Integrating the OHIF Viewer into XNAT: Achievements, Challenges and Prospects for Quantitative Imaging Studies
Abstract
:1. Introduction
1.1. Conceptual Differences between XNAT and PACS
- Research data should be normally curated in “projects” that reflect identifiable academic activities (e.g., clinical trial, PhD project, blinded image review, online analysis “challenge”) each of which may have an individual Data Management Plan.
- Researchers from many different organisations (e.g., hospital, academia, industry) may need to access the platform.
- Unlike PACS, where any clinical user might need to access images for any patient, user permissions are customised per project according to ethical protocols, data transfer agreements, collaborations and time-limited embargoes.
- Academic principal/chief investigators may demand a high degree of autonomy, with the ability to curate, structure and manage their own information assets.
- Research data require anonymisation prior to introduction into the academic workflow and this may need to be tailored to individual studies.
- The repository platform should make data findable, accessible, interoperable and reusable (FAIR) [4]. This is typically achieved by equipping platforms with Representational State Transfer (REST) Application Programming Interfaces (APIs), thus enabling integration with a diverse range of end-user tools [5].
- Projects may combine DICOM with clinical, digital pathology, multi-omic and other non-DICOM data. Each project may also be associated with its own bespoke analysis software and data formats.
- Arbitrary processing outputs are frequently created by external data analysis tools and need to be stored back on the repository platform with appropriate provenance.
- Data enrichment via expert annotation should be exportable and should use standardised, portable formats, rather than be “locked into” a given vendor’s image display platform.
- Most importantly, for the remainder of this article, the processing and data visualisation methods used are often the subject of the research itself. Hence, the image viewer configuration needs to be agile, with the potential for incorporation of novel software that is, by definition, experimental and has not undergone regulatory approval for clinical use.
1.2. Image Viewing in XNAT, the OHIF Viewer and Other Web-Based Solutions
1.3. Quantitative Imaging Motivation for Development of the OHIF Viewer within XNAT
1.4. Image Annotation
2. Materials and Methods
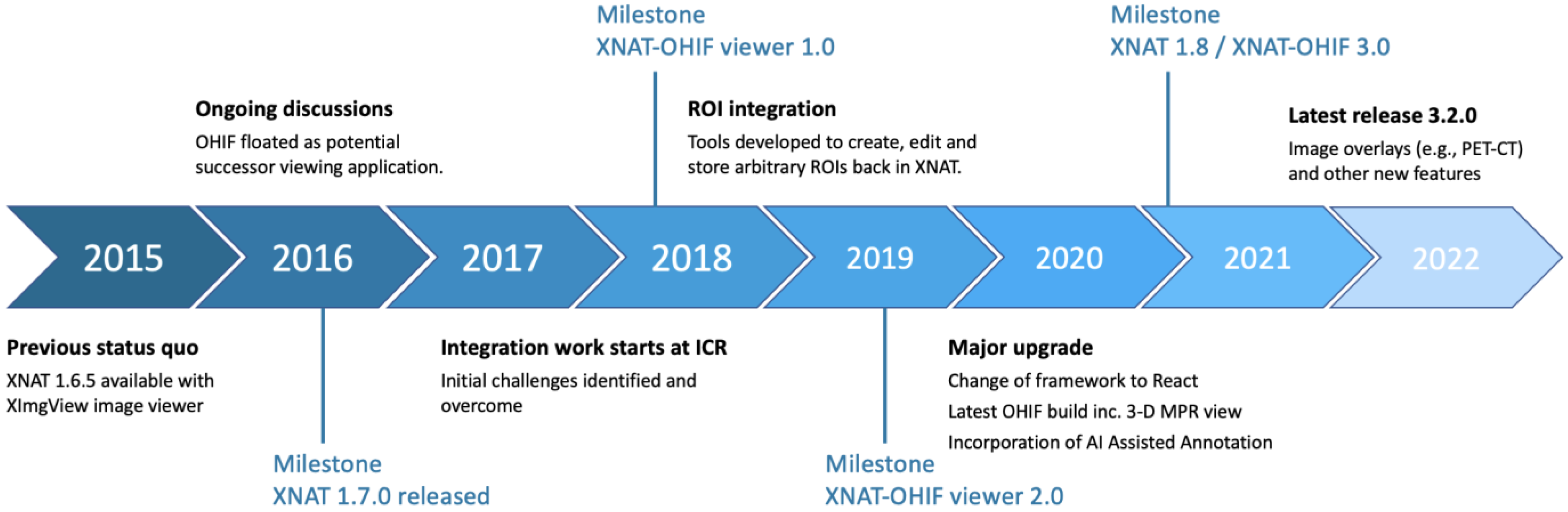
2.1. Integration Timeline
2.2. Initial Challenges
- Accessibility of images is governed by the XNAT security model.
