibiotic compounds used in the agricultural food industry [47]. Moreover, previous studies indicate that the age and diet of cattle appeared to play a significant role in the acquisition and development of AMR commensal microflora, compared to the subtherapeutic use of antimicrobials [40].
Moreover, due to potential biases introduced by variations in study design, sample size, geographic location, and sampling methodology among the included studies, the global prevalence of resistance must be interpreted with caution. The wide 95% CI for this estimate (0.17–0.48) indicates a high degree of uncertainty, which likely reflects the substantial heterogeneity of the included studies. These findings highlight the complex and multifactorial nature of AMR development, necessitating additional exhaustive research. The observed higher heterogeneity across the studies included in this meta-analysis might be indicative of a multitude of interacting variables influencing the prevalence of tetracycline resistance in E. coli isolated from beef cattle. These could span from genetic and environmental factors to livestock management practices and variations in antimicrobial administration. Enhanced heterogeneity suggests that the outcomes of individual studies are not merely the result of sampling error, but are also influenced by these different factors, each contributing to the overall resistance prevalence [48]. The high heterogeneity, therefore, suggests a complex interplay of factors that is difficult to put into a single aggregated measure. This realization places a spotlight on the necessity for caution when interpreting the overall pooled resistance prevalence and the possibility of not adequately reflecting the specific contexts of individual studies. Consequently, it prompts a call for more focused, context-specific research that can better account for this inherent variability and offer a more nuanced understanding of tetracycline resistance in differing beef cattle populations.
The lack of studies employing advanced molecular resistance determination techniques, such as PCR or WGS, was one of the limitations of our investigation. In addition, the increased heterogeneity observed among the studies highlights the urgent need for standardization of methodologies and reporting practices within the field of AMR research. This substantial variation between studies has the potential to obscure nuanced interpretations and dilute specific findings, thereby potentially diminishing the significance of the aggregated results. Uniformity in methodological design and reporting will ensure more reliable comparability and enhance the efficacy of meta-analyses in elucidating clear AMR trends and patterns. This call to action for the harmonization of research emphasizes the significance of international collaboration and shared guidelines in the fight against AMR.
5. Conclusions
This meta-analysis has shed light on the magnitude of tetracycline resistance in E. coli isolated from beef cattle and highlighted the complex interplay of variables driving AMR in beef cattle. Regardless of whether beef cattle were directly exposed to tetracycline during the feeding period or not, the prevalence of tetracycline resistance among feedlot cattle was comparable. The notion that subtherapeutic use of antimicrobial in livestock leads to widespread resistance may be outdated. The current findings call for a thorough review and improvement of antimicrobial research to better understand what drives AMR in general and in beef cattle in particular. A deeper understanding of AMR mechanisms allows the development of strategies tailored to the beef industry’s specific challenges. Harmonization and standardization of studies reporting AMR are urgently required.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/vetsci10070479/s1, Supplementary Figure S1. Prevalence of tetracycline resistance in Escherichia coli isolates obtained from beef cattle without any antibiotic intervention, after ‘Alexander et al., 2008’ [26] and ‘Benedict et al., 2015′ [32] were removed as outliers. Samples were collected upon the entry of cattle into the feedlots (‘Entry’) and at the time of their exit (‘Control’). It’s important to note that some of the studies classified under the ‘Control’ subgroup were part of case-control studies, while others originated from cohort observational studies [4,24,25,27,28,29,30,31,33,34,35,36]. Supplementary Figure S2. Prevalence of tetracycline resistance in Escherichia coli isolates sourced from beef cattle subjected to antibiotic intervention After ‘Alexander et al., 2008′ [26] and ‘Lefebvre et al., 2006′ [34] were removed as outliers [24,25,27,28,29,30,37]. Supplementary Figure S3. (A) Funnel Plot of Publication bias for a group of studies with intervention. (B) Funnel Plot of Publication bias for a group Studies without Intervention.
