1. Introduction
The magnetization prepared rapid gradient echo (MPRAGE) sequence has become the standard for structural
T1-weighted (
T1-w) 3D imaging. The
T1 contrast is obtained by an inversion pulse followed by a rapid gradient echo (RAGE) readout with optional delays for free recovery before and after the readout [
1]. At ultra-high field (UHF) MRI (7 T or above), the signal-to-noise ratio (SNR) is improved through the increased polarization of nuclear spins, which can be translated into either increased spatial resolution or faster scan times. The former alternative allows for visualization of substructures, unfeasible at lower field strengths [
2]. A major challenge of UHF is the increased inhomogeneity of the radiofrequency (RF)
B1 field, which varies based on subject, positioning, and coil [
3]. Because of the unpredictability of the
B1 field, pixel values from images acquired at different scanning sessions are generally not reproducible. This means that an important requirement for longitudinal or multi-site studies is not fulfilled. The increased inhomogeneity applies to both the transmit (
B1+) and the receive sensitivity components, which combine to form a total bias field, that diminishes the uniformity of the image. To correct for this intensity bias, it was suggested to acquire a reference gradient-recalled echo (GRE) in conjunction with the MPRAGE sequence [
4]. Through a simple division of the two acquisitions, a normalized MPRAGE image is obtained where signal variations due to the shared receive coils are eliminated, thus improving image quality as well as reproducibility.
B1+ affects the MPRAGE and the GRE pulse sequences differently through the local flip angle and, therefore, the related bias is reduced but not removed in the normalized MPRAGE. The division also removes the influence of proton density (PD) and
T2*, thereby creating a purely
T1-w image with improved tissue contrast. As the pixel values depend on the sequence, the technique is considered to be “semi-quantitative”. The reference GRE can be acquired either separately or interleaved with the MPRAGE at a longer inversion time (TI), and the latter approach has been popularized as the MP2RAGE (magnetization prepared two rapid acquisition gradient echoes) sequence [
5].
In this work, we describe an implementation where MPRAGE and reference are acquired in a non-interleaved, sequential fashion to produce the normalized MPRAGE. This approach is an accessible alternative to the interleaved MP2RAGE which, initially, was not readily available on our 7T MR system. Although the non-interleaved variant is expected to be more susceptible to inter-scan subject motion, the reference GRE can be accelerated through enlarged acquisition voxels, allowing for shorter scan time. The optimization procedure focused on the reference GRE, specifically on the flip angle and voxel size, in an effort to improve contrast, SNR, residual
B1+ bias, and scan time. The protocol was implemented at three different spatial resolutions, and normalized MPRAGE images acquired at a higher versus a lower spatial resolution were compared. The intra-subject and inter-subject reproducibility (crucial for a semi-quantitative protocol) were also investigated. Finally, the feasibility of calculating
T1 maps from a look-up table (LUT) of the normalized signal, obtained by forward modelling, was explored. The latter approach extends beyond the semi-quantitative domain and results in fully quantitative maps which would allow for a more direct biophysical interpretation in terms of, for instance, myelination [
6]. It is analogous to the approach described for the interleaved MP2RAGE by Marques et al. [
5]. The work presented here documents an optimized protocol for bias field-corrected structural imaging at 7 T, easily implemented and interpretable by radiologists.
2. Materials and Methods
2.1. Theory
The influence of receive sensitivity, PD, and
T2* were removed through division of the MPRAGE signal,
SMP, by the reference GRE signal,
SGRE, to yield the normalized MPRAGE signal,
SMP/GRE. This rationale is illustrated by paraphrasing Equation (3) in [
4]:
where
denotes PD,
is the longitudinal magnetization per unit PD,
is a factor accounting for the spatial dependence of receive sensitivity (here, a weighted linear combination of individual channels). The arguments of the sine functions are the local flip angles, i.e.,
, where
takes account of the transmit field (
B1+) inhomogeneity. Finally,
is the effective transverse relaxation rate. Note that
is acquired under transient conditions towards a driven equilibrium,
, with an increased rate
[
7]:
where
is the magnetization at thermal equilibrium
is
and
. If full relaxation within one cycle is not obtained,
will attain a dynamic steady-state between cycles, usually occurring after a few cycles [
8]. The degree of agreement between
and
when the central k-space line is acquired is a function of TI and
as well as of the local flip angle and thus
. Note that, without an inversion pulse,
is always acquired under steady-state conditions so that
Thus, transmit field related bias may not be removed completely for as Equation (1) might imply.
2.2. Equipment
The protocols were implemented on an actively shielded 7T MR system (Achieva, Philips Healthcare, Best, The Netherlands), using a head coil with two transmitter channels and 32 receive channels (Nova Medical, Wilmington, MA, USA). Healthy subjects were scanned after giving informed written consent and the study was approved by the regional Ethical Review Board. Dielectric pads were used in the experiments [
9].
2.3. MPRAGE Acquisition
The MPRAGE protocol was built upon the standard protocol for structural MRI available at the research site. Isotropic voxel sizes of either 0.7
3, 0.8
3, or 0.9
3 mm
3 were acquired with a slab-selective excitation, a readout flip angle of α
MP = 8°, TR = 8 ms, fat-water in-phase TE = 1.97 ms and a bandwidth/px of 503 Hz/px. For inversion, an adiabatic pulse with duration 22 ms and a maximal
B1 amplitude of 15 μT was used. The delay from inversion to the central k-space readout (linear phase encoding) was TI = 1200 ms and the time between inversions was T
cycle = 3500 ms. These timings (i) allowed for a period of free relaxation after the readout train (increasing dynamic range) and (ii) ensured that the
Mz of cerebrospinal fluid (CSF) was close to the zero-crossing during acquisition of the center of k-space. Both (i) and (ii) will improve
T1 contrast in magnitude images. After each inversion, a single 2D plane of k-space was acquired so that the turbo factor (TF) was identical to the acquisition matrix size in the inner loop phase-encoded direction,
Ny. Parallel imaging in the form of sensitivity encoding (SENSE) was applied in the outer loop (right–left direction) with a reduction factor of 2.5 [
10]. The inner loop corresponded to the anterior-posterior (AP) direction. Switching between the different resolutions will alter TF and thus affect the
T1 contrast (see Results 4.3). For (0.7 mm)
3 resolution, a small inner loop SENSE reduction factor of 1.11 had to be applied to fit the readout train within TI = 1200 ms (TF =
Ny/1.11). Note that the FOV in the outer loop (FOV
FH,AP,RL = 230 × 230 × 180 mm
3) can be enlarged without affecting contrast. SENSE-related wrap-around artifacts in the AP direction were avoided by an oversampling margin (default setting) which did not affect the acquisition time (
Tacq). Lastly, to explore the possibility of further reducing
Tacq, the protocol was implemented with elliptical k-space sampling in the phase encoding directions. A prerequisite for this kind of readout is a multi-shot acquisition combined with a zigzag k-space trajectory involving both phase encoding directions during the RAGE readout. This switching between the inner and outer phase encoding loops is performed in such a way that overall contrast is unaffected for constant TF. The acquisition matrix (
Nx,y,z), TF and
Tacq for all spatial resolutions with/without elliptical k-space phase encoding are listed in
Table 1.
