Post-Processing Bias Field Inhomogeneity Correction for Assessing Background Parenchymal Enhancement on Breast MRI as a Quantitative Marker of Treatment Response
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Cohort
2.2. Pathological Response Assessment
2.3. MRI Data Acquisition
2.4. FGT Segmentation and BPE Calculation
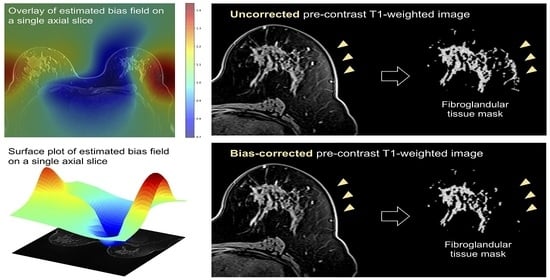
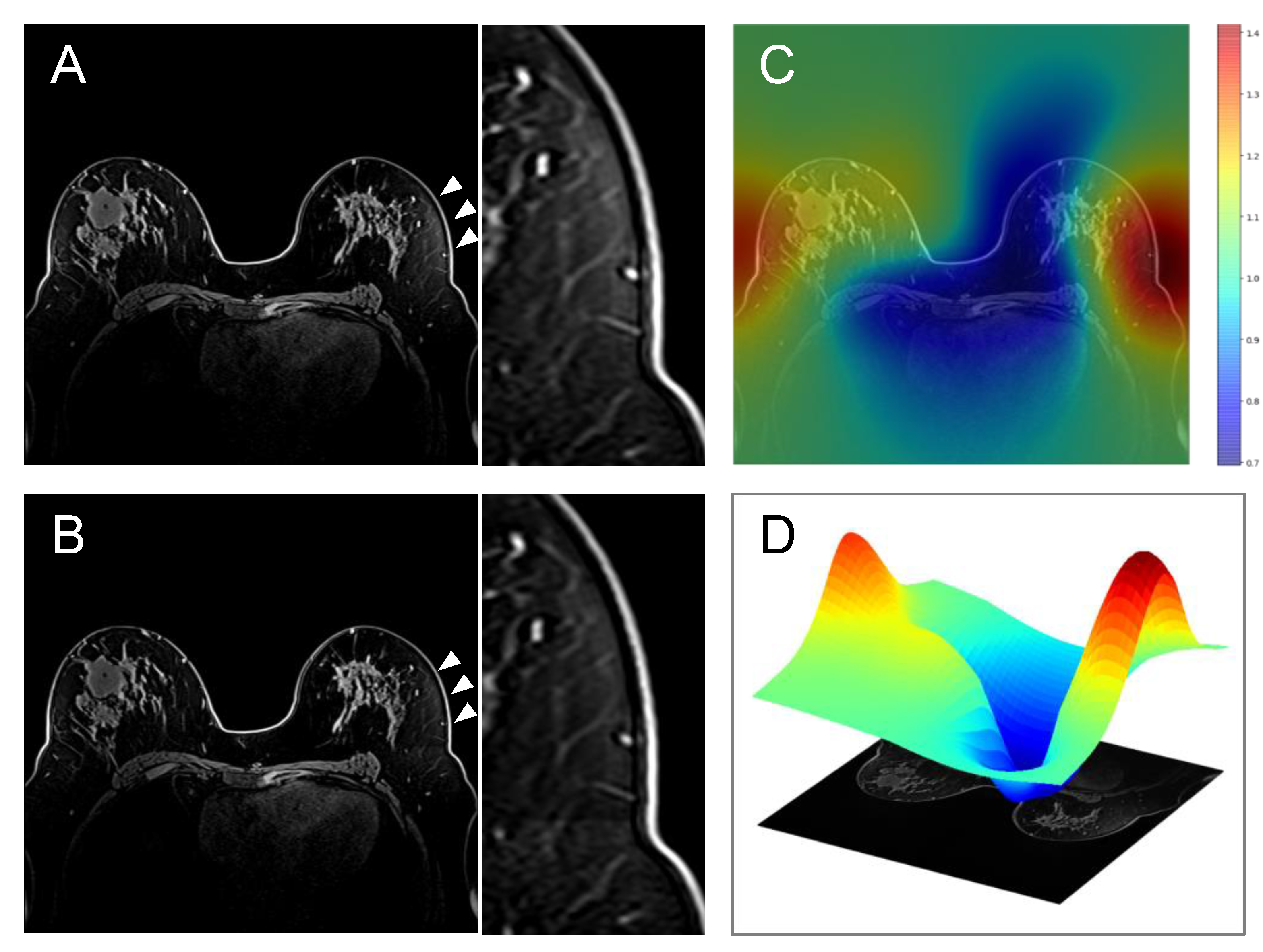
2.5. Bias Correction
2.6. Quantitative Comparison of Uncorrected vs. Bias-Corrected BPE Masks
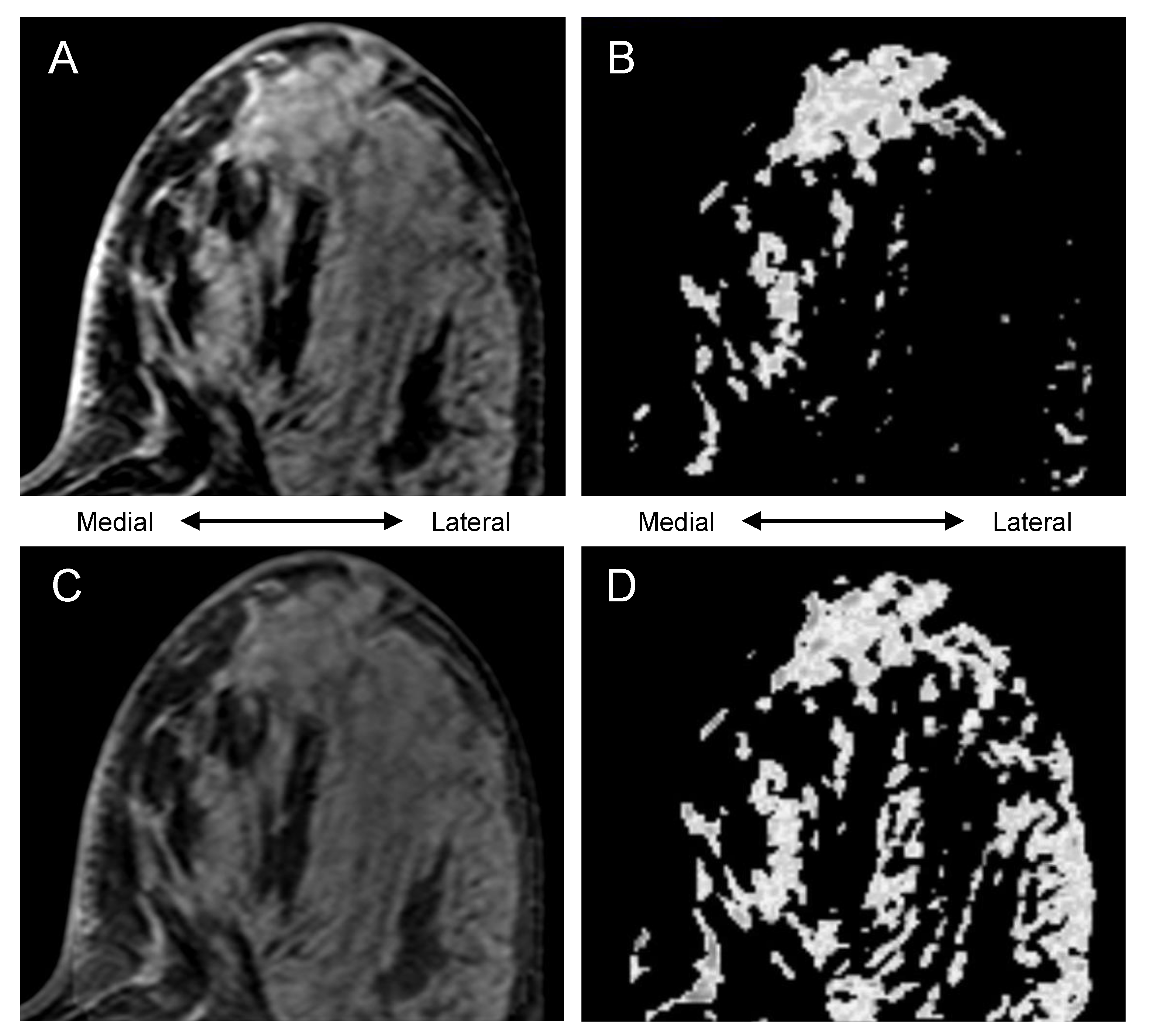
2.7. Visual Comparison of Uncorrected vs. Bias-Corrected BPE Masks
2.8. Statistical Analysis
3. Results
3.1. Patient Characteristics
3.2. Quantitatively Evaluated Effect of Bias Correction
3.3. Visually Evaluated Effect of Bias Correction
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Liao, G.J.; Henze Bancroft, L.C.; Strigel, R.M.; Chitalia, R.D.; Kontos, D.; Moy, L.; Partridge, S.C.; Rahbar, H. Background parenchymal enhancement on breast MRI: A comprehensive review. J. Magn. Reson. Imaging 2019, 51, 43–61. [Google Scholar] [CrossRef] [PubMed]
- Morris, E.A.; Comstock, C.E.; Lee, C.H. ACR BI-RADS® Magnetic Resonance Imaging. In ACR BI-RADS® Atlas, Breast Imaging Reporting and Data System; American College of Radiology: Reston, VA, USA, 2013. [Google Scholar]
- Chen, J.H.; Yu, H.J.; Hsu, C.; Mehta, R.S.; Carpenter, P.M.; Su, M.Y. Background Parenchymal Enhancement of the Contralateral Normal Breast: Association with Tumor Response in Breast Cancer Patients Receiving Neoadjuvant Chemotherapy. Transl. Oncol. 2015, 8, 204–209. [Google Scholar] [CrossRef] [PubMed]
- You, C.; Peng, W.; Zhi, W.; He, M.; Liu, G.; Xie, L.; Jiang, L.; Hu, X.; Shen, X.; Gu, Y. Association between background parenchymal enhancement and pathologic complete remission throughout the neoadjuvant chemotherapy in breast cancer patients. Transl. Oncol. 2017, 10, 786–792. [Google Scholar] [CrossRef]