Author Contributions
Conceptualization, K.P., Y.E.M. and G.M.W.; methodology, Y.E.M. and G.M.W.; formal analysis, G.M.W.; investigation, Y.E.M. and G.M.W.; writing—original draft preparation, G.M.W. and Y.E.M.; writing—review and editing, G.M.W., Y.E.M. and K.P.; supervision, K.P.; project administration, K.P.; fund acquisition, K.P. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data can be available upon request to authors.
Acknowledgments
The authors extend their heartfelt gratitude to Roy Kirkwood for his insightful feedback and invaluable contributions towards refining the manuscript. The authors are indebted to Caitlin Wringe for her initial efforts and groundwork, which significantly paved the way for the successful completion of this article.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Kasimanickam, V.; Kasimanickam, M.; Kasimanickam, R. Antibiotics Use in Food Animal Production: Escalation of Antimicrobial Resistance: Where Are We Now in Combating AMR? Med. Sci. 2021, 9, 14. [Google Scholar] [CrossRef] [PubMed]
- Nicola, I.; Cerutti, F.; Grego, E.; Bertone, I.; Gianella, P.; D’Angelo, A.; Peletto, S.; Bellino, C. Characterization of the upper and lower respiratory tract microbiota in Piedmontese calves. Microbiome 2017, 5, 152. [Google Scholar] [CrossRef] [PubMed]
- Snowder, G.D.; Van Vleck, L.D.; Cundiff, L.V.; Bennett, G.L. Bovine respiratory disease in feedlot cattle: Environmental, genetic, and economic factors. J. Anim. Sci. 2006, 84, 1999–2008. [Google Scholar] [CrossRef] [PubMed]
- Noyes, N.R.; Benedict, K.M.; Gow, S.P.; Booker, C.W.; Hannon, S.J.; McAllister, T.A.; Morley, P.S. Mannheimia haemolytica in feedlot cattle: Prevalence of recovery and associations with antimicrobial use, resistance, and health outcomes. J. Vet. Intern. Med. 2015, 29, 705–713. [Google Scholar] [CrossRef] [PubMed]
- Maples, W.E.; Brorsen, B.W.; Peel, D.; Hicks, B. Observational study of the effect of metaphylaxis treatment on feedlot cattle productivity and health. Front. Vet. Sci. 2022, 9, 947585. [Google Scholar] [CrossRef]
- Urban-Chmiel, R.; Grooms, D. Prevention and control of bovine respiratory disease. J. Livest. Sci. 2012, 3, 27–36. [Google Scholar]
- Ives, S.E.; Richeson, J.T. Use of antimicrobial metaphylaxis for the control of bovine respiratory disease in high-risk cattle. Vet. Clin. Food Anim. Pract. 2015, 31, 341–350. [Google Scholar] [CrossRef]
- Nickell, J.S.; White, B.J. Metaphylactic antimicrobial therapy for bovine respiratory disease in stocker and feedlot cattle. Vet. Clin. Food Anim. Pract. 2010, 26, 285–301. [Google Scholar] [CrossRef]
- Horton, L.M.; Depenbusch, B.E.; Dewsbury, D.M.; McAtee, T.B.; Betts, N.B.; Renter, D.G. Comprehensive outcomes affected by antimicrobial metaphylaxis of Feedlot Calves at Medium-Risk for bovine respiratory disease from a randomized controlled trial. Vet. Sci. 2023, 10, 67. [Google Scholar] [CrossRef]
- Granados-Chinchilla, F.; Rodriguez, C. Tetracyclines in Food and Feedingstuffs: From Regulation to Analytical Methods, Bacterial Resistance, and Environmental and Health Implications. J. Anal. Methods Chem. 2017, 2017, 1315497. [Google Scholar] [CrossRef]
