Evaluation of a Deep Learning Algorithm for Automated Spleen Segmentation in Patients with Conditions Directly or Indirectly Affecting the Spleen
Abstract
:1. Introduction
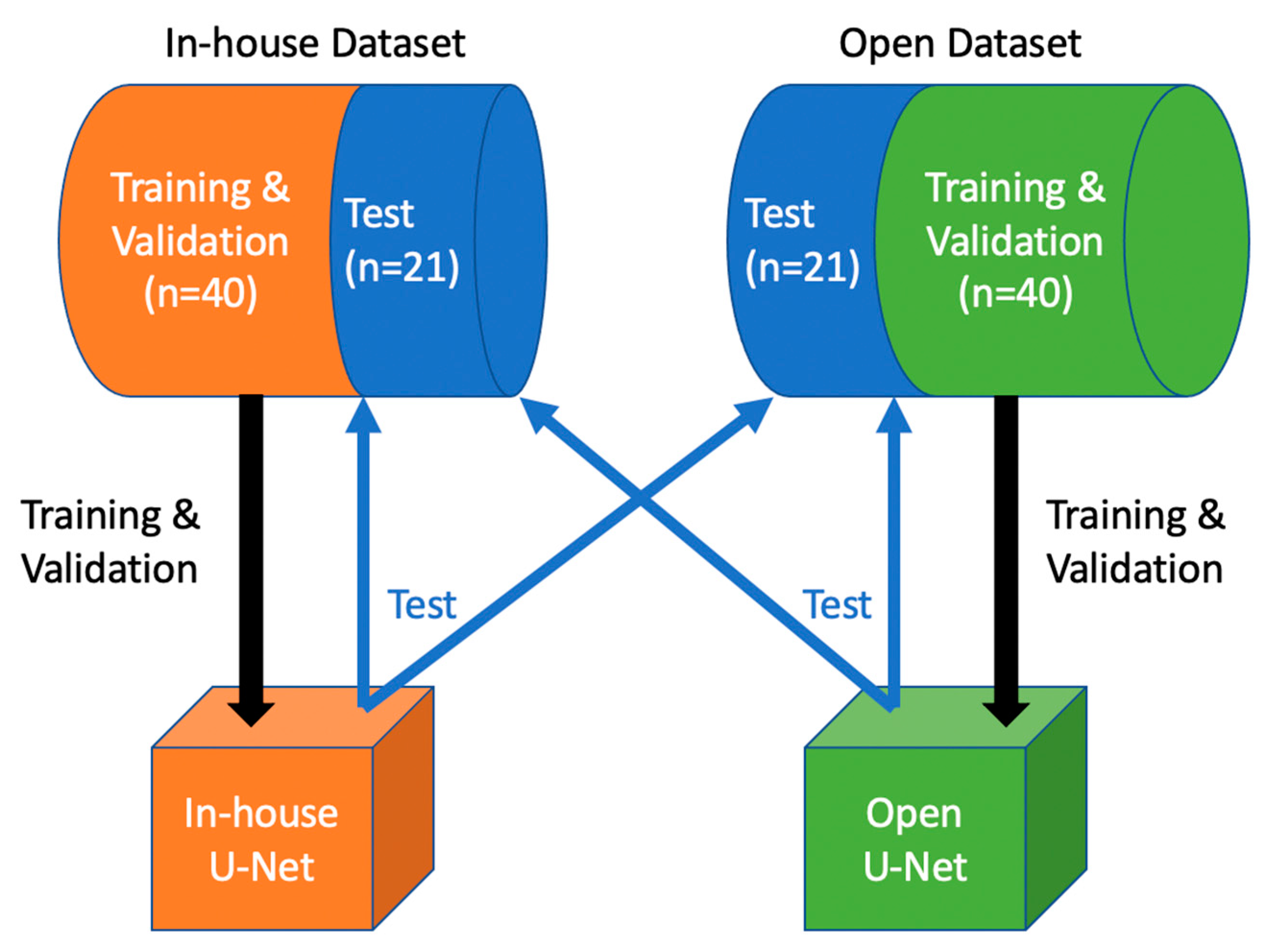
2. Materials and Methods
2.1. In-House Dataset
2.2. Medical Segmentation Decathlon Dataset (Open Dataset)
2.3. Image Preprocessing and Model Architecture
2.4. Network Training
2.5. Image Postprocessing
2.6. Statistical Analysis and Evaluation
3. Results
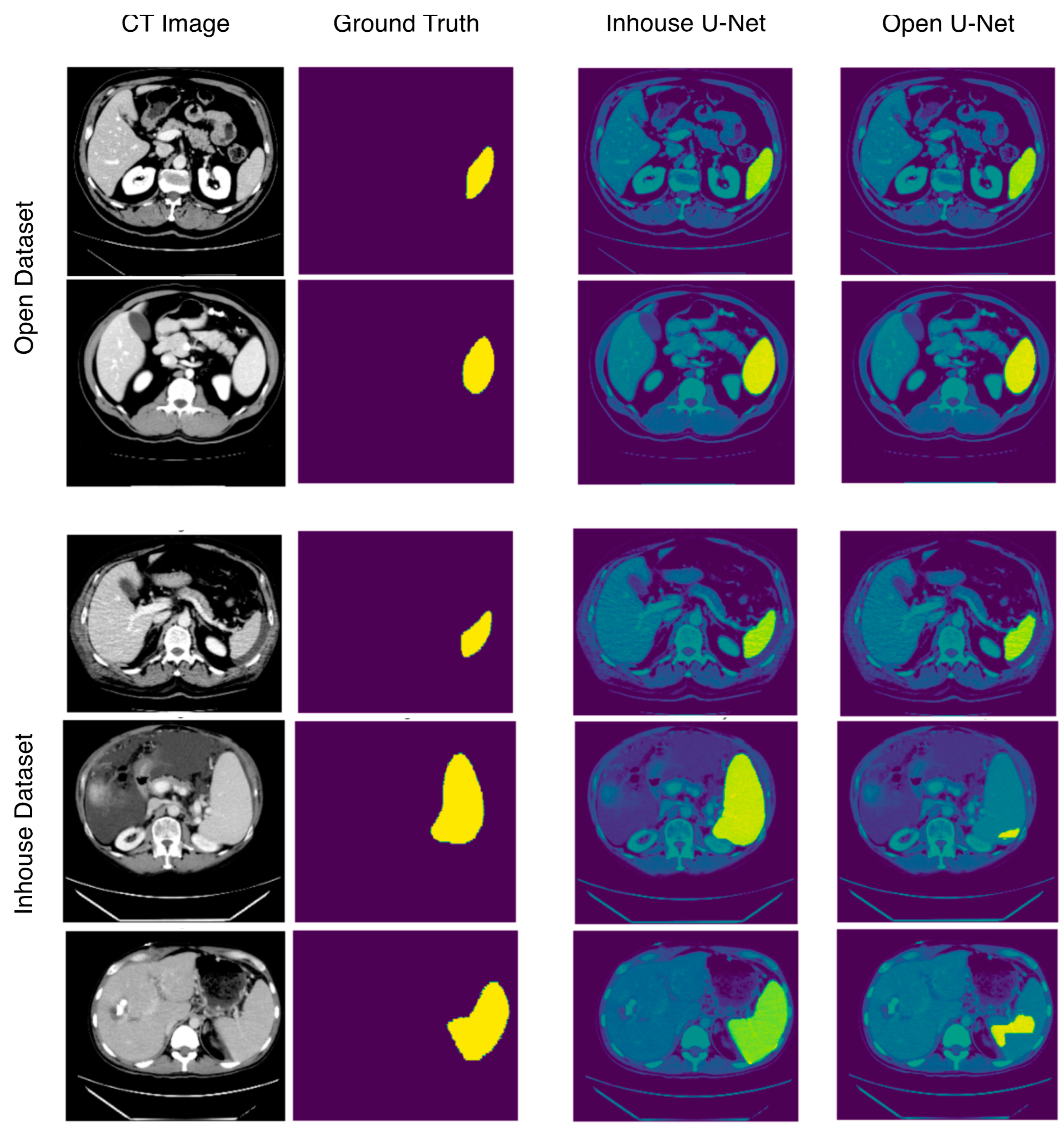
3.1. Segmentation Performance in the Open Test Dataset
3.2. Segmentation Performance in the In-House Test Dataset
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bronte, V.; Pittet, M.J. The spleen in local and systemic regulation of immunity. Immunity 2013, 39, 806–818. [Google Scholar] [CrossRef] [Green Version]
- Saboo, S.S.; Krajewski, K.M.; O’Regan, K.N.; Giardino, A.; Brown, J.R.; Ramaiya, N.; Jagannathan, J.P. Spleen in haematological malignancies: Spectrum of imaging findings. Br. J. Radiol. 2012, 85, 81–92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Stasi, M.; Cavanna, L.; Fornari, F.; Vallisa, D.; Buscarini, E.; Civardi, G.; Rossi, S.; Bertè, R.; Tansini, P.; Buscarini, L. Splenic lesions in Hodgkin’s and non-Hodgkin’s lymphomas. An ultrasonographic study. Eur. J. Ultrasound 1995, 2, 117–124. [Google Scholar] [CrossRef]
- Curovic Rotbain, E.; Lund Hansen, D.; Schaffalitzky de Muckadell, O.; Wibrand, F.; Meldgaard Lund, A.; Frederikse, H. Splenomegaly—Diagnostic validity, work-up, and underlying causes. PLoS ONE 2017, 12, e0186674. [Google Scholar] [CrossRef] [Green Version]
- Siliézar, M.M.; Muñoz, C.C.; Solano-Iturri, J.D.; Ortega-Comunian, L.; Mollejo, M.; Montes-Moreno, S.; Piris, M.A. Spontaneously ruptured spleen samples in patients with infectious mononucleosisanalysis of histology and lymphoid subpopulations. Am. J. Clin. Pathol. 2018, 150, 310–317. [Google Scholar] [CrossRef] [PubMed]
- Karalilova, R.; Doykova, K.; Batalov, Z.; Doykov, D.; Batalov, A. Spleen elastography in patients with Systemic sclerosis. Rheumatol. Int. 2021, 41, 633–641. [Google Scholar] [CrossRef]
- Yang, Y.; Tang, Y.; Gao, R.; Bao, S.; Huo, Y.; McKenna, M.T.; Savona, M.R.; Abramson, R.G.; Landman, B.A. Validation and estimation of spleen volume via computer-assisted segmentation on clinically acquired CT scans. J. Med. Imaging 2021, 8, 014004. [Google Scholar] [CrossRef] [PubMed]