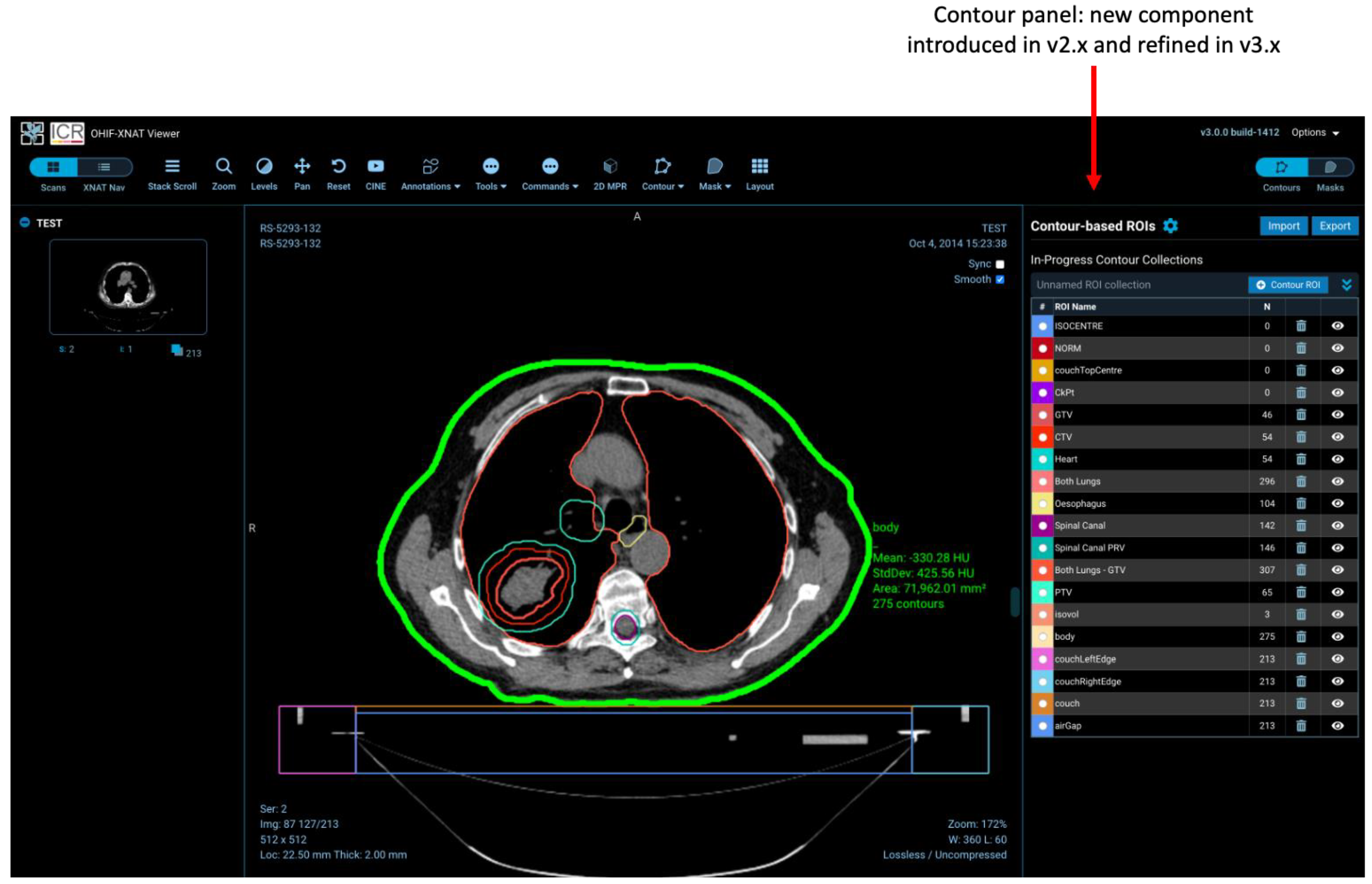
- Non-image DICOM series (e.g., ROIs) are removed from the study thumbnail list and handled by separate elements of the user interface (the contour and mask panels).
- Storage of ROIs uses XNAT’s REST API, creating new session resources and XNAT assessors, rather than simply adding contours and masks as new DICOM series in the study.
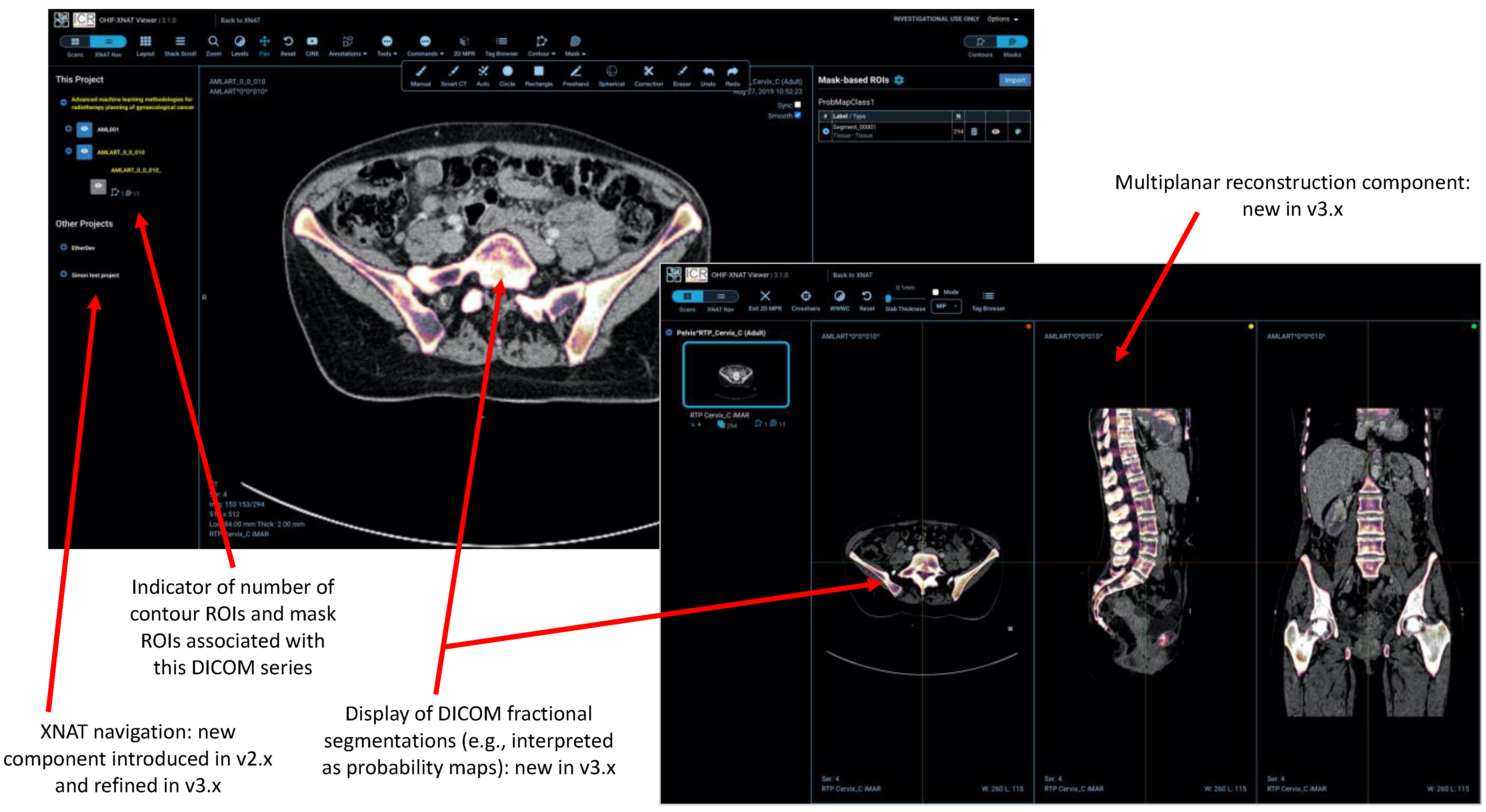
- Several other tools (e.g., the XNAT navigation sidebar and integration of the AIAA server) make calls to the XNAT REST API.