- Lam, D.L.; Hippe, D.S.; Kitsch, A.E.; Partridge, S.C.; Rahbar, H. Assessment of quantitative magnetic resonance imaging background parenchymal enhancement parameters to improve determination of individual breast cancer risk. J. Comput. Assist. Tomogr. 2019, 43, 85–92. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Jiang, L.; Li, Q.; Gu, Y. Quantitative assessment of background parenchymal enhancement in breast magnetic resonance images predicts the risk of breast cancer. Oncotarget 2017, 8, 10620–10627. [Google Scholar] [CrossRef][Green Version]
- van der Velden, B.H.M.; Sutton, E.J.; Carbonaro, L.A.; Pijnappel, R.M.; Morris, E.A.; Gilhuijs, K.G.A. Contralateral parenchymal enhancement on dynamic contrast-enhanced MRI reproduces as a biomarker of survival in ER-positive/HER2-negative breast cancer patients. Eur. Radiol. 2018, 28, 4705–4716. [Google Scholar] [CrossRef]
- Ragusi, M.A.A.; Loo, C.E.; van der Velden, B.H.M.; Wesseling, J.; Linn, S.C.; Beets-Tan, R.G.; Elias, S.G.; Gilhuijs, K.G.A. Contralateral parenchymal enhancement on breast MRI before and during neoadjuvant endocrine therapy in relation to the preoperative endocrine prognostic index. Eur. Radiol. 2020, 30, 6740–6748. [Google Scholar] [CrossRef]
- van der Velden, B.H.M.; Dmitriev, I.; Loo, C.E.; Pijnappel, R.M.; Gilhuijs, K.G.A. Association between Parenchymal Enhancement of the Contralateral Breast in Dynamic Contrast-enhanced MR Imaging and Outcome of Patients with Unilateral Invasive Breast Cancer. Radiology 2015, 276, 675–685. [Google Scholar] [CrossRef]
- Luo, J.; Johnston, B.S.; Kitsch, A.E.; Hippe, D.S.; Korde, L.A.; Javid, S.; Lee, J.M.; Peacock, S.; Lehman, C.D.; Partridge, S.C.; et al. Ductal Carcinoma in Situ: Quantitative Preoperative Breast MR Imaging Features Associated with Recurrence after Treatment. Radiology 2017, 285, 788–797. [Google Scholar] [CrossRef]
- Wu, S.; Weinstein, S.P.; DeLeo, M.J.; Conant, E.F.; Chen, J.; Domchek, S.M.; Kontos, D. Quantitative assessment of background parenchymal enhancement in breast MRI predicts response to risk-reducing salpingo-oophorectomy: Preliminary evaluation in a cohort of BRCA1/2 mutation carriers. Breast Cancer Res. 2015, 17, 67. [Google Scholar] [CrossRef]