- Breiman, R.S.; Beck, J.W.; Korobkin, M.; Glenny, R.; Akwari, O.E.; Heaston, D.K.; Moore, A.V.; Ram, P.C. Volume determinations using computed tomography. Am. J. Roentgenol. 1982, 138, 329–333. [Google Scholar] [CrossRef] [Green Version]
- Harris, A.; Kamishima, T.; Hao, H.Y.; Kato, F.; Omatsu, T.; Onodera, Y.; Terae, S.; Shirato, H. Splenic volume measurements on computed tomography utilizing automatically contouring software and its relationship with age, gender, and anthropometric parameters. Eur. J. Radiol. 2010, 75, e97–e101. [Google Scholar] [CrossRef]
- Linguraru, M.G.; Sandberg, J.K.; Li, Z.; Shah, F.; Summers, R.M. Automated segmentation and quantification of liver and spleen from CT images using normalized probabilistic atlases and enhancement estimation. Med. Phys. 2010, 37, 771–783. [Google Scholar] [CrossRef] [Green Version]
- Sykes, J. Reflections on the current status of commercial automated segmentation systems in clinical practice. J. Med. Radiat. Sci. 2014, 61, 131–134. [Google Scholar] [CrossRef] [PubMed]
- Hosny, A.; Parmar, C.; Quackenbush, J.; Schwartz, L.H.; Aerts, H.J.W.L. Artificial intelligence in radiology. Nat. Rev. Cancer 2018, 18, 500–510. [Google Scholar] [CrossRef]
- Nowak, S.; Faron, A.; Luetkens, J.A.; Geißler, H.L.; Praktiknjo, M.; Block, W.; Thomas, D.; Sprinkart, A.M. Fully automated segmentation of connective tissue compartments for CT-based body composition analysis: A deep learning approach. Investig. Radiol. 2020, 55, 357–366. [Google Scholar] [CrossRef] [PubMed]
- Uthoff, J.; Stephens, M.J.; Newell, J.D.; Hoffman, E.A.; Larson, J.; Koehn, N.; De Stefano, F.A.; Lusk, C.M.; Wenzlaff, A.S.; Watza, D.; et al. Machine learning approach for distinguishing malignant and benign lung nodules utilizing standardized perinodular parenchymal features from CT. Med. Phys. 2019, 46, 3207–3216. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Liu, X.; Li, C.; Xu, Z.; Ruan, J.; Zhu, H.; Meng, T.; Li, K.; Huang, N.; Zhang, S. A noise-robust framework for automatic segmentation of COVID-19 pneumonia lesions from CT images. IEEE Trans. Med. Imaging 2020, 39, 2653–2663. [Google Scholar] [CrossRef] [PubMed]
- Shah, P.; Bakrola, V.; Pati, S. Optimal Approach for Image Recognition Using Deep Convolutional Architecture; Springer: Singapore, 2019. [Google Scholar]