2.4. Reference GRE Acquisition
The steady-state GRE sequence was acquired with TR, TE, and outer loop SENSE factor identical to the MPRAGE sequence, but with 50% zero-filling in the outer loop to reduce scan time. In other words, voxel dimensions in the right–left direction were 1.4, 1.6, or 1.8 mm for the (0.7)3, (0.8)3, and (0.9)3 mm3 protocols, respectively. When determining the in-plane voxel size (see Results 3.2), the bandwidth/pixel was changed accordingly so that the absolute fat signal displacement was constant between GRE and MPRAGE. Receiver gain and flip angle were calibrated for MPRAGE and then kept constant during the GRE. After acquisition, the volume was reconstructed to the same matrix size as the MPRAGE through zero-filling.
2.5. Data Post-processing
Images in the Digital Imaging and Communications in Medicine (DICOM) file format were exported, pseudo-anonymized, and converted to the Neuroimaging Informatics Technology Initiative (NIfTI) file format using an in-house modification of the dcm2niix tool [
11]. The platform-dependent scaling of signal intensities was reverted from stored values to 32-bit floating point/1000 (to obtain pixel values in the range of 0–1000) [
12]. Spatial dimensions were re-ordered to transverse orientation according to radiological convention (right–left). Rigid co-registration of the reference GRE to the MPRAGE volume was performed using the FMRIB Linear Image Registration Tool (FLIRT) [
13,
14], where after the normalization was performed as in Equation (1). A mask of the brain was obtained by applying the Brain Extraction Tool (BET) to the PD-w reference GRE [