- Masse, D.I.; Saady, N.M.; Gilbert, Y. Potential of Biological Processes to Eliminate Antibiotics in Livestock Manure: An Overview. Animals 2014, 4, 146–163. [Google Scholar] [CrossRef] [PubMed]
- Andersson, D.I.; Hughes, D. Microbiological effects of sublethal levels of antibiotics. Nat. Rev. Microbiol. 2014, 12, 465–478. [Google Scholar] [CrossRef] [PubMed]
- Samtiya, M.; Matthews, K.R.; Dhewa, T.; Puniya, A.K. Antimicrobial Resistance in the Food Chain: Trends, Mechanisms, Pathways, and Possible Regulation Strategies. Foods 2022, 11, 2966. [Google Scholar] [CrossRef]
- Gallo, G.F.; Berg, J.L. Efficacy of a feed-additive antibacterial combination for improving feedlot cattle performance and health. Can. Vet. J. 1995, 36, 223–229. [Google Scholar]
- Serwecińska, L. Antimicrobials and Antibiotic-Resistant Bacteria: A Risk to the Environment and to Public Health. Water 2020, 12, 3313. [Google Scholar] [CrossRef]
- Prestinaci, F.; Pezzotti, P.; Pantosti, A. Antimicrobial resistance: A global multifaceted phenomenon. Pathog. Glob. Health 2015, 109, 309–318. [Google Scholar] [CrossRef] [PubMed]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. Int. J. Surg. 2021, 88, 105906. [Google Scholar] [CrossRef]
- Viechtbauer, W. Conducting Meta-Analyses inRwith themetaforPackage. J. Stat. Softw. 2010, 36, 25. [Google Scholar] [CrossRef]
- Olkin, I.; Dahabreh, I.J.; Trikalinos, T.A. GOSH—A graphical display of study heterogeneity. Res. Synth. Methods 2012, 3, 214–223. [Google Scholar] [CrossRef]
- Ester, M.; Kriegel, H.-P.; Sander, J.; Xu, X. A density-based algorithm for discovering clusters in large spatial databases with noise. In Proceedings of the KDD, Portland, OR, USA, 2–4 August 1996; pp. 226–231. [Google Scholar]
- Viechtbauer, W.; Cheung, M.W. Outlier and influence diagnostics for meta-analysis. Res. Synth. Methods 2010, 1, 112–125. [Google Scholar] [CrossRef]
- Peters, J.L.; Sutton, A.J.; Jones, D.R.; Abrams, K.R.; Rushton, L. Comparison of two methods to detect publication bias in meta-analysis. JAMA 2006, 295, 676–680. [Google Scholar] [CrossRef]
- Sterne, J.A.; Egger, M.; Moher, D. Addressing reporting biases. In Cochrane Handbook for Systematic Reviews of Interventions; Cochrane Book Series; John Wiley & Sons: Chichester, UK, 2008; pp. 297–333. [Google Scholar]
- Alexander, T.W.; Inglis, G.D.; Yanke, L.J.; Topp, E.; Read, R.R.; Reuter, T.; McAllister, T.A. Farm-to-fork characterization of Escherichia coli associated with feedlot cattle with a known history of antimicrobial use. Int. J. Food Microbiol. 2010, 137, 40–48. [Google Scholar] [CrossRef]
- Alexander, T.W.; Reuter, T.; Sharma, R.; Yanke, L.J.; Topp, E.; McAllister, T.A. Longitudinal characterization of resistant Escherichia coli in fecal deposits from cattle fed subtherapeutic levels of antimicrobials. Appl. Environ. Microbiol. 2009, 75, 7125–7134. [Google Scholar] [CrossRef]
- Alexander, T.W.; Yanke, L.J.; Topp, E.; Olson, M.E.; Read, R.R.; Morck, D.W.; McAllister, T.A. Effect of subtherapeutic administration of antibiotics on the prevalence of antibiotic-resistant Escherichia coli bacteria in feedlot cattle. Appl. Environ. Microbiol. 2008, 74, 4405–4416. [Google Scholar] [CrossRef] [PubMed]
