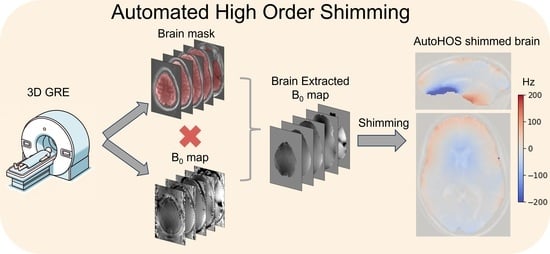
Automated High-Order Shimming for Neuroimaging Studies
Abstract
:1. Introduction
2. Materials and Methods
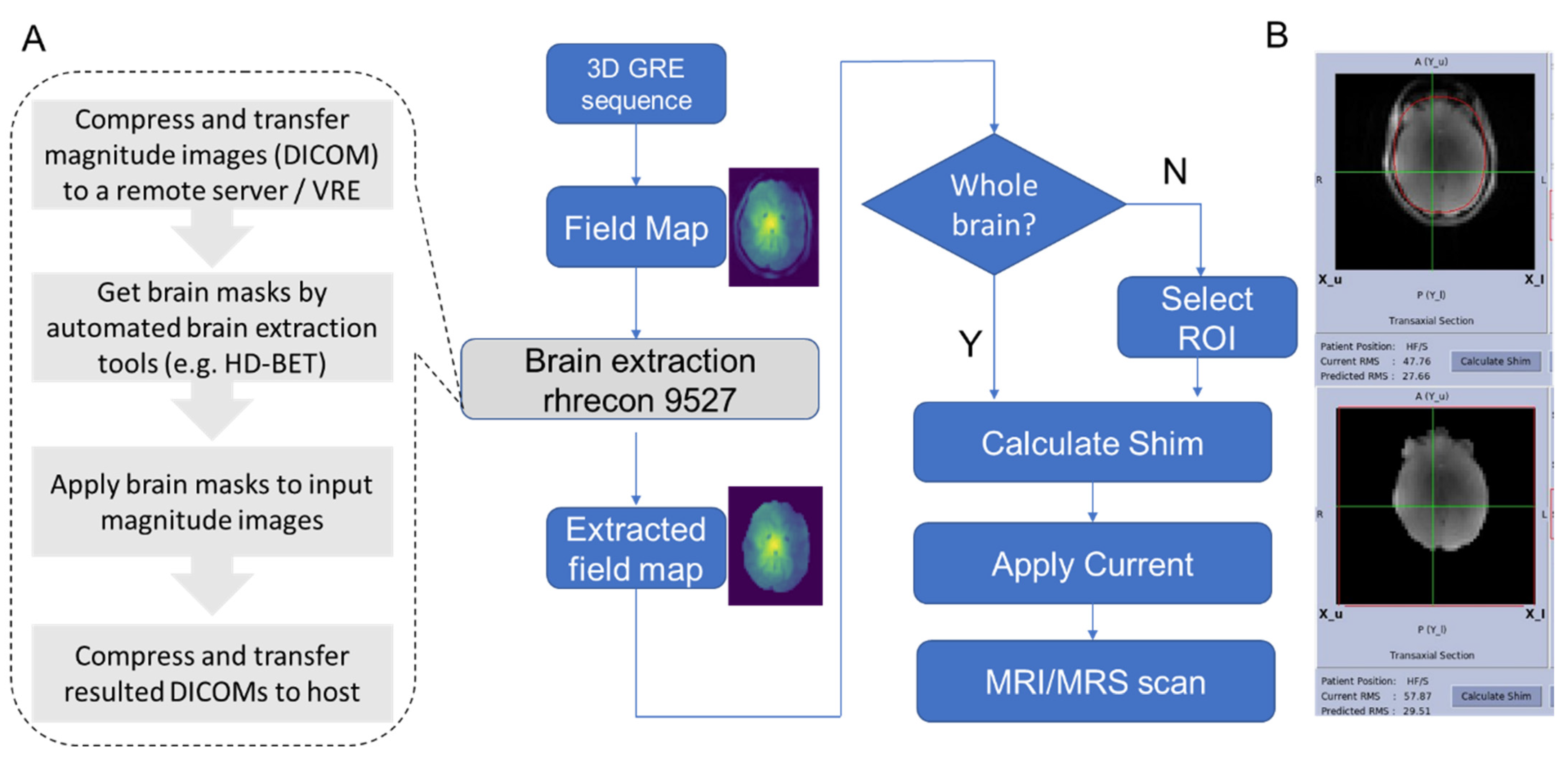
2.1. Automated HOS Pipeline and Implementation
2.2. Participants and Experimental Protocol
2.3. 3T MR Imaging Protocol
2.4. 7T MR Imaging Protocol
2.5. Image Analysis
3. Results
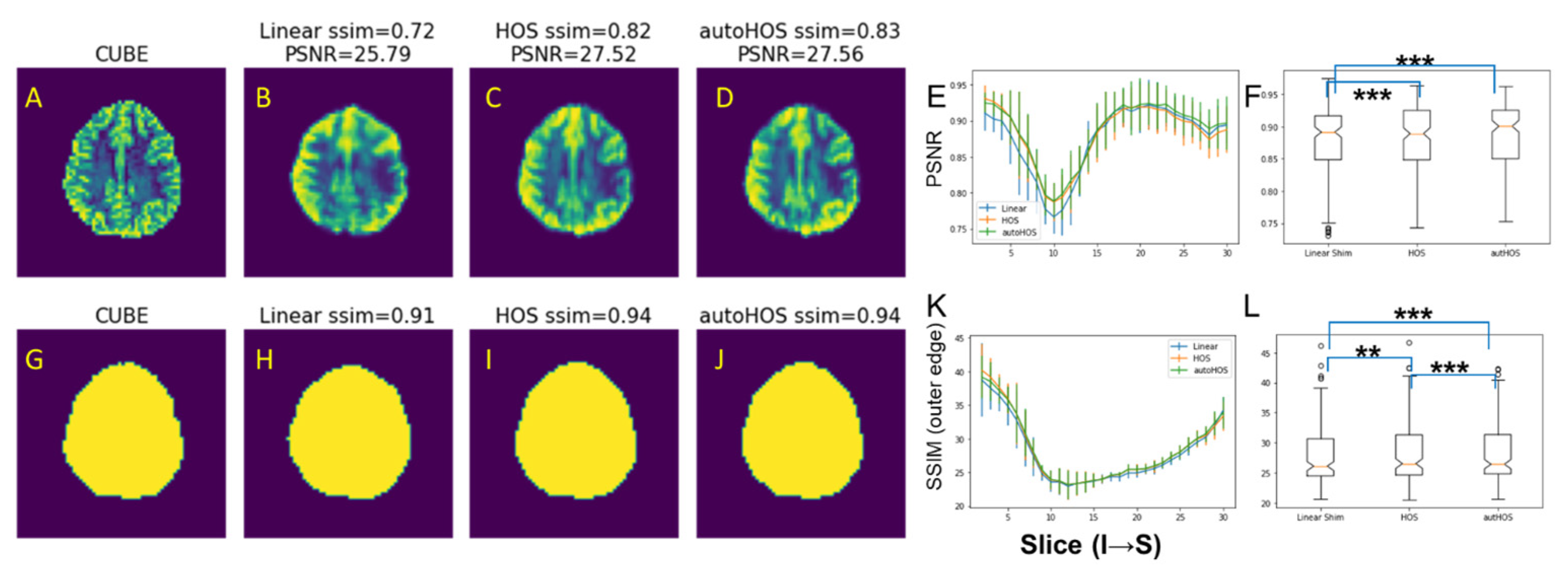
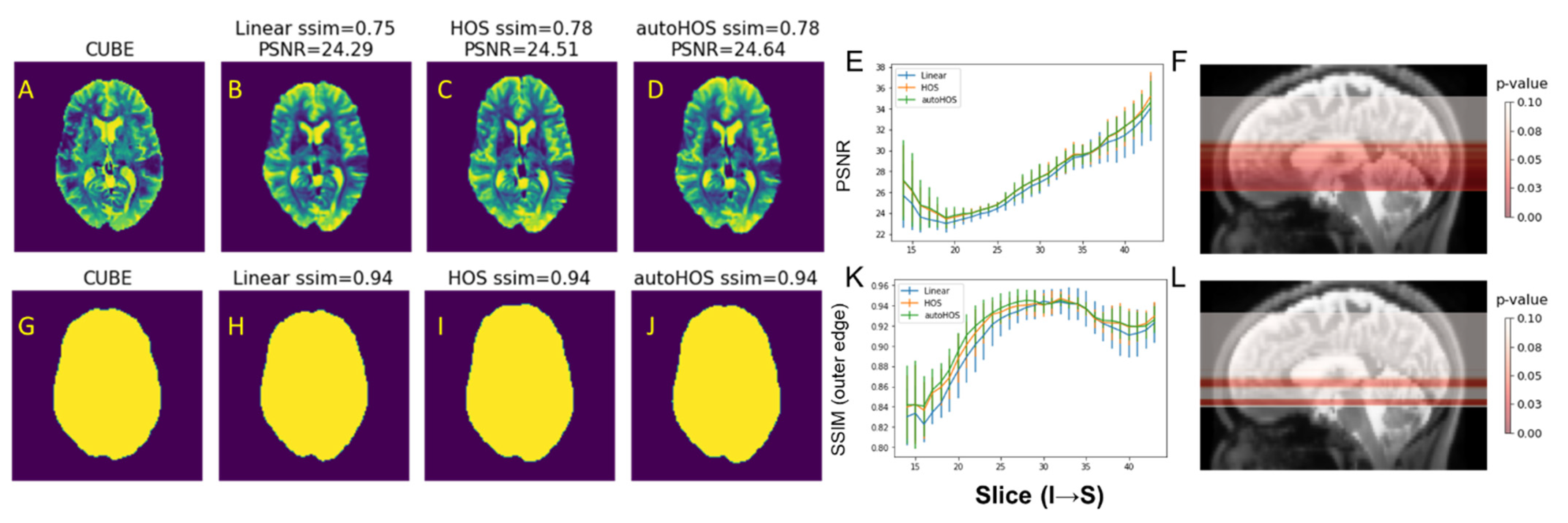
3.1. Comparison of B0 Homogeneities by Shimming Techniques
3.2. Comparison of EPI Distortion by Shimming Techniques
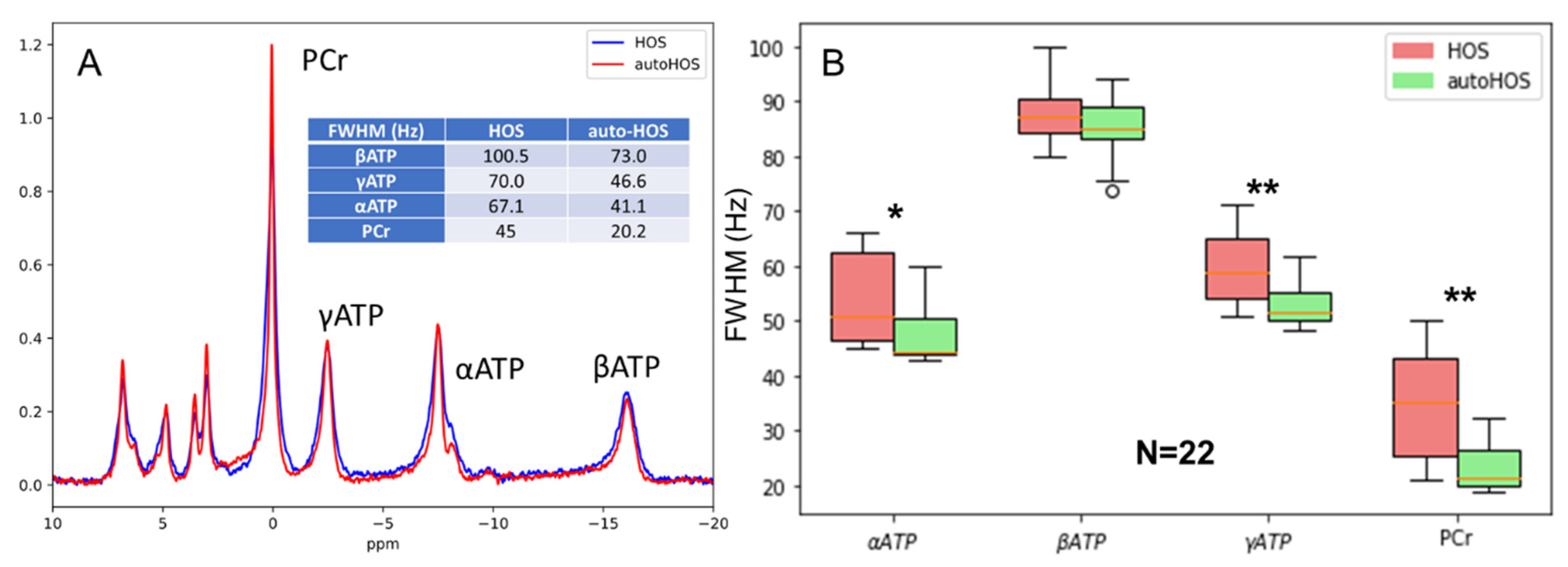
3.3. Superiority of autoHOS in MRS Spectral Linewidths
4. Discussion
4.1. High-Order Shimming Techniques Improve B0 Homogeneity Similarly at 3T and 7T
4.2. HOS and AutoHOS Reduces EPI Distortions
4.3. AutoHOS Significantly Improves Nonlocalized 31P MRS
4.4. Limitations
4.5. Future Work
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Juchem, C.; Cudalbu, C.; de Graaf, R.A.; Gruetter, R.; Henning, A.; Hetherington, H.P.; Boer, V.O. B0 shimming for in vivo magnetic resonance spectroscopy: Experts’ consensus recommendations. NMR Biomed. 2021, 34, e4350. [Google Scholar] [CrossRef] [PubMed]