15]. Segmentation of the three major tissue classes, white matter (WM), gray matter (GM) and CSF, was performed using the FMRIB Automated Segmentation Tool (FAST) [
16]. Improvement in spatial homogeneity, after signal normalization, for different parameter settings of the reference GRE was analyzed using the coefficient-of-variation (
CV) over the tissue classes in the normalized MPRAGE. The
CV within a small WM ROI was used to evaluate relative changes in SNR; the rationale for this approach was that the
CV in a spatially restricted ROI of homogenous tissue should be unaffected by
B1+ and dominated by SNR. Average contrast between WM and GM was defined as
where
and
is the average pixel value of the respective segmented tissue type.
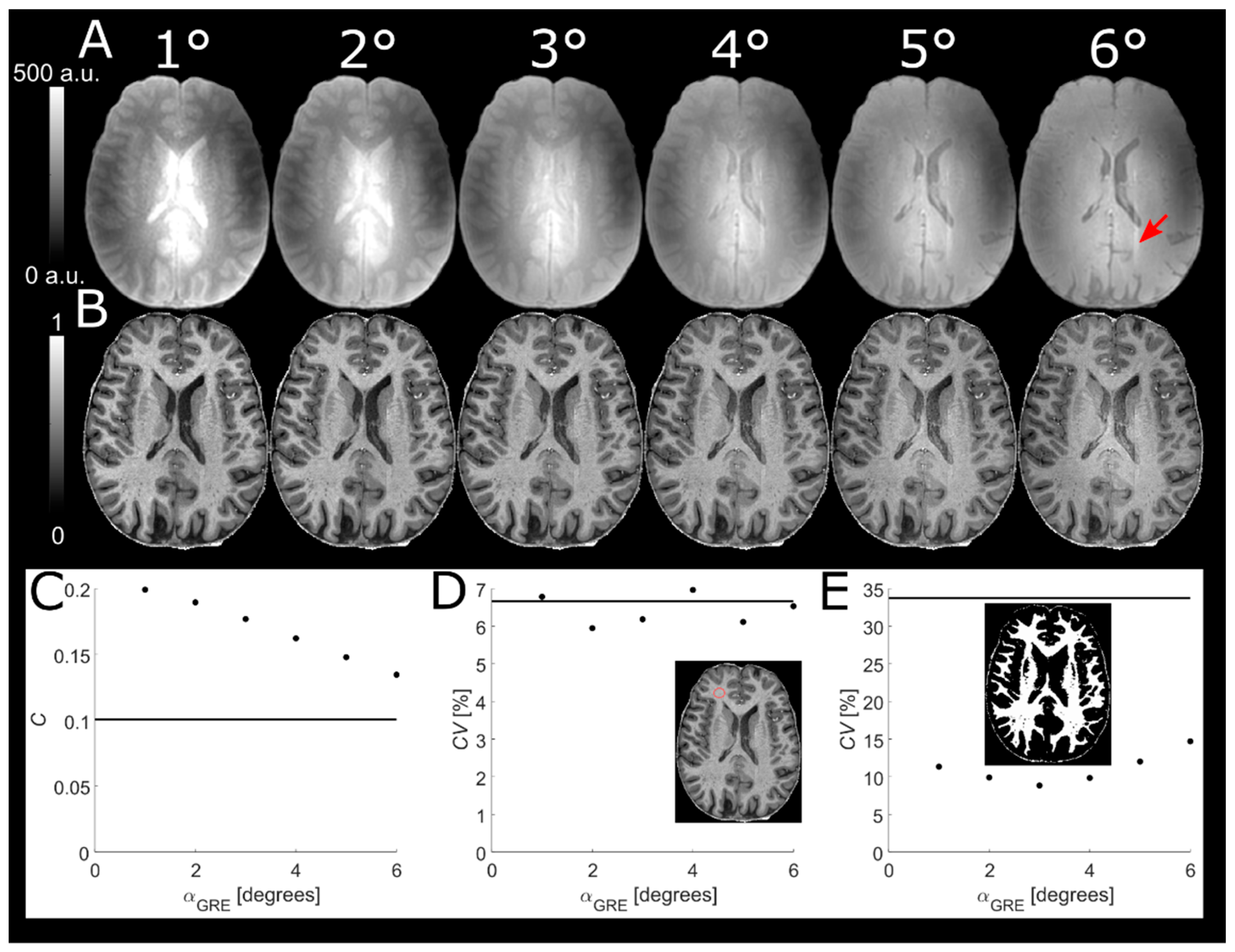
2.6. Readout Flip Angle of Reference GRE
A higher α
GRE will increase the
T1-w of the predominantly PD-w reference GRE, thus reducing tissue contrast in the normalized MPRAGE image. On the other hand, reducing α
GRE below the Ernst angle will decrease SNR. To find a compromise between SNR and tissue contrast in the normalized MPRAGE, α
GRE was varied from 1° to 6° in increments of 1° in a single subject. In order to obtain comparable pixel values, reference GREs were scaled by a global factor giving a scaled signal
where a single
(corresponding to expected WM
) was used to approximate the saturation of
Mz [
17,
18]. The scaling only served to facilitate identical windowing of the images and does not affect the analysis itself. The contrast between segmented WM and GM as well as the
CV of segmented WM was plotted as a function of α