- Nguyen, A.A.-T.; Arasu, V.A.; Strand, F.; Li, W.; Onishi, N.; Gibbs, J.; Jones, E.F.; Joe, B.N.; Esserman, L.J.; Newitt, D.C.; et al. Comparison of segmentation methods in assessing background parenchymal enhancement as a biomarker for response to neoadjuvant therapy. Tomography 2020, 6, 101–110. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Newitt, D.C.; Gibbs, J.; Wilmes, L.J.; Jones, E.F.; Arasu, V.A.; Strand, F.; Onishi, N.; Nguyen, A.A.-T.; Kornak, J.; et al. Predicting breast cancer response to neoadjuvant treatment using multi-feature MRI: Results from the I-SPY 2 TRIAL. NPJ Breast Cancer 2020, 6, 63. [Google Scholar] [CrossRef] [PubMed]
- Onishi, N.; Li, W.; Newitt, D.C.; Harnish, R.J.; Strand, F.; Nguyen, A.A.-T.; Arasu, V.A.; Gibbs, J.; Jones, E.F.; Wilmes, L.J.; et al. Breast MRI during Neoadjuvant Chemotherapy: Lack of Background Parenchymal Enhancement Suppression and Inferior Treatment Response. Radiology 2021, 301, 295–308. [Google Scholar] [CrossRef] [PubMed]
- Tustison, N.J.; Avants, B.B.; Cook, P.A.; Zheng, Y.; Egan, A.; Yushkevich, P.A.; Gee, J.C. N4ITK: Improved N3 bias correction. IEEE Trans. Med. Imaging 2010, 29, 1310–1320. [Google Scholar] [CrossRef]
- Klifa, C.; Carballido-Gamio, J.; Wilmes, L.; Laprie, A.; Lobo, C.; Demicco, E.; Watkins, M.; Shepherd, J.; Gibbs, J.; Hylton, N. Quantification of breast tissue index from MR data using fuzzy clustering. In Proceedings of the 26th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, San Francisco, CA, USA, 1–5 September 2004; Volume 3, pp. 1667–1670. [Google Scholar]
- Avants, B.B.; Tustison, N.J.; Song, G.; Cook, P.A.; Klein, A.; Gee, J.C. A reproducible evaluation of ANTs similarity metric performance in brain image registration. Neuroimage 2011, 54, 2033–2044. [Google Scholar] [CrossRef]
- Swift, A.J.; Wild, J.M.; Fichele, S.; Woodhouse, N.; Fleming, S.; Waterhouse, J.; Lawson, R.A.; Paley, M.N.J.; Van Beek, E.J.R. Emphysematous changes and normal variation in smokers and COPD patients using diffusion 3He MRI. Eur. J. Radiol. 2005, 54, 352–358. [Google Scholar] [CrossRef]