2.3. Development Environment
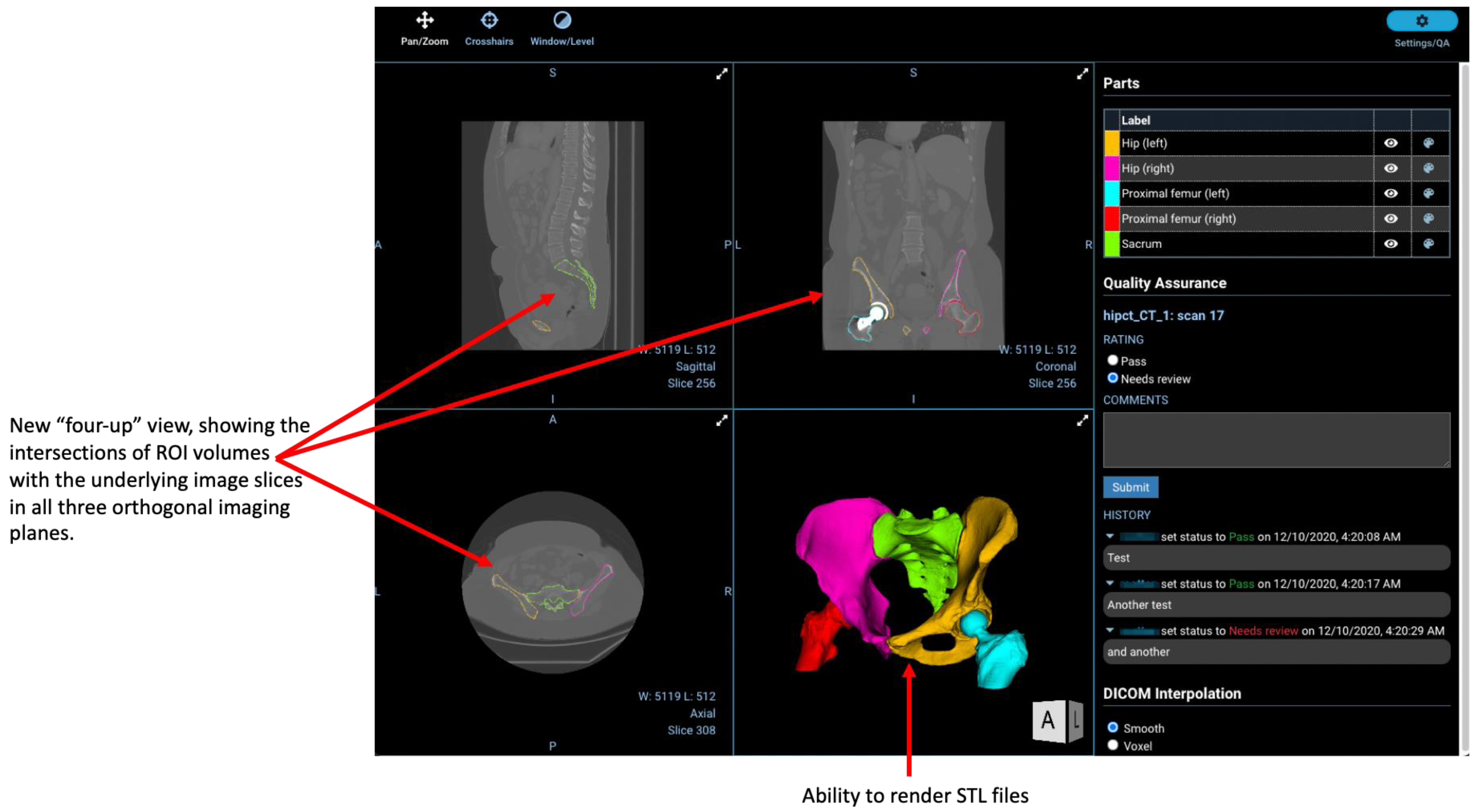
2.4. Regions-of-Interest
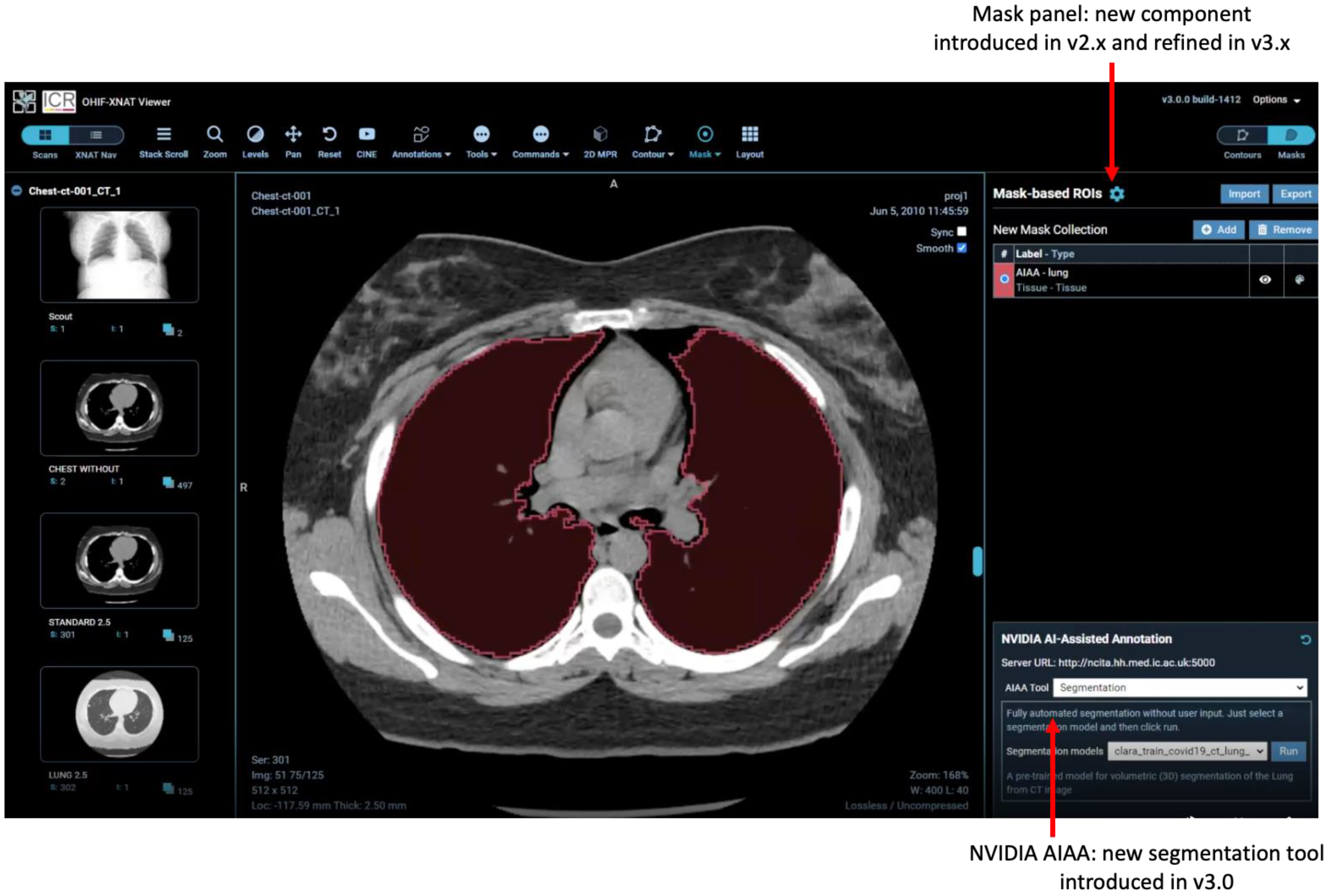
2.5. AI-Assisted Annotation
- Fully automated “Segmentation Models” return multi-label segmentations without any user input.
- Semi-automated “Annotation Models” use a minimum of six clicks from the user to define the bounding box of a structure and return the segmentation.
- “Deep Grow Models” are interactive, taking a point of reference and using successive “foreground” and/or “background” clicks to refine the model’s inference, by including or excluding regions.
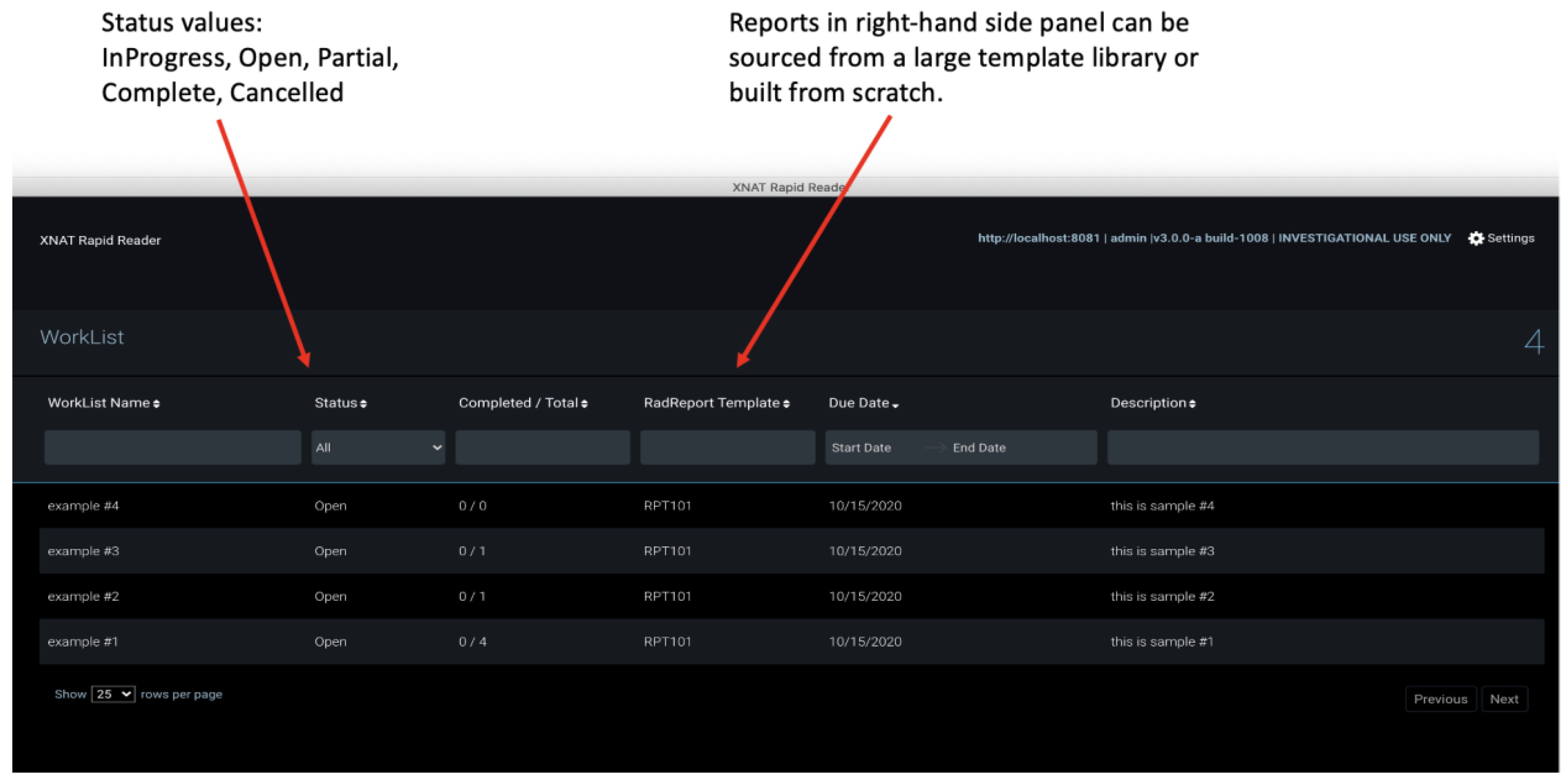
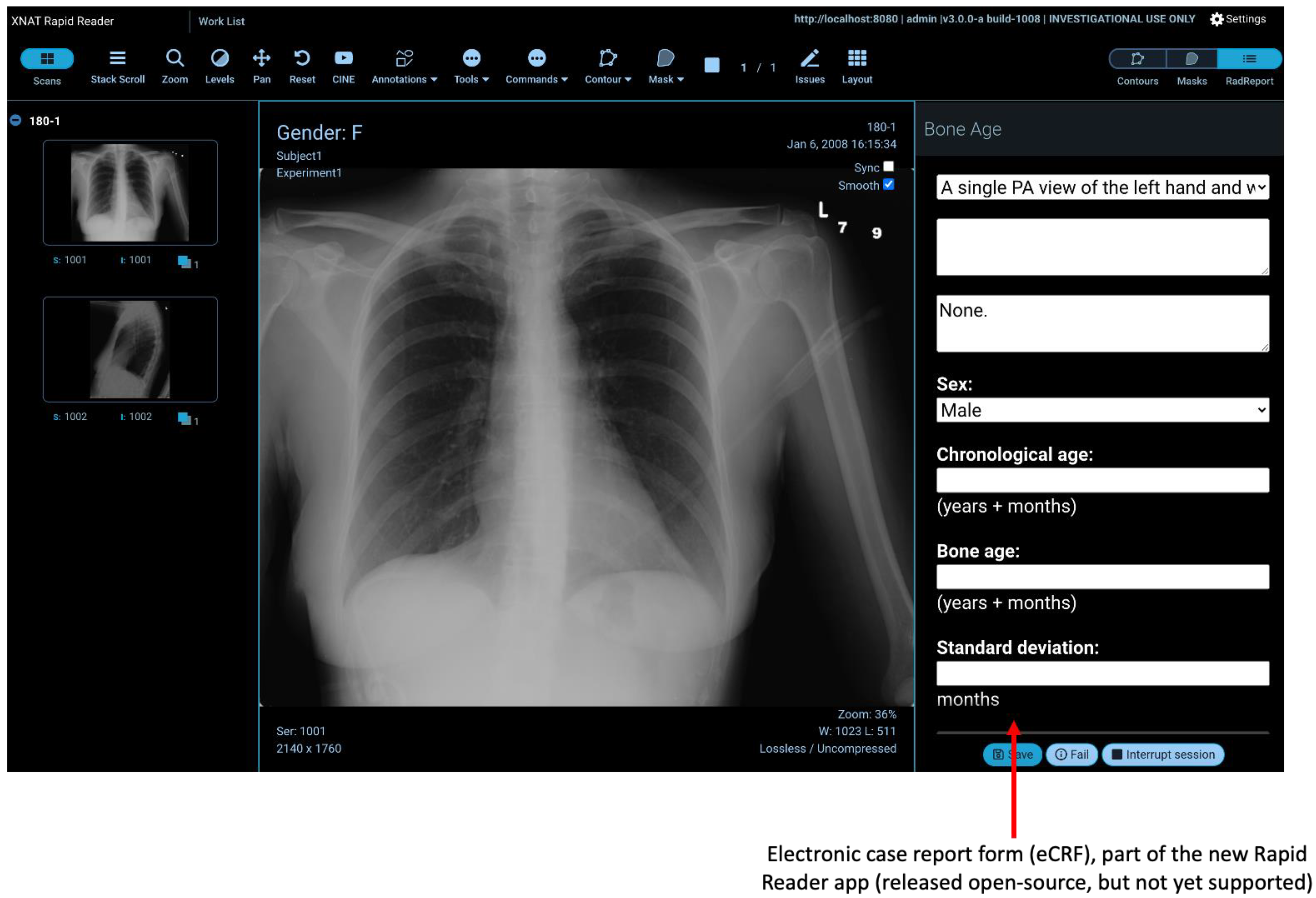
2.6. Rapid Reader
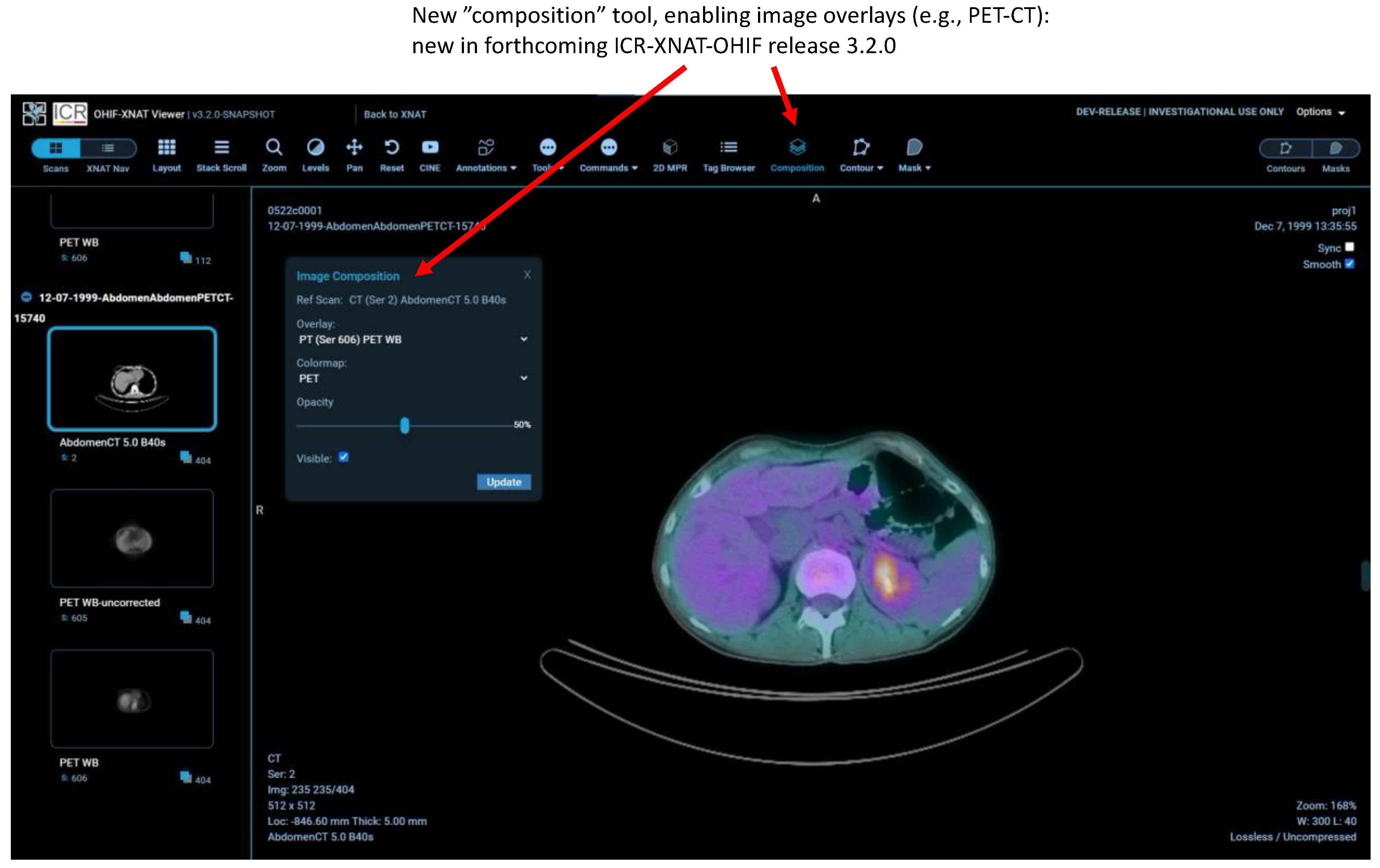
2.7. Other New Features
3. Results
4. Discussion
- There is no overhead of local software installation, the only prerequisite being a standard web browser. Although the user experience is better when running that browser on a high-spec desktop machine, the software performs well even on older computers with more modest capabilities. The viewer can be used very successfully on a tablet and, indeed, using a large tablet together with an appropriate stylus represents a potentially optimum combination for manual annotation of ROIs. Images can even be viewed on a mobile phone, subject to obvious limitations in screen real