- Checkley, S.L.; Campbell, J.R.; Chirino-Trejo, M.; Janzen, E.D.; McKinnon, J.J. Antimicrobial resistance in generic fecal Escherichia coli obtained from beef cattle on arrival at the feedlot and prior to slaughter, and associations with volume of total individual cattle antimicrobial treatments in one western Canadian feedlot. Can. J. Vet. Res. 2008, 72, 101–108. [Google Scholar] [PubMed]
- Checkley, S.L.; Campbell, J.R.; Chirino-Trejo, M.; Janzen, E.D.; Waldner, C.L. Associations between antimicrobial use and the prevalence of antimicrobial resistance in fecal Escherichia coli from feedlot cattle in western Canada. Can. Vet. J. 2010, 51, 853–861. [Google Scholar]
- Lefebvre, B.; Diarra, M.S.; Giguere, K.; Roy, G.; Michaud, S.; Malouin, F. Antibiotic resistance and hypermutability of Escherichia coli O157 from feedlot cattle treated with growth-promoting agents. J. Food Prot. 2005, 68, 2411–2419. [Google Scholar] [CrossRef]
- Sharma, R.; Munns, K.; Alexander, T.; Entz, T.; Mirzaagha, P.; Yanke, L.J.; Mulvey, M.; Topp, E.; McAllister, T. Diversity and distribution of commensal fecal Escherichia coli bacteria in beef cattle administered selected subtherapeutic antimicrobials in a feedlot setting. Appl. Environ. Microbiol. 2008, 74, 6178–6186. [Google Scholar] [CrossRef]
- Becker, J.; Perreten, V.; Steiner, A.; Stucki, D.; Schupbach-Regula, G.; Collaud, A.; Rossano, A.; Wuthrich, D.; Muff-Hausherr, A.; Meylan, M. Antimicrobial susceptibility in E. coli and Pasteurellaceae at the beginning and at the end of the fattening process in veal calves: Comparing ‘outdoor veal calf’ and conventional operations. Vet. Microbiol. 2022, 269, 109419. [Google Scholar] [CrossRef]
- Benedict, K.M.; Gow, S.P.; McAllister, T.A.; Booker, C.W.; Hannon, S.J.; Checkley, S.L.; Noyes, N.R.; Morley, P.S. Antimicrobial Resistance in Escherichia coli Recovered from Feedlot Cattle and Associations with Antimicrobial Use. PLoS ONE 2015, 10, e0143995. [Google Scholar] [CrossRef]
- Gay, E.; Bour, M.; Cazeau, G.; Jarrige, N.; Martineau, C.; Madec, J.Y.; Haenni, M. Antimicrobial Usages and Antimicrobial Resistance in Commensal Escherichia coli From Veal Calves in France: Evolution During the Fattening Process. Front. Microbiol. 2019, 10, 792. [Google Scholar] [CrossRef] [PubMed]
- Lefebvre, B.; Malouin, F.; Roy, G.; Giguere, K.; Diarra, M.S. Growth performance and shedding of some pathogenic bacteria in feedlot cattle treated with different growth-promoting agents. J. Food Prot. 2006, 69, 1256–1264. [Google Scholar] [CrossRef] [PubMed]
- Messele, Y.E.; Alkhallawi, M.; Veltman, T.; Trott, D.J.; McMeniman, J.P.; Kidd, S.P.; Low, W.Y.; Petrovski, K.R. Phenotypic and Genotypic Analysis of Antimicrobial Resistance in Escherichia coli Recovered from Feedlot Beef Cattle in Australia. Animals 2022, 12, 2256. [Google Scholar] [CrossRef] [PubMed]
- Rao, S.; Van Donkersgoed, J.; Bohaychuk, V.; Besser, T.; Song, X.M.; Wagner, B.; Hancock, D.; Renter, D.; Dargatz, D.; Morley, P.S. Antimicrobial drug use and antimicrobial resistance in enteric bacteria among cattle from Alberta feedlots. Foodborne Pathog. Dis. 2010, 7, 449–457. [Google Scholar] [CrossRef]