- Juchem, C.; de Graaf, R.A. B(0) magnetic field homogeneity and shimming for in vivo magnetic resonance spectroscopy. Anal. Biochem. 2017, 529, 17–29. [Google Scholar] [CrossRef] [PubMed]
- Stockmann, J.P.; Wald, L.L. In vivo B(0) field shimming methods for MRI at 7T. Neuroimage 2018, 168, 71–87. [Google Scholar] [CrossRef] [PubMed]
- Fillmer, A.; Kirchner, T.; Cameron, D.; Henning, A. Constrained image-based B0 shimming accounting for “local minimum traps” in the optimization and field inhomogeneities outside the region of interest. Magn. Reson. Med. 2015, 73, 1370–1380. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Rycyna, R.E.; Rothman, D.L. Improvements on an in vivo automatic shimming method [FASTERMAP]. Magn. Reson. Med. 1997, 38, 834–839. [Google Scholar] [CrossRef] [PubMed]
- Gruetter, R.; Tkac, I. Field mapping without reference scan using asymmetric echo-planar techniques. Magn. Reson. Med. 2000, 43, 319–323. [Google Scholar] [CrossRef]
- Landheer, K.; Juchem, C. FAMASITO: FASTMAP Shim Tool towards user-friendly single-step B(0) homogenization. NMR Biomed. 2021, 34, e4486. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Adalsteinsson, E.; Glover, G.H.; Spielman, D.M. Regularized higher-order in vivo shimming. Magn. Reson. Med. 2002, 48, 715–722. [Google Scholar] [CrossRef] [PubMed]
- Hetherington, H.P.; Chu, W.J.; Gonen, O.; Pan, J.W. Robust fully automated shimming of the human brain for high-field 1H spectroscopic imaging. Magn. Reson. Med. 2006, 56, 26–33. [Google Scholar] [CrossRef] [PubMed]
- Isensee, F.; Schell, M.; Pflueger, I.; Brugnara, G.; Bonekamp, D.; Neuberger, U.; Wick, A.; Schlemmer, H.P.; Heiland, S.; Wick, W.; et al. Automated brain extraction of multisequence MRI using artificial neural networks. Hum. Brain Mapp. 2019, 40, 4952–4964. [Google Scholar] [CrossRef] [PubMed]
- Kalavathi, P.; Prasath, V.B. Methods on Skull Stripping of MRI Head Scan Images-a Review. J. Digit. Imaging 2016, 29, 365–379. [Google Scholar] [CrossRef] [PubMed]
- Juchem, C.; Nixon, T.W.; Diduch, P.; Rothman, D.L.; Starewicz, P.; de Graaf, R.A. Dynamic Shimming of the Human Brain at 7 Tesla. Concepts Magn. Reson. Part. B Magn. Reson. Eng. 2010, 37B, 116–128. [Google Scholar] [CrossRef] [PubMed]