GRE. The latter was used as a proxy to evaluate any residual influence of
B1+ inhomogeneities.
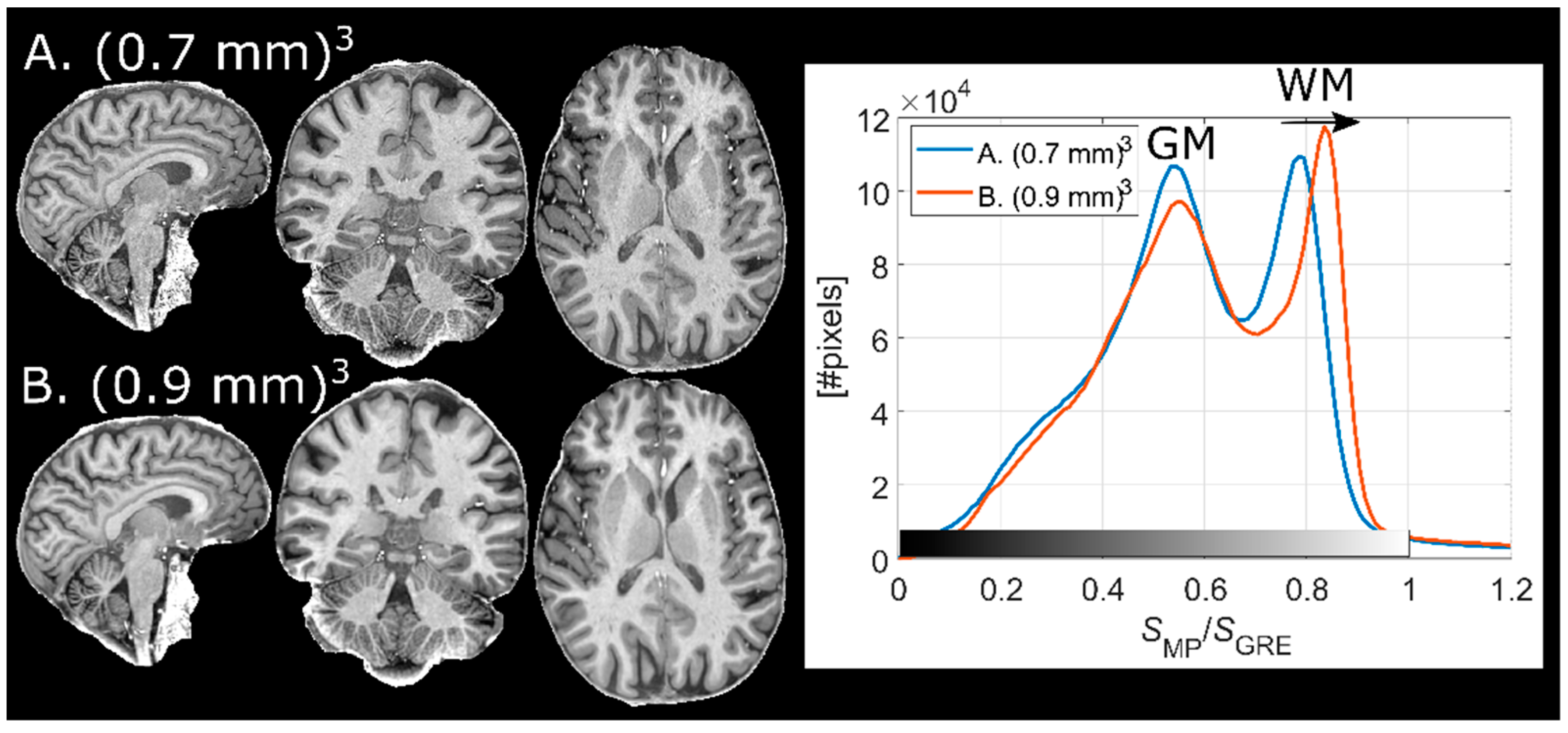
2.7. Voxel Size of Reference GRE
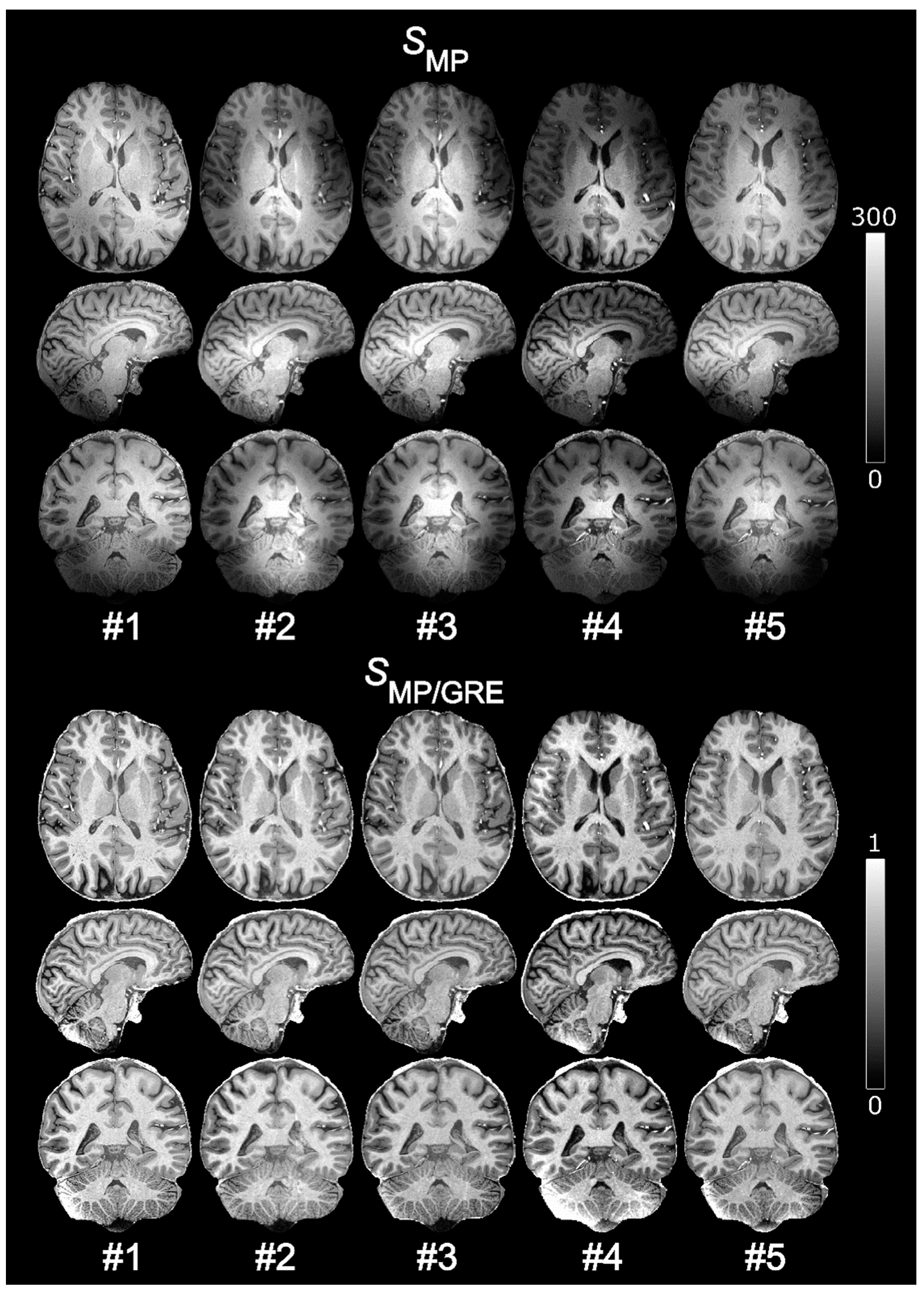
The B1-related intensity bias (of both receive and transmit effects) comprises mostly low spatial frequencies. Thus, a reference GRE with low spatial resolution is sufficient to correct for B1 inhomogeneity. On the other hand, this is not the case for PD and T2* contrast. If the normalized MPRAGE is purposed to produce only semi-quantitative images with a greatly reduced intensity field bias, some dilution of the “pure” T1 contrast could be acceptable to reduce scan time. In an effort to further reduce scan time and evaluate the effect on the resulting image quality, an MPRAGE volume with 0.7 mm isotropic resolution was normalized by a reference GRE for which the voxel size, Vref, was varied in-plane as 0.70 × 0.70, 1.05 × 1.05, 1.40 × 1.40, 2.10 × 2.10, and 2.80 × 2.80 mm2 (i.e., ×1, ×1.5, ×2, ×3, ×4 the MPRAGE resolution) in a single subject. The voxel dimension in the outer loop (right–left direction) was constant at 1.40 mm resulting in acquisition times of Tacq = 2:21, 1:35, 1:10, 0:49, and 0:37 min, respectively.
2.8. Implementation at Different Resolutions
Two finalized protocols (Results 3.1–3.2) with voxel sizes of (0.7 mm)3 and (0.9 mm)3 were used on a single subject to compare the effect of the corresponding TF on the contrast. Further, the potential increase of interpolation artifacts at different spatial resolutions (especially in the reference GRE) was of interest to assess.