- Falk, T.; Mai, D.; Bensch, R.; Çiçek, Ö.; Abdulkadir, A.; Marrakchi, Y.; Böhm, A.; Deubner, J.; Jäckel, Z.; Seiwald, K.; et al. U-Net: Deep learning for cell counting, detection, and morphometry. Nat. Methods 2019, 16, 67–70. [Google Scholar] [CrossRef]
- Roth, H.R.; Oda, H.; Zhou, X.; Shimizu, N.; Yang, Y.; Hayashi, Y.; Oda, M.; Fujiwara, M.; Misawa, K.; Mori, K. An application of cascaded 3D fully convolutional networks for medical image segmentation. Comput. Med. Imaging Graph. 2018, 66, 90–99. [Google Scholar] [CrossRef] [Green Version]
- Gibson, E.; Giganti, F.; Hu, Y.; Bonmati, E.; Bandula, S.; Gurusamy, K.; Davidson, B.; Pereira, S.P.; Clarkson, M.J.; Barratt, D.C. Automatic multi-organ segmentation on abdominal CT with dense V-networks. IEEE Trans. Med. Imaging 2018, 37, 1822–1834. [Google Scholar] [CrossRef] [Green Version]
- Su, T.-Y.; Fang, Y.-H. Future Trends in Biomedical and Health Informatics and Cybersecurity in Medical Devices. In Proceedings of the International Conference on Biomedical and Health Informatics, ICBHI 2019, Taipei, Taiwan, 17–20 April 2019; pp. 33–41. [Google Scholar] [CrossRef]
- Moon, H.; Huo, Y.; Abramson, R.G.; Peters, R.A.; Assad, A.; Moyo, T.K.; Savona, M.R.; Landman, B.A. Acceleration of spleen segmentation with end-to-end deep learning method and automated pipeline. Comput. Biol. Med. 2019, 107, 109–117. [Google Scholar] [CrossRef]
- The MONAI Consortium. Project MONAI 2021. MONAI Core v0.5.0. Available online: https://zenodo.org/record/5728262#.Ya9ZA7oo9PY (accessed on 8 December 2021).
- Simpson, A.L.; Antonelli, M.; Bakas, S.; Bilello, M.; Farahani, K.; van Ginneken, B.; Kopp-Schneider, A.; Landman, B.A.; Litjens, G.; Menze, B.; et al. A large annotated medical image dataset for the development and evaluation of segmentation algorithms. arXiv 2019, arXiv:1902.0906. [Google Scholar]
- Fedorov, A.; Beichel, R.; Kalpathy-Cramer, J.; Finet, J.; Fillion-Robin, J.-C.; Pujol, S.; Bauer, C.; Jennings, D.; Fennessy, F.; Sonka, M.; et al. 3D slicer as an image computing platform for the quantitative imaging network. Magn. Reson. Imaging 2012, 30, 1323–1341. [Google Scholar] [CrossRef] [Green Version]
- Koo, T.K.; Li, M.Y. A guideline of selecting and reporting intraclass correlation coefficients for reliability research. J. Chiropr. Med. 2016, 15, 155–163. [Google Scholar] [CrossRef] [Green Version]