- Rehman, H.Z.U.; Hwang, H.; Lee, S. Conventional and Deep Learning Methods for Skull Stripping in Brain MRI. Appl. Sci. 2020, 10, 1773. [Google Scholar] [CrossRef]
- Zhao, X.; Zhao, X.M. Deep learning of brain magnetic resonance images: A brief review. Methods 2021, 192, 131–140. [Google Scholar] [CrossRef] [PubMed]
- Cox, R.W. AFNI: Software for analysis and visualization of functional magnetic resonance neuroimages. Comput. Biomed. Res. 1996, 29, 162–173. [Google Scholar] [CrossRef] [PubMed]
- Avants, B.B.; Tustison, N.J.; Song, G.; Cook, P.A.; Klein, A.; Gee, J.C. A reproducible evaluation of ANTs similarity metric performance in brain image registration. Neuroimage 2011, 54, 2033–2044. [Google Scholar] [CrossRef] [PubMed]
- van der Walt, S.; Schonberger, J.L.; Nunez-Iglesias, J.; Boulogne, F.; Warner, J.D.; Yager, N.; Gouillart, E.; Yu, T.; the scikit-image contributors. Scikit-image: Image processing in Python. PeerJ 2014, 2, e453. [Google Scholar] [CrossRef] [PubMed]
- Helmus, J.J.; Jaroniec, C.P. Nmrglue: An open source Python package for the analysis of multidimensional NMR data. J. Biomol. NMR 2013, 55, 355–367. [Google Scholar] [CrossRef] [PubMed]