- Nalawade, S.; Murugesan, G.K.; Vejdani-Jahromi, M.; Fisicaro, R.A.; Bangalore Yogananda, C.G.; Wagner, B.; Mickey, B.; Maher, E.; Pinho, M.C.; Fei, B.; et al. Classification of brain tumor isocitrate dehydrogenase status using MRI and deep learning. J. Med. Imaging (Bellingham) 2019, 6, 046003. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Oh, S.J.; Chae, E.Y.; Cha, J.H.; Shin, H.J.; Choi, W.J.; Kim, H.H. Relationship between background parenchymal enhancement on breast MRI and pathological tumor response in breast cancer patients receiving neoadjuvant chemotherapy. Br. J. Radiol. 2018, 91, 20170550. [Google Scholar] [CrossRef] [PubMed]
- Preibsch, H.; Wanner, L.; Bahrs, S.D.; Wietek, B.M.; Siegmann-Luz, K.C.; Oberlecher, E.; Hahn, M.; Staebler, A.; Nikolaou, K.; Wiesinger, B. Background parenchymal enhancement in breast MRI before and after neoadjuvant chemotherapy: Correlation with tumour response. Eur. Radiol. 2016, 26, 1590–1596. [Google Scholar] [CrossRef]
- You, C.; Gu, Y.; Peng, W.; Li, J.; Shen, X.; Liu, G.; Peng, W. Decreased background parenchymal enhancement of the contralateral breast after two cycles of neoadjuvant chemotherapy is associated with tumor response in HER2-positive breast cancer. Acta Radiol. 2018, 59, 806–812. [Google Scholar] [CrossRef] [PubMed]
- Rella, R.; Bufi, E.; Belli, P.; Contegiacomo, A.; Giuliani, M.; Rosignuolo, M.; Rinaldi, P.; Manfredi, R. Background parenchymal enhancement in breast magnetic resonance imaging: A review of current evidences and future trends. Diagn. Interv. Imaging 2018, 99, 815–826. [Google Scholar] [CrossRef] [PubMed]
- Bauer, E.; Levy, M.S.; Domachevsky, L.; Anaby, D.; Nissan, N. Background parenchymal enhancement and uptake as breast cancer imaging biomarkers: A state-of-the-art review. Clin. Imaging 2022, 83, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Bignotti, B.; Signori, A.; Valdora, F.; Rossi, F.; Calabrese, M.; Durando, M.; Mariscotto, G.; Tagliafico, A. Evaluation of background parenchymal enhancement on breast MRI: A systematic review. Br. J. Radiol. 2017, 90, 20160542. [Google Scholar] [CrossRef]