- Paszke, A.; Gross, S.; Massa, F.; Lerer, A.; Bradbury, J.; Chanan, G.; Killeen, T.; Lin, Z.; Gimelshein, N.; Antiga, L.; et al. PyTorch: An Imperative Style, High-Performance Deep Learning Library. In Proceedings of the 33rd Conference on Neural Information Processing Systems (NeurIPS 2019), Vancouver, BC, Canada, 3 December 2019. [Google Scholar]
- Harris, C.R.; Millman, K.J.; van der Walt, S.J.; Gommers, R.; Virtanen, P.; Cournapeau, D.; Wieser, E.; Taylor, J.; Berg, S. Array Programming with NumPy. Nature 2020, 585, 357–362. [Google Scholar] [CrossRef]
- Hunter, J.D. Matplotlib: A 2D Graphics Environment. Comput. Sci. Eng. 2007, 9, 90–95. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-Learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Seabold, S.; Perktold, J. Statsmodels: Econometric and Statistical Modeling with Python. In Proceedings of the 9th Python in Science Conference, Austin, TX, USA, 28 June 2010; pp. 92–96. [Google Scholar] [CrossRef] [Green Version]
- Krasoń, A.; Woloshuk, A.; Spinczyk, D. Segmentation of abdominal organs in computed tomography using a generalized statistical shape model. Comput. Med. Imaging Graph. 2019, 78, 101672. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Heath, D.G.; Fishman, E.K. Abdominal image segmentation using three-dimensional deformable models. Investig. Radiol. 1998, 33, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Bobo, M.F.; Bao, S.; Huo, Y.; Yao, Y.; Virostko, J.; Plassard, A.J.; Lyu, I.; Assad, A.; Abramson, R.G.; Hilmes, M.A.; et al. Fully Convolutional Neural Networks Improve Abdominal Organ Segmentation. In Proceedings of the SPIE Medical Imaging 2018: Image Processing, Houston, TX, USA, 10–15 February 2018; Volume 10574, p. 105742V. [Google Scholar] [CrossRef]
- Humpire-Mamani, G.E.; Bukala, J.; Scholten, E.T.; Prokop, M.; van Ginneken, B.; Jacobs, C. Fully automatic volume measurement of the spleen at ct using deep learning. Radiol. Artif. Intell. 2020, 2, e190102. [Google Scholar] [CrossRef]
- Ahn, Y.; Yoon, J.S.; Lee, S.S.; Suk, H.-I.; Son, J.H.; Sung, Y.S.; Lee, Y.; Kang, B.-K.; Kim, H.S. Deep learning algorithm for automated segmentation and volume measurement of the liver and spleen using portal venous phase computed tomography images. Korean J. Radiol. 2020, 21, 987–997. [Google Scholar] [CrossRef]