estate. The web-based approach is well aligned with the needs of “opening” multi-centre trials with standardised software and procedures in organisations with diverse hardware and varying levels of local technical support.
- Computationally intensive tasks (e.g., real-time ML model inference) can be handled server-side, reducing the need for high-performance web clients and bringing advanced techniques within the reach of all centres participating in a trial, thus “democratising” the use of AI.
- Researchers gain immediate access to all images granted by their XNAT permissions, without any need to “import” data into an application or hold them locally on a workstation. For large archives, the accessible data might comprise tens of terabytes, representing hundreds of thousands of subjects.
- By contrast, each time an image is reviewed, it needs to be retrieved from the server in real-time. Depending on data volume and internet speeds, this might be slow, leading to “data-buffering“ delays of tens of seconds prior to large 3D images being fully available for viewing. This is a major disadvantage of the zero-footprint design compared with a more traditional desktop application where data are fully resident on a local disk. It currently degrades the viewing experience for radiologists, leading to reluctant uptake in some quarters. The work targets a different use case to a PACS and, at present, OHIF would not be a suitable replacement clinically. However, we envisage future improvements via plausible mitigations to: (i) improve backend efficiency in querying the data (e.g., store each DICOM series as a single compressed file); (ii) employ smarter caching so that images are stored in the most easily displayable representations; (iii) use a “progressive” DICOM codec such as HTJ2K [37,38]; (iv) use server-side image rendering and transmit only the final, rendered image to the browser; (v) employ novel solutions such as blockchain-enabled distributed storage [39] to bring the data closer to users.
- Browser memory restrictions limit the complexity of data displayed (for example, the number of active 3D DICOM segmentation objects loaded in MPR view). This problem currently has no easy solution.
- enhanced support for radiotherapy objects (e.g., DICOM RT-DOSE);
- enhanced facilities for duplicating ROIs both between DICOM series within the same study and, via registration, between different imaging sessions;
- support for versioning of ROIs;
- enhanced annotation workflows;
- creation of hanging protocols;
- further development of the Rapid Reader;
- full support within XNAT for the OHIF microscopy extension.
- transition to core OHIF-v3.
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AIAA | Artificial Intelligence Assisted Annotation |