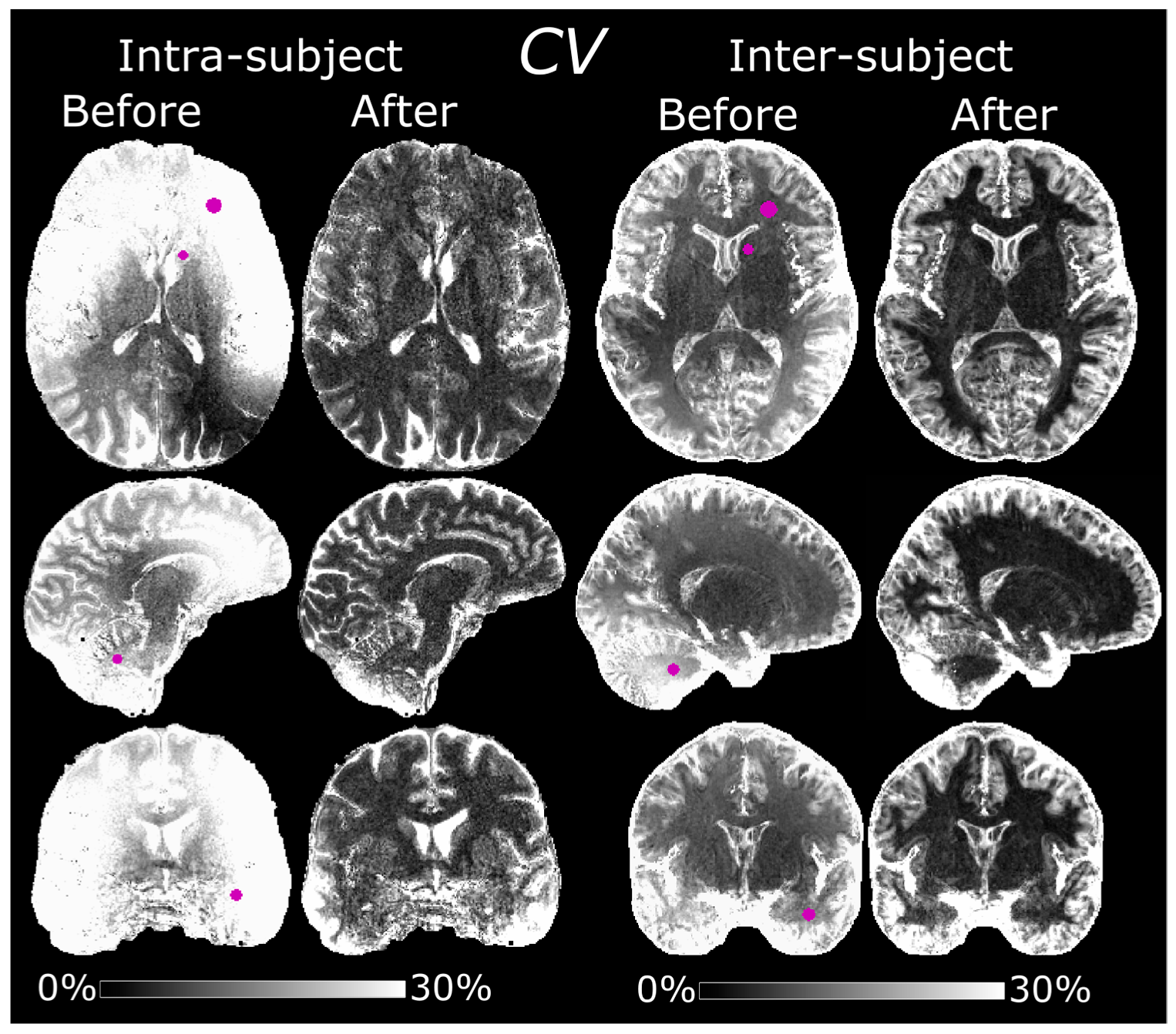
2.9. Intra-Subject Variability
One subject (male, 51 years old, body mass index (BMI) of 23.6) was scanned on five separate occasions over a period of about 8 months using the normalized MPRAGE protocol with 0.7 mm isotropic resolution. Maps of CV were compared before and after normalization and the average CVs in WM, GM, and CSF were calculated. Regions of interest (ROIs) were manually delineated in the CV maps to study local variability. The ROIs were defined in left frontal WM, the left caudate head, the cerebellum, and the left temporal lobe.
2.10. Inter-Subject Variability
The 0.8 mm isotropic resolution protocol was previously used in a separate work to study correlations between cortical morphology and language-learning aptitude [
19]. From the underlying subject population of this work, a subset of 10 randomly chosen volunteers (7 female, 22 ± 1.9 years old) were used to examine the inter-subject variability of the normalized MPRAGE images. The subjects were all of normal build, that is, height, weight, and BMI were perceived to fall within the 5th and 95th percentiles. Images were diffeomorphically registered to the Montreal Neurological Institute (MNI) space using the diffeomorphic anatomical registration using exponentiated Lie algebra (DARTEL) algorithm as embedded in the statistical parameter mapping (SPM)-based histological MRI (hMRI) toolbox [
20,
21]. As in the previous subsection, maps of the
CV were then compared before and after normalization and the average
CVs in WM, GM, and CSF were calculated. ROIs were likewise defined in MNI space in the same four areas as in the previous subsection, i.e., left frontal WM, left caudate head, cerebellum, and the left temporal lobe. The average
CVs in WM, GM and CSF were also calculated based on the average pixel intensities of individual segmentations rather than one common segmentation in MNI space.