- Gauriau, R.; Ardon, R.; Lesage, D.; Bloch, I. Multiple Template Deformation Application to Abdominal Organ Segmentation. In Proceedings of the IEEE 12th International Symposium on Biomedical Imaging (ISBI), New York, NY, USA, 16–19 April 2015; pp. 359–362. [Google Scholar] [CrossRef]
- Wood, A.; Soroushmehr, S.M.R.; Farzaneh, N.; Fessell, D.; Ward, K.R.; Gryak, J.; Kahrobaei, D.; Na, K. Fully Automated Spleen Localization and Segmentation Using Machine Learning and 3D Active Contours. In Proceedings of the 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Honolulu, HI, USA, 17–21 July 2018; pp. 53–56. [Google Scholar] [CrossRef]
- Gloger, O.; Tönnies, K.; Bülow, R.; Völzke, H. Automatized spleen segmentation in non-contrast-enhanced MR volume data using subject-specific shape priors. Phys. Med. Biol. 2017, 62, 5861–5883. [Google Scholar] [CrossRef] [PubMed]
- Son, J.H.; Lee, S.S.; Lee, Y.; Kang, B.-K.; Sung, Y.S.; Jo, S.; Yu, E. assessment of liver fibrosis severity using computed tomography–based liver and spleen volumetric indices in patients with chronic liver disease. Eur. Radiol. 2020, 30, 3486–3496. [Google Scholar] [CrossRef] [PubMed]
- Iranmanesh, P.; Vazquez, O.; Terraz, S.; Majno, P.; Spahr, L.; Poncet, A.; Morel, P.; Mentha, G.; Toso, C. Accurate computed tomography-based portal pressure assessment in patients with hepatocellular carcinoma. J. Hepatol. 2014, 60, 969–974. [Google Scholar] [CrossRef] [PubMed]
- Karimi, D.; Warfield, S.K.; Gholipour, A. transfer learning in medical image segmentation: New insights from analysis of the dynamics of model parameters and learned representations. Artif. Intell. Med. 2021, 116, 102078. [Google Scholar] [CrossRef] [PubMed]
- Gong, M.; Chen, S.; Chen, Q.; Zeng, Y.; Zhang, Y. Generative adversarial networks in medical image processing. Curr. Pharm. Des. 2021, 27, 1856–1868. [Google Scholar] [CrossRef] [PubMed]

| Training and Validation Dataset | Testing Dataset | |
|---|---|---|
| Number of Patients | 41 | 20 |
| Female | 23 (56%) | 5 (25%) |
| Age * | 62.6 ± 16.2 | 60.4 ± 15.1 |
| Splenomegaly | 21 (51.2%) | 7 (35%) |
| Liver cirrhosis | 11 (26.8%) | 3 (15%) |
| Lymphoma | 6 (15.6%) | 2 (10%) |
| No pathology | 4 (9.8%) | 1 (5%) |
| Other ** | 21 (51.2%) | 14 (70%) |
| Model | Dataset | DSC | RAVD * (%) | ASSD (mm) | Hausdorff (mm) | |