- Vanhamme, L.; van den Boogaart, A.; Van Huffel, S. Improved method for accurate and efficient quantification of MRS data with use of prior knowledge. J. Magn. Reson. 1997, 129, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Foxall, D.L.; Prins, W.M.U.S. A1 Shimming of MRI Scanner Involving Fat Suppression and/or Black Blood Preparation. Patent No. 0164082, 2006. [Google Scholar]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, J.; Yang, B.; Kelley, D.; Magnotta, V.A. Automated High-Order Shimming for Neuroimaging Studies. Tomography 2023, 9, 2148-2157. https://doi.org/10.3390/tomography9060168
Xu J, Yang B, Kelley D, Magnotta VA. Automated High-Order Shimming for Neuroimaging Studies. Tomography. 2023; 9(6):2148-2157. https://doi.org/10.3390/tomography9060168
Chicago/Turabian StyleXu, Jia, Baolian Yang, Douglas Kelley, and Vincent A. Magnotta. 2023. "Automated High-Order Shimming for Neuroimaging Studies" Tomography 9, no. 6: 2148-2157. https://doi.org/10.3390/tomography9060168
APA StyleXu, J., Yang, B., Kelley, D., & Magnotta, V. A. (2023). Automated High-Order Shimming for Neuroimaging Studies. Tomography, 9(6), 2148-2157. https://doi.org/10.3390/tomography9060168