2.11. Comparison to MP2RAGE
The finalized (0.7 mm)
3 sequential protocol was compared to an interleaved MP2RAGE protocol with the same spatial resolution in one subject [
22]. The pulse sequence parameters of the MP2RAGE protocol was as follows:
Tcycle/
TI1/
TI2 = 5/0.9/2.75 s, α
1/α
2 = 5/3 degrees, TF = 256, TR/TE = 6.8/2.4 ms, bandwidth/px = 365 Hz,
Nx,y,z = 320 × 320 × 256,
Tacq = 8:20 min with a partial Fourier acquisition of 0.75, and a SENSE-factor of 2 in the outer loop right–left direction as well as elliptical k-space phase encoding. The same inversion pulse as in the sequential protocol was used. The MP2RAGE image was calculated using complex data as described in ref. [
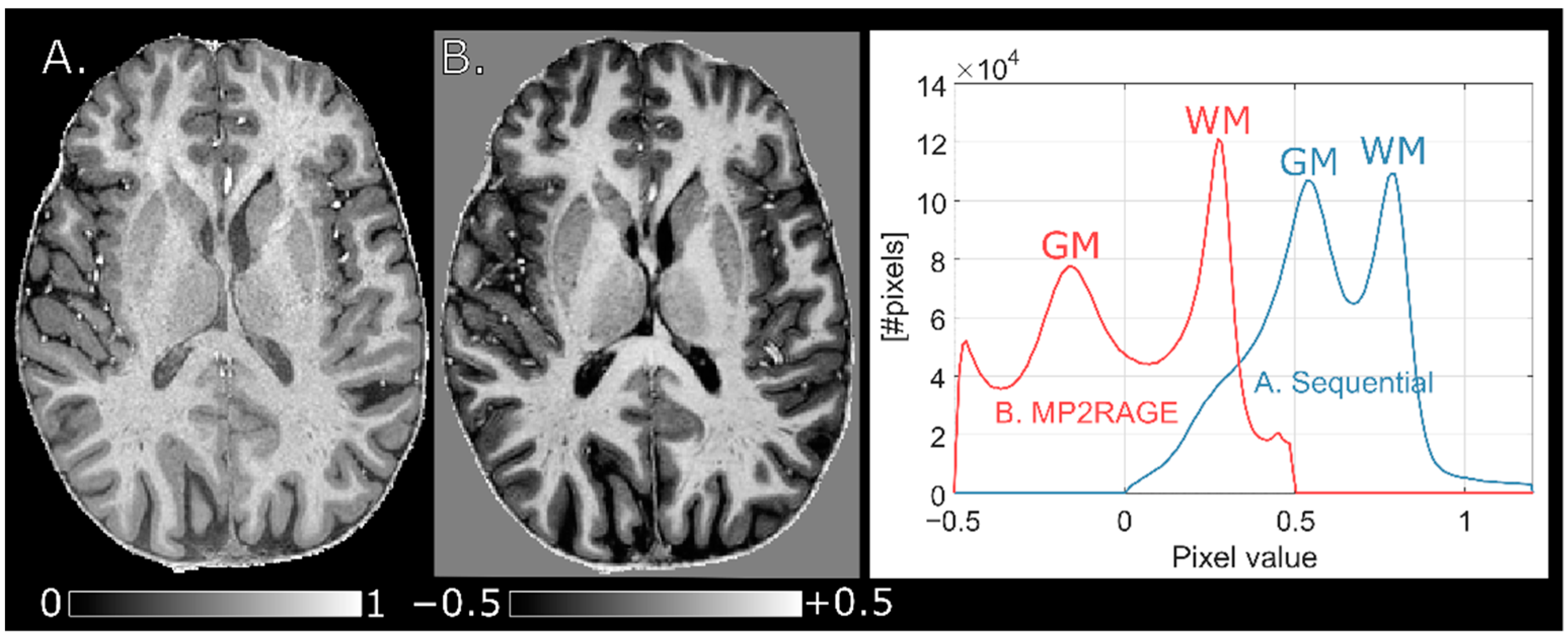
5] and rigidly co-registered to the sequentially normalized MPRAGE image.
2.12. T1 Calculation
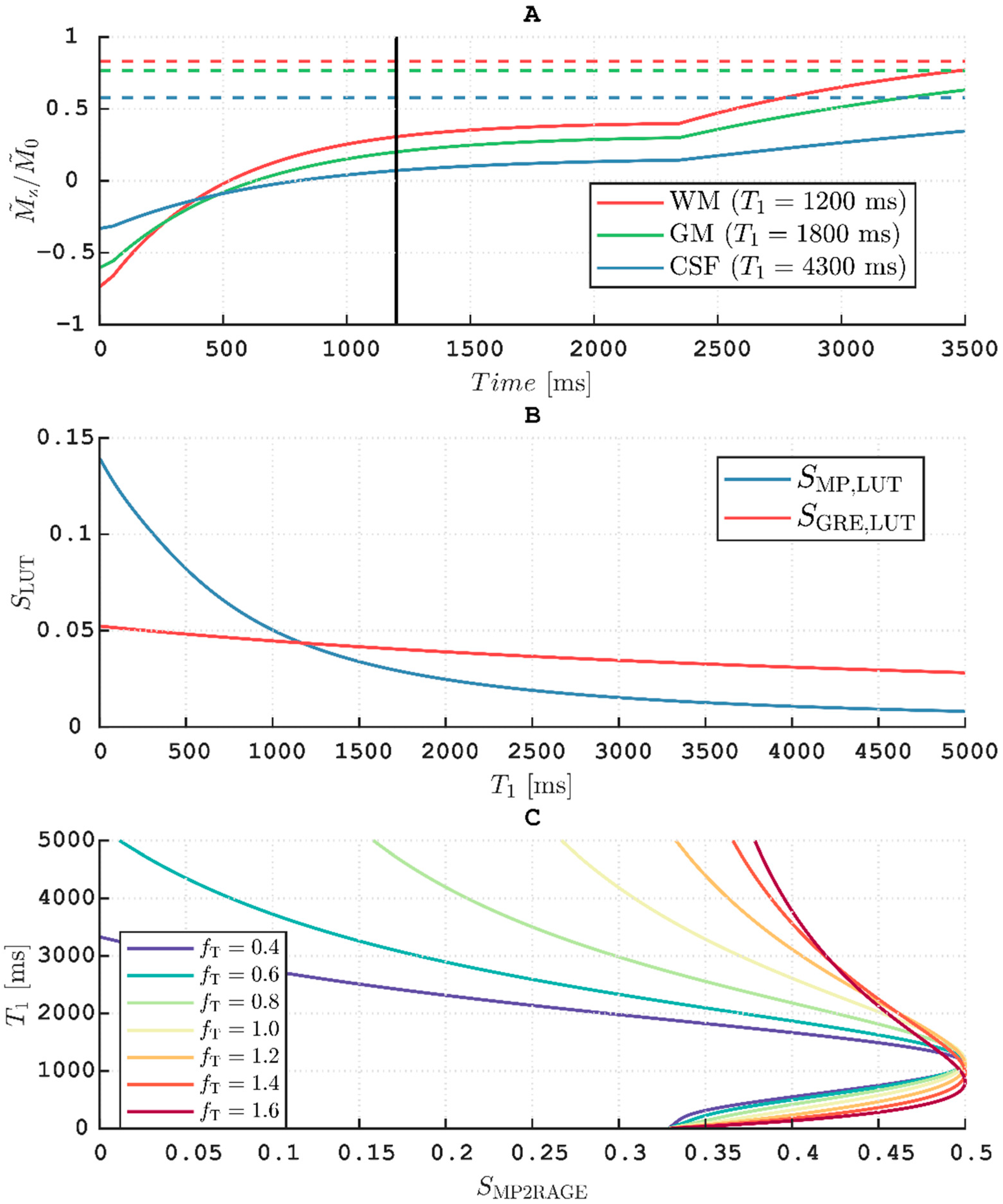
For proof-of-principle,
T1-mapping using a LUT-based approach was performed on a healthy subject using the (0.8 mm)
3 resolution protocol together with a DREAM flip angle map [
23]. First, the evolution of the longitudinal magnetization,
Mz, was simulated for the MPRAGE sequence with imaging parameters as described above, i.e., with α
MP = 8°, TR = 8 ms, TI = 1200 ms and TF = 288.