|---|---|---|---|---|---|---|
| In-house U-Net | In-house testing dataset | Mean ±SD | 0.941 ±0.021 | 4.203 | 0.772 ±0.274 | 7.137 ±5.440 |
| 95% CI | 0.932–0.951 | 2.313–6.094 | 0.644–0.900 | 4.591–9.683 | ||
| Open testing dataset | Mean ±SD | 0.906 ±0.071 | 9.690 | 0.999 ±0.657 | 8.787 ±6.889 | |
| 95% CI | 0.873–0.939 | 3.877–15.504 | 0.692–1.307 | 5.563–12.011 | ||
| Open U-Net | In-house testing dataset | Mean ±SD | 0.648 ±0.289 | 42.255 | 5.158 ±5.881 | 30.085 ±30.885 |
| 95% CI | 0.513–0.784 | 26.503–58.008 | 2.406–7.911 | 15.630–44.539 | ||
| Open testing dataset | Mean ±SD | 0.897 ±0.082 | 11.488 | 0.982 ±0.618 | 7.569 ±5.242 | |
| 95% CI | 0.859–0.935 | 5.323–17.653 | 0.693–1.272 | 5.115–10.023 |
| Method | DSC | Modality | Abnormalities |
|---|---|---|---|
| Gauriau et al. [36] | 0.870 ± 0.150 | Abd. CT | No |
| Wood et al. [37] | 0.873 | Abd. CT | Yes |
| Gloger et al. [38] | 0.906 ± 0.037 | MRI | No |
| In-house U-Net | 0.941 ± 0.021 | Abd. CT | Yes |
| Gibson et al. [19] | 0.950 | Abd. CT | No |
| Linguraru et al. [10] | 0.952 ± 0.014 | Abd. CT | Yes |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Meddeb, A.; Kossen, T.; Bressem, K.K.; Hamm, B.; Nagel, S.N. Evaluation of a Deep Learning Algorithm for Automated Spleen Segmentation in Patients with Conditions Directly or Indirectly Affecting the Spleen. Tomography 2021, 7, 950-960. https://doi.org/10.3390/tomography7040078
Meddeb A, Kossen T, Bressem KK, Hamm B, Nagel SN. Evaluation of a Deep Learning Algorithm for Automated Spleen Segmentation in Patients with Conditions Directly or Indirectly Affecting the Spleen. Tomography. 2021; 7(4):950-960. https://doi.org/10.3390/tomography7040078
Chicago/Turabian StyleMeddeb, Aymen, Tabea Kossen, Keno K. Bressem, Bernd Hamm, and Sebastian N. Nagel. 2021. "Evaluation of a Deep Learning Algorithm for Automated Spleen Segmentation in Patients with Conditions Directly or Indirectly Affecting the Spleen" Tomography 7, no. 4: 950-960. https://doi.org/10.3390/tomography7040078
APA StyleMeddeb, A., Kossen, T., Bressem, K. K., Hamm, B., & Nagel, S. N. (2021). Evaluation of a Deep Learning Algorithm for Automated Spleen Segmentation in Patients with Conditions Directly or Indirectly Affecting the Spleen. Tomography, 7(4), 950-960. https://doi.org/10.3390/tomography7040078