| AIM | Annotation and Imaging Markup |
| API | Application Programming Interface |
| DICOM | Digital Imaging and Communications in Medicine |
| IOD | Information Object Definition |
| MONAI | Medical Open Network for AI |
| NIfTI | Neuroimaging Informatics Technology Initiative |
| OHIF | Open Health Imaging Foundation |
| PACS | Picture Archiving and Communications Systems |
| P10 | DICOM Part 10 |
| REST | Representational State Transfer |
| ROI | Region-of-Interest |
| STL | Standard Triangle Language (or Standard Tesselation Language) |
| UI | User interface |
| VR | Value Representation |
| XNAT | eXtensible Neuroimaging Archive Toolkit (Note that XNAT was originally an acronym with this definition. However, recognition by the core XNAT team that its remit runs far beyond neuroimaging has led to the current practice of regarding XNAT as a name, not an acronym any longer requiring definition) |
References
- Ziegler, E.; Urban, T.; Brown, D.; Petts, J.; Pieper, S.D.; Lewis, R.; Hafey, C.; Harris, G.J. Open Health Imaging Foundation Viewer: An Extensible Open-Source Framework for Building Web-Based Imaging Applications to Support Cancer Research. JCO Clin. Cancer Inform. 2020, 4, 336–345. [Google Scholar] [CrossRef]
- The Open Health Imaging Foundation. Available online: www.ohif.org (accessed on 14 July 2021).