The evolution of
Mz during
Tcycle in the outer loop steady-state (occurring after 2–3 cycles) was simulated using Equations (2) and (3) in the intervals where readout occurred and using normal
T1 relaxation where readout did not occur. The simulations can be performed with
(i.e., per unit
). The inversion efficiency applied at the end of each cycle was assumed to be
finv = 0.96 [
5]. The LUT-derived signal was then calculated for a constant
but over a range of
as:
The reference GRE was assumed to always be in the inner loop steady-state. Hence,
and consequently the LUT-derived GRE signal,
, is constant for a constant
and
:
Thus, 2D (
) LUTs of
and
are obtained for a range of
(step size of 1 ms,
) and
(step size of 0.01,
). The LUTs of the two simulated signals were then combined as follows [
5]:
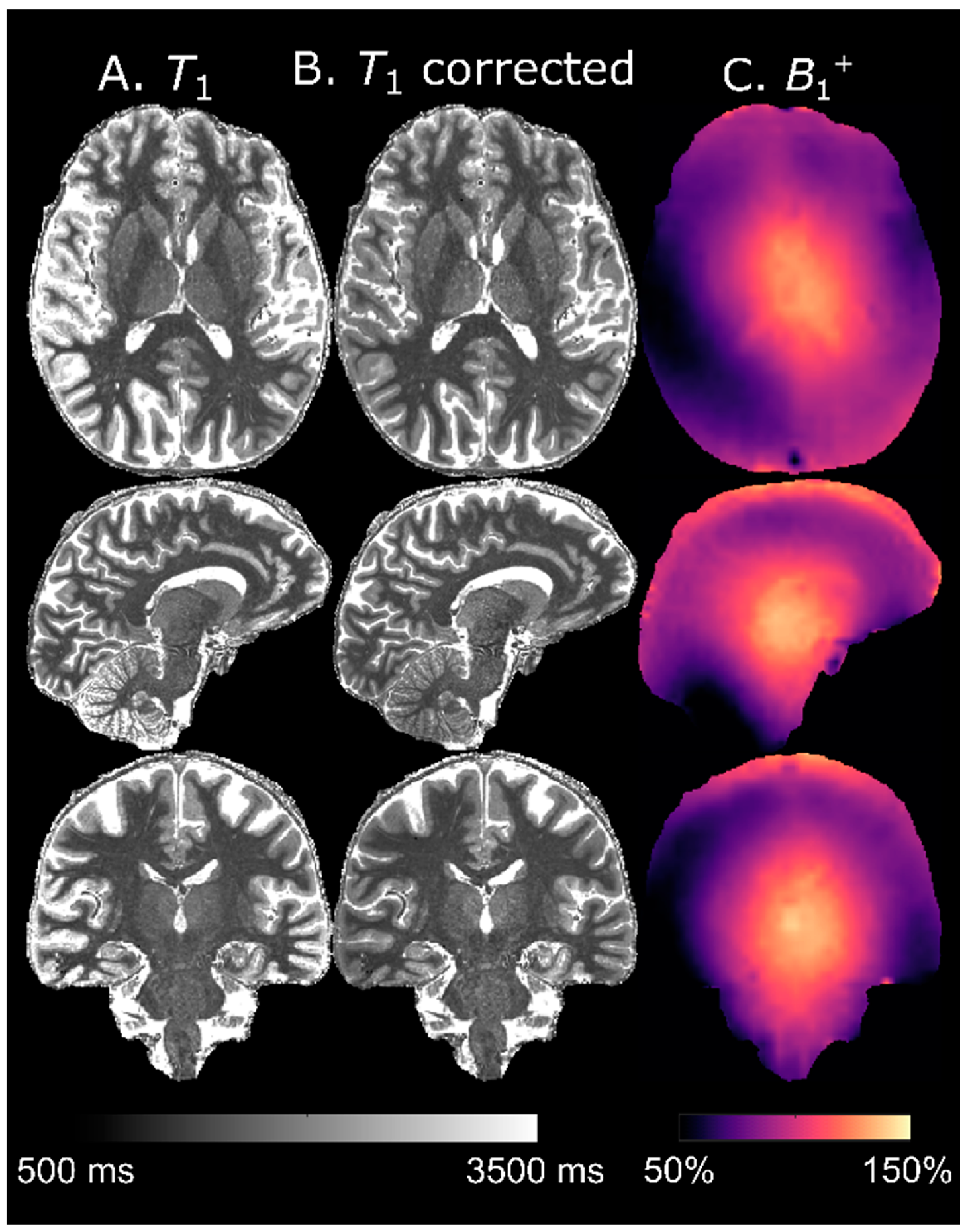
Thus, the values are limited to in the final LUT for comparison with calculated from the measured magnitude signals. Note that the measured is limited to positive values for this implementation, since it is not possible to relate the phases of two signals measured sequentially. The for which was minimal was calculated pixelwise either for or as determined by the DREAM flip angle map.
4. Discussion
In this study, we describe the process of implementing a sequential protocol for bias field correction of MPRAGE images at 7 T. Effects of varying the flip angle, different spatial resolutions and acquisition voxel size of the reference GRE were studied, mainly to improve WM-GM contrast and to minimize scan time without introducing biases. The main purpose of the protocols was to obtain semi-quantitative images with “pure” T1 contrast and improved reproducibility. Improved intra-subject reproducibility was demonstrated by a decreased CV of 7.9 ± 3.3% in segmented WM after normalization compared to 20 ± 7.8% before. Likewise, an improved inter-subject reproducibility was demonstrated by the CV in segmented WM of 10 subjects which decreased from 13 ± 7.8% to 7.6 ± 7.6%. Local improvement could be higher, for instance 39 ± 3.1% to 7.6 ± 2.2% in frontal WM in the intra-subject experiment and 22 ± 1.4% to 4.4 ± 0.91% in the cerebellum in the inter-subject experiment.