- Kanwar, N.; Scott, H.M.; Norby, B.; Loneragan, G.H.; Vinasco, J.; Cottell, J.L.; Chalmers, G.; Chengappa, M.M.; Bai, J.; Boerlin, P. Impact of treatment strategies on cephalosporin and tetracycline resistance gene quantities in the bovine fecal metagenome. Sci. Rep. 2014, 4, 5100. [Google Scholar] [CrossRef]
- Hu, X.; Zhang, Y.; Chen, Z.; Gao, Y.; Teppen, B.; Boyd, S.A.; Zhang, W.; Tiedje, J.M.; Li, H. Tetracycline accumulation in biofilms enhances the selection pressure on Escherichia coli for expression of antibiotic resistance. Sci. Total Environ. 2023, 857, 159441. [Google Scholar] [CrossRef]
- Zhang, Y.; Pei, M.; Zhang, B.; He, Y.; Zhong, Y. Changes of antibiotic resistance genes and bacterial communities in the advanced biological wastewater treatment system under low selective pressure of tetracycline. Water Res. 2021, 207, 117834. [Google Scholar] [CrossRef]
- Mirzaagha, P.; Louie, M.; Sharma, R.; Yanke, L.J.; Topp, E.; McAllister, T.A. Distribution and characterization of ampicillin- and tetracycline-resistant Escherichia coli from feedlot cattle fed subtherapeutic antimicrobials. BMC Microbiol. 2011, 11, 78. [Google Scholar] [CrossRef]
- Galland, J.C.; Hyatt, D.R.; Crupper, S.S.; Acheson, D.W. Prevalence, antibiotic susceptibility, and diversity of Escherichia coli O157:H7 isolates from a longitudinal study of beef cattle feedlots. Appl. Environ. Microbiol. 2001, 67, 1619–1627. [Google Scholar] [CrossRef] [PubMed]
- Morris, C.; Wickramasingha, D.; Abdelfattah, E.M.; Pereira, R.V.; Okello, E.; Maier, G. Prevalence of antimicrobial resistance in fecal Escherichia coli and Enterococcus spp. isolates from beef cow-calf operations in northern California and associations with farm practices. Front. Microbiol. 2023, 14, 1086203. [Google Scholar] [CrossRef]
- Gaze, W.H.; Krone, S.M.; Larsson, D.G.; Li, X.Z.; Robinson, J.A.; Simonet, P.; Smalla, K.; Timinouni, M.; Topp, E.; Wellington, E.M.; et al. Influence of humans on evolution and mobilization of environmental antibiotic resistome. Emerg. Infect. Dis. 2013, 19, e120871. [Google Scholar] [CrossRef]
- Anderson, G.G.; O’Toole, G.A. Innate and induced resistance mechanisms of bacterial biofilms. Curr. Top. Microbiol. Immunol. 2008, 322, 85–105. [Google Scholar] [CrossRef]
- Tao, S.; Chen, H.; Li, N.; Wang, T.; Liang, W. The Spread of Antibiotic Resistance Genes In Vivo Model. Can. J. Infect. Dis. Med. Microbiol. 2022, 2022, 3348695. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.; Wang, B.; Huang, K.; Yin, J.; Ren, X.; Wang, Z.; Zhang, X.-X. Correlations among antibiotic resistance genes, mobile genetic elements and microbial communities in municipal sewage treatment plants revealed by high-throughput sequencing. Int. J. Environ. Res. Public Health 2023, 20, 3593. [Google Scholar] [CrossRef] [PubMed]
- Wales, A.D.; Davies, R.H. Co-Selection of Resistance to Antibiotics, Biocides and Heavy Metals, and Its Relevance to Foodborne Pathogens. Antibiotics 2015, 4, 567–604. [Google Scholar] [CrossRef] [PubMed]
- Stogiannis, D.; Siannis, F.; Androulakis, E. Heterogeneity in meta-analysis: A comprehensive overview. Int. J. Biostat. 2023. online ahead of print. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).