- Klifa, C.; Suzuki, S.; Aliu, S.; Singer, L.; Wilmes, L.; Newitt, D.; Joe, B.; Hylton, N. Quantification of background enhancement in breast magnetic resonance imaging. J. Magn. Reson. Imaging 2011, 33, 1229–1234. [Google Scholar] [CrossRef] [PubMed]
- Fiaschetti, V.; Pistolese, C.A.; Funel, V.; Rascioni, M.; Claroni, G.; Della Gatta, F.; Cossu, E.; Perretta, T.; Simonetti, G. Breast MRI artefacts: Evaluation and solutions in 630 consecutive patients. Clin. Radiol. 2013, 68, e601–e608. [Google Scholar] [CrossRef]
- Harvey, J.A.; Hendrick, R.E.; Coll, J.M.; Nicholson, B.T.; Burkholder, B.T.; Cohen, M.A. Breast MR imaging artifacts: How to recognize and fix them. Radiographics 2007, 27 (Suppl. 1), S131–S145. [Google Scholar] [CrossRef]
- Yitta, S.; Joe, B.N.; Wisner, D.J.; Price, E.R.; Hylton, N.M. Recognizing artifacts and optimizing breast MRI at 1.5 and 3 T. AJR Am. J. Roentgenol. 2013, 200, W673–W682. [Google Scholar] [CrossRef]
- Giess, C.S.; Yeh, E.D.; Raza, S.; Birdwell, R.L. Background parenchymal enhancement at breast MR imaging: Normal patterns, diagnostic challenges, and potential for false-positive and false-negative interpretation. Radiographics 2014, 34, 234–247. [Google Scholar] [CrossRef]




| Assessment | Label for UC | Label for BC | Category |
|---|---|---|---|
| A >> B | A | B | −2: worse agreement for BC than UC with substantial difference * |
| A > B | A | B | −1: worse agreement for BC than UC |
| A = B | A | B | 0: BC and UC showed equivalent agreement with the gold standard |
| A < B | A | B | 1: better agreement for BC than UC |
| A << B | A | B | 2: better agreement for BC than UC with substantial difference * |
| A >> B | B | A | 2: better agreement for BC than UC with substantial difference * |
| A > B | B | A | 1: better agreement for BC than UC |
| A = B | B | A | 0: BC and UC showed equivalent agreement with the gold standard |
| A < B | B | A | −1: worse agreement for BC than UC |
| A << B | B | A | −2: worse agreement for BC than UC with substantial difference * |
| Parameter | Whole Cohort (n = 735) | High-Quality Cohort (n = 340) | Non-High-Quality Patients (n = 395) | p Value | |