The multiplicative spatial intensity bias imposed by the combination of receive coil signals was removed by normalization. Due to a smaller readout flip angle in the reference GRE compared to the MPRAGE, and to the rather small flip angles overall, the effect of the inhomogeneous transmit field (
B1+) was also mostly removed. Since the analytical description of the MPRAGE signal is too complicated to evaluate [
25], we estimated
T1 maps through a LUT-based approach analogous to MP2RAGE [
5]. As expected, accuracy was improved by using a separate
B1+ map.
The protocol was not designed with
T1 calculation, as featured by MP2RAGE, in mind. Thus, at the rather long TI = 1200 ms the protocol fails to effectively exploit the dynamic range obtained when inverting fully relaxed longitudinal magnetization (cf.
Figure 7, panel A). This results in quite “saturated” images when normalizing using Equation (8) (pixels close to 0.5), leading to a loss of precision in the
T1 calculation. Further, although whole-brain histograms of
T1 were very similar when using different
Vref, it is most likely prudent to use identical voxel sizes of MPRAGE and the reference GRE if accurate
T1 estimation is of interest, especially at the cortical boundaries that are most susceptible to PVEs. It is important to note that the LUT-based approach assumes that all differences in pixel values are solely caused by variations in
T1 (i.e., that there is “pure”
T1 contrast). By design, the influence of
B1+ inhomogeneities on the
T1 calculation is decreased by the normalization of signals. The choice of α
MP = 2.7α
GRE (8° vs. 3°) appeared to minimize residual effects of
B1+ inhomogeneity on the normalized MPRAGE (cf.
Figure 1) and thus also on the
T1 calculation. The largest residual effect of
B1+ inhomogeneity was found at α
MP = 1.3α
GRE (8° vs. 6°). This is in concordance with the work of Van de Moortele et al. in which a choice of α
MP = α
GRE resulted in a much stronger residual
B1+ dependence than α
MP = 2α
GRE [
4]. To increase accuracy, a separately acquired flip angle map is still recommended if
T1-mapping is of interest [
26]. This is particularly important for longer
T1 values where the
B1+ influence is stronger (cf. panel C,
Figure 7). The loss of contrast observed in the cerebellum (cf. panel C,
Figure 9) occurs when
B1+ decreases below the threshold required for the inversion pulse to fulfil the adiabatic condition which cannot be resolved by flip angle mapping [
24].
The obvious benefit of an interleaved acquisition is that identical scanning conditions, such as RF power calibration, is guaranteed, as well as increased robustness against inter-scan movement. Inter-scan motion can be corrected by offline rigid coregistration, however. Further, the reduced duration of any one acquisition will reduce the total risk of intra-scan subject movement. The risk of introducing
T1 contrast in the GRE reference due to poor timings (mainly TI and/or TF×TR being too short) is also removed [
4]. Importantly, the option to increase
Vref facilitates the possibility to have a shorter total scan time than needed for the interleaved MP2RAGE.
A protocol with an MPRAGE acquisition voxel size of (0.6 mm)
3 was also explored (data not shown). However, the SNR in the MPRAGE was deemed unacceptably low. Hence, the (0.7 mm)
3 protocol here represents the upper limit of the spatial resolution imposed by SNR. It should be noted that noise propagation will moderately decrease the SNR in the normalized MPRAGE,
, relative to the unnormalized,
, by
, thereby somewhat adversely affecting obtainable spatial resolution [
4]. Decreasing the acquisition voxel size also entails increasing TF and thus the duration of the readout train and eventually of TI. This limitation can be circumvented by introducing a SENSE factor in the inner loop, as shown here for (0.7 mm)
3. The employed zigzag k-space trajectory (see the view-ordering schemes presented for turbo spin echoes in ref. [
27]) allows the use of a TF larger than the number of inner loop k-space lines (
Ny), which is an effective way to decrease scan time. It further allows the enabling of an elliptical k-space phase encoding which decreases the acquisition time by a factor of approximately
(cf.
Table 1). To our knowledge, this kind of k-space trajectory is only readily available on Philips’s systems. Hence, the total scan time of the protocols are supplied with/without elliptical phase encoding.
Correction of RF field-induced bias is recommended at field strengths of 7 T and above. However, normalization may also be necessary to correct for intensity field bias at 3 T, as the wavelength of the
B1 field is approximately 25 cm in brain tissue and thus of the same order of magnitude as the imaged object [
28].
5. Limitations
We did not attempt to optimize the point-spread function (PSF) of the MPRAGE sequence, although this will be broadened when signal is acquired during a transient state [
8]. At increasing resolution, readouts are appended at the beginning and end of the RAGE trains (cf.
Figure 7, panel A). These will compromise the PSF, so the effective spatial resolution will not increase as much as the nominal spatial resolution. The increased change in
Mz during acquisition may also influence the WM-GM contrast (cf.
Figure 3). In this work, we focused on the reference GRE for normalization, which is acquired entirely in a steady-state and thus does not suffer from such PSF distortions. Further, the PSF cannot be modelled for an elliptical k-space phase encoding, since the trajectory is proprietary.
Like in MP2RAGE, the loss of
T1 contrast at very low
B1+ (below the threshold for adiabatic inversion) cannot be mended by normalization [
22].
Although subdural ringing artifacts did not noticeably increase when using a protocol with lower spatial resolution (cf.
Figure 3), curved ringing artifacts were occasionally observed. These were more evident in the reference GRE but could also be seen in MPRAGE. The artifacts appeared to be correlated to subject movement, but this was not confirmed. These artifacts are believed to be related to interpolation of the low spatial resolution (5.5 × 7.4 × 4.0 mm
3) SENSE reference scan, acquired prior to the MPRAGE and reference GRE. The artifact was very similar to the “streaky-linear” artifact “type A” described by Sartoretti et al. and showcased in
Figure 3, panels (g), (h), and (i) [
29].