- Marcus, D.S.; Olsen, T.R.; Ramaratnam, M.; Buckner, R.L. The extensible neuroimaging archive toolkit. Neuroinformatics 2007, 5, 11–33. [Google Scholar] [CrossRef]
- Schulz, S.; Wilkinson, M.D.; Aalbersberg, I.J. Faculty Opinions recommendation of The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data 2018, 3, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Fielding, R.T.; Taylor, R.N. Principled design of the modern Web architecture. ACM Trans. Internet Technol. 2002, 2, 115–150. [Google Scholar] [CrossRef]
- Marcus, D.S.; Harms, M.; Snyder, A.Z.; Jenkinson, M.; Wilson, J.A.; Glasser, M.F.; Barch, D.M.; Archie, K.A.; Burgess, G.C.; Ramaratnam, M.; et al. Human Connectome Project informatics: Quality control, database services, and data visualization. NeuroImage 2013, 80, 202–219. [Google Scholar] [CrossRef] [Green Version]
- Xtk. Available online: www.github.com/xtk (accessed on 14 July 2021).
- Wadali, J.S.; Sood, S.P.; Kaushish, R.; Syed-Abdul, S.; Khosla, P.K.; Bhatia, M. Evaluation of Free, Open-source, Web-based DICOM Viewers for the Indian National Telemedicine Service (eSanjeevani). J. Digit. Imaging 2020, 33, 1499–1513. [Google Scholar] [CrossRef]
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G.; PRISMA Group. Preferred reporting items for systematic reviews and meta-analyses: The PRISMA statement. PLoS Med. 2009, 6, e1000097. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DWV (DICOM Web Viewer). Available online: https://github.com/ivmartel/dwv (accessed on 1 February 2022).
- Moreira, D.A.; Hage, C.; Luque, E.F.; Willrett, D.; Rubin, D.L. 3D markup of radiological images in ePAD, a web-based image annotation tool. In Proceedings of the 2015 IEEE 28th International Symposium on Computer-Based Medical Systems, Sao Carlos, Brazil, 22–25 June 2015. [Google Scholar]
- Open Source Clinical Image and Object Management. Available online: https://www.dcm4che.org/ (accessed on 1 February 2022).
- Annotation and Image Markup. Available online: https://github.com/NCIP/annotation-and-image-markup (accessed on 14 July 2021).
- Wild, D.; Weber, M.; Egger, J. Client/server based online environment for manual segmentation of medical images. arXiv 2019, arXiv:1904.08610. [Google Scholar]
- Lösel, P.D.; Van De Kamp, T.; Jayme, A.; Ershov, A.; Faragó, T.; Pichler, O.; Jerome, N.T.; Aadepu, N.; Bremer, S.; Chilingaryan, S.A.; et al. Introducing Biomedisa as an open-source online platform for biomedical image segmentation. Nat. Commun. 2020, 11, 5577. [Google Scholar] [CrossRef]
- Abid, A.; Abdalla, A.; Abid, A.; Khan, D.; Alfozan, A.; Zou, J. An online platform for interactive feedback in biomedical machine learning. Nat. Mach. Intell. 2020, 2, 86–88. [Google Scholar] [CrossRef]
- McAteer, M.; O’Connor, J.P.B.; Koh, D.M.; Leung, H.Y.; Doran, S.J.; Jauregui-Osoro, M.; Muirhead, N.; Brew-Graves, C.; Plummer, E.R.; Sala, E.; et al. Introduction to the National Cancer Imaging Translational Accelerator (NCITA): A UK-wide infrastructure for multicentre clinical translation of cancer imaging biomarkers. Br. J. Cancer 2021, 125, 1462–1465. [Google Scholar] [CrossRef] [PubMed]
- O’Connor, J.P.B.; Aboagye, E.; Adams, J.E.; Aerts, H.J.W.L.; Barrington, S.F.; Beer, A.J.; Boellaard, R.; Bohndiek, S.; Brady, M.; Brown, G.; et al. Imaging biomarker roadmap for cancer studies. Nat. Rev. Clin. Oncol. 2016, 14, 169–186. [Google Scholar] [CrossRef] [PubMed]
- van Griethuysen, J.J.M.; Fedorov, A.; Parmar, C.; Hosny, A.; Aucoin, N.; Narayan, V.; Beets-Tan, R.G.H.; Fillion-Robin, J.-C.; Pieper, S.; Aerts, H.J.W.L. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res. 2017, 77, e104–e107. [Google Scholar] [CrossRef] [Green Version]
- Meteor. Available online: https://meteor.com (accessed on 14 July 2021).
- React: A JavaScript Library for Building User Interfaces. Available online: https://reactjs.org (accessed on 14 July 2021).
- ICR Imaging Informatics Open Source Repository. Available online: https://bitbucket.org/icrimaginginformatics/ohif-viewer-xnat-plugin/downloads/ (accessed on 14 July 2021).
- Kahn, C.E., Jr.; Langlotz, C.P.; Channin, D.S.; Rubin, D.L. Informatics in radiology: An information model of the DICOM standard. Radiographics 2011, 31, 295–304. [Google Scholar] [CrossRef] [PubMed]
- DICOMweb Standard. Available online: https://www.dicomstandard.org/dicomweb (accessed on 14 July 2021).
- OHIF Extensions. Available online: https://docs.ohif.org/extensions/ (accessed on 14 July 2021).
- Modes: Overview. Available online: https://v3-docs.ohif.org/platform/modes/index/ (accessed on 6 December 2021).