|---|---|---|---|---|---|
| Age (y) | |||||
| Mean ± SD | 49 ± 11 | 49 ± 10 | 49 ± 11 | 0.898 | |
| Range | 24–77 | 24–77 | 25–73 | ||
| Menopausal status | 0.942 | ||||
| Pre-menopausal | 342 (47) | 153 (45) | 189 (48) | ||
| Peri-menopausal | 26 (4) | 12 (4) | 14 (4) | ||
| Post-menopausal | 223 (30) | 105 (31) | 118 (30) | ||
| Unclear * | 95 (13) | 47 (14) | 48 (12) | ||
| No data | 49 (7) | 23 (7) | 26 (7) | ||
| Race | 0.359 | ||||
| White | 597 (81) | 281 (83) | 316 (80) | ||
| African American | 78 (11) | 28 (8) | 50 (13) | ||
| Asian | 47 (6) | 23 (7) | 24 (6) | ||
| American Indian or Alaska Native | 3 (0) | 2 (1) | 1 (0) | ||
| Native Hawaiian or Pacific Islander | 4 (1) | 2 (1) | 2 (1) | ||
| Mix | 6 (1) | 4 (1) | 2 (1) | ||
| Immunohistochemical subtype | 0.667 | ||||
| HR+/HER2– | 299 (41) | 140 (41) | 159 (40) | ||
| HR+/HER2+ | 112 (15) | 57 (17) | 55 (14) | ||
| HR–/HER2+ | 61 (8) | 27 (8) | 34 (9) | ||
| HR–/HER2– | 263 (36) | 116 (34) | 147 (37) | ||
| Assigned chemotherapy | 0.720 | ||||
| Standard chemotherapy | 158 (21) | 71 (21) | 87 (22) | ||
| Experimental chemotherapy | 577 (79) | 269 (79) | 308 (78) | ||
| Treatment response | 0.353 | ||||
| pCR | 258 (35) | 113 (33) | 145 (37) | ||
| non-pCR | 477 (65) | 227 (67) | 250 (63) | ||
| Cohort and Timepoint | Voxel Count for UC BPE Mask | Voxel Count for BC BPE Mask | Difference of Voxel Count * | ||
|---|---|---|---|---|---|
| Estimated Pseudo-Median | 95% CI | p-Value | |||
| Whole cohort | |||||
| T0 | 62,791 [39,090, 92,646] | 62,372 [37,478, 94,399] | 493.5 | −357, 1374.5 | 0.251 |
| T1 | 58,831 [34,199, 89,447] | 58,343 [35,580, 91,010] | 693.5 | −100.5, 1493 | 0.086 |
| T2 | 53,996 [33,076, 85,252] | 52,834 [33,168, 83,312] | 2.5 | −739.5, 770.5 | 0.995 |
| High-quality cohort | |||||
| T0 | 59,190 [37,981, 87,995] | 60,343 [37,233, 87,997] | −310.5 | −1293.5, 731 | 0.565 |
| T1 | 56,245 [36,219, 83,899] | 55,510 [35,830, 83,418] | 326.48 | −657, 1305.5 | 0.519 |
| T2 | 51,346 [34,779, 77,317] | 51,124 [33,884, 80,426] | −348 | −1247.5, 629 | 0.455 |