- Jodogne, S. The Orthanc Ecosystem for Medical Imaging. J. Digit. Imaging 2018, 31, 341–352. [Google Scholar] [CrossRef] [Green Version]
- Segal, E.; Sirlin, C.; Ooi, C.; Adler, A.; Gollub, J.; Chen, X.; Chan, B.K.; Matcuk, G.; Barry, C.T.; Chang, H.Y.; et al. Decoding global gene expression programs in liver cancer by noninvasive imaging. Nat. Biotechnol. 2007, 25, 675–680. [Google Scholar] [CrossRef]
- Gillies, R.J.; Kinahan, P.E.; Hricak, H. Radiomics: Images Are More than Pictures, They Are Data. Radiology 2016, 278, 563–577. [Google Scholar] [CrossRef] [Green Version]
- DICOM SR for Communicating Planar Annotations: An Imaging Data Commons (IDC) White Paper. Available online: https://docs.google.com/document/d/1bR6m7foTCzofoZKeIRN5YreBrkjgMcBfNA7r9wXEGR4/edit#heading=h.vdjcb712p7rz (accessed on 28 April 2021).
- Cardobi, N.; Palù, A.D.; Pedrini, F.; Beleù, A.; Nocini, R.; De Robertis, R.; Ruzzenente, A.; Salvia, R.; Montemezzi, S.; D’Onofrio, M. An Overview of Artificial Intelligence Applications in Liver and Pancreatic Imaging. Cancers 2021, 13, 2162. [Google Scholar] [CrossRef]
- NVIDIA Clara: An Application Framework Optimized for Healthcare and Life Sciences Developers. Available online: https://developer.nvidia.com/clara (accessed on 14 July 2021).
- MONAI: Medical Open Network for AI. Available online: https://monai.io/ (accessed on 14 July 2021).
- Validating QIN Tools. Available online: https://imaging.cancer.gov/programs_resources/specialized_initiatives/qin/tools/default.htm (accessed on 7 December 2021).
- Fedorov, A.; Beichel, R.; Kalpathy-Cramer, J.; Finet, J.; Fillion-Robin, J.-C.; Pujol, S.; Bauer, C.; Jennings, D.; Fennessy, F.; Sonka, M.; et al. 3D Slicer as an image computing platform for the Quantitative Imaging Network. Magn. Reson. Imaging 2012, 30, 1323–1341. [Google Scholar] [CrossRef] [Green Version]
- Kagadis, G.C.; Kloukinas, C.; Moore, K.; Philbin, J.; Papadimitroulas, P.; Alexakos, C.; Nagy, P.G.; Visvikis, D.; Hendee, W.R. Cloud computing in medical imaging. Med. Phys. 2013, 40, 070901. [Google Scholar] [CrossRef] [Green Version]
- Taubman, D.; Naman, A.; Mathew, R.; Smith, M.; Watanabe, O. High Throughput JPEG 2000 (HTJ2K): Algorithm, Performance and Potential; White Paper to facilitate assessment and deployment of ITU-T Rec T.814|IS 15444-15; International Telecommunications Union (ITU): Geneva, Switzerland, 2019. [Google Scholar]
- Taubman, D.; Naman, A.; Mathew, R. High throughput block coding in the HTJ2K compression standard. In Proceedings of the 2019 IEEE International Conference on Image Processing (ICIP), Taipei, Taiwan, 22–25 September 2019. [Google Scholar]
- Jabarulla, M.Y.; Lee, H.-N. Blockchain-Based Distributed Patient-Centric Image Management System. Appl. Sci. 2020, 11, 196. [Google Scholar] [CrossRef]
- Li, X.; Morgan, P.S.; Ashburner, J.; Smith, J.; Rorden, C. The first step for neuroimaging data analysis: DICOM to NIfTI conversion. J. Neurosci. Methods 2016, 264, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Gorgolewski, K.J.; Auer, T.; Calhoun, V.D.; Craddock, R.C.; Das, S.; Duff, E.P.; Flandin, G.; Ghosh, S.S.; Glatard, T.; Halchenko, Y.O.; et al. The brain imaging data structure, a format for organizing and describing outputs of neuroimaging experiments. Sci. Data 2016, 3, 160044. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Doran, S.J.; Al Sa’d, M.; Petts, J.A.; Darcy, J.; Alpert, K.; Cho, W.; Escudero Sanchez, L.; Alle, S.; El Harouni, A.; Genereaux, B.; et al. Integrating the OHIF Viewer into XNAT: Achievements, Challenges and Prospects for Quantitative Imaging Studies. Tomography 2022, 8, 497-512. https://doi.org/10.3390/tomography8010040
Doran SJ, Al Sa’d M, Petts JA, Darcy J, Alpert K, Cho W, Escudero Sanchez L, Alle S, El Harouni A, Genereaux B, et al. Integrating the OHIF Viewer into XNAT: Achievements, Challenges and Prospects for Quantitative Imaging Studies. Tomography. 2022; 8(1):497-512. https://doi.org/10.3390/tomography8010040
Chicago/Turabian StyleDoran, Simon J., Mohammad Al Sa’d, James A. Petts, James Darcy, Kate Alpert, Woonchan Cho, Lorena Escudero Sanchez, Sachidanand Alle, Ahmed El Harouni, Brad Genereaux, and et al. 2022. "Integrating the OHIF Viewer into XNAT: Achievements, Challenges and Prospects for Quantitative Imaging Studies" Tomography 8, no. 1: 497-512. https://doi.org/10.3390/tomography8010040
APA StyleDoran, S. J., Al Sa’d, M., Petts, J. A., Darcy, J., Alpert, K., Cho, W., Escudero Sanchez, L., Alle, S., El Harouni, A., Genereaux, B., Ziegler, E., Harris, G. J., Aboagye, E. O., Sala, E., Koh, D.-M., & Marcus, D. (2022). Integrating the OHIF Viewer into XNAT: Achievements, Challenges and Prospects for Quantitative Imaging Studies. Tomography, 8(1), 497-512. https://doi.org/10.3390/tomography8010040