| Cohort and Timepoint | BPE Measurement for UC BPE Mask | BPE Measurement for BPE Mask | Difference of BPE Measurement * | ||
|---|---|---|---|---|---|
| Estimated Pseudo-Median | 95% CI | p Value | |||
| Whole cohort | |||||
| T0 | 23.3 [16.3, 34.3] | 24.0 [16.6, 35.1] | 0.64 | 0.52, 0.76 | <0.001 ** |
| T1 | 19.1 [13.5, 27.4] | 19.9 [14.1, 28.6] | 0.58 | 0.48, 0.69 | <0.001 ** |
| T2 | 17.1 [12.3, 23.4] | 17.6 [12.6, 24.3] | 0.48 | 0.38, 0.58 | <0.001 ** |
| High-quality cohort | |||||
| T0 | 23.2 [16.5, 35.1] | 23.4 [16.3, 35.1] | 0.43 | 0.29, 0.58 | <0.001 ** |
| T1 | 19.7 [14.5, 27.7] | 19.9 [15.2, 27.6] | 0.41 | 0.28, 0.55 | <0.001 ** |
| T2 | 17.7 [13.4, 24.1] | 18.1 [13.7, 25.3] | 0.36 | 0.24, 0.49 | <0.001 ** |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nguyen, A.A.-T.; Onishi, N.; Carmona-Bozo, J.; Li, W.; Kornak, J.; Newitt, D.C.; Hylton, N.M. Post-Processing Bias Field Inhomogeneity Correction for Assessing Background Parenchymal Enhancement on Breast MRI as a Quantitative Marker of Treatment Response. Tomography 2022, 8, 891-904. https://doi.org/10.3390/tomography8020072
Nguyen AA-T, Onishi N, Carmona-Bozo J, Li W, Kornak J, Newitt DC, Hylton NM. Post-Processing Bias Field Inhomogeneity Correction for Assessing Background Parenchymal Enhancement on Breast MRI as a Quantitative Marker of Treatment Response. Tomography. 2022; 8(2):891-904. https://doi.org/10.3390/tomography8020072
Chicago/Turabian StyleNguyen, Alex Anh-Tu, Natsuko Onishi, Julia Carmona-Bozo, Wen Li, John Kornak, David C. Newitt, and Nola M. Hylton. 2022. "Post-Processing Bias Field Inhomogeneity Correction for Assessing Background Parenchymal Enhancement on Breast MRI as a Quantitative Marker of Treatment Response" Tomography 8, no. 2: 891-904. https://doi.org/10.3390/tomography8020072
APA StyleNguyen, A. A.-T., Onishi, N., Carmona-Bozo, J., Li, W., Kornak, J., Newitt, D. C., & Hylton, N. M. (2022). Post-Processing Bias Field Inhomogeneity Correction for Assessing Background Parenchymal Enhancement on Breast MRI as a Quantitative Marker of Treatment Response. Tomography, 8(2), 891-904. https://doi.org/10.3390/tomography8020072