Using Constrained-Disorder Principle-Based Systems to Improve the Performance of Digital Twins in Biological Systems
Abstract
1. Introduction
2. The Constrained-Disorder Principle Defines Noise as Inherent to Biological Systems
3. Bioengineering Needs to Account for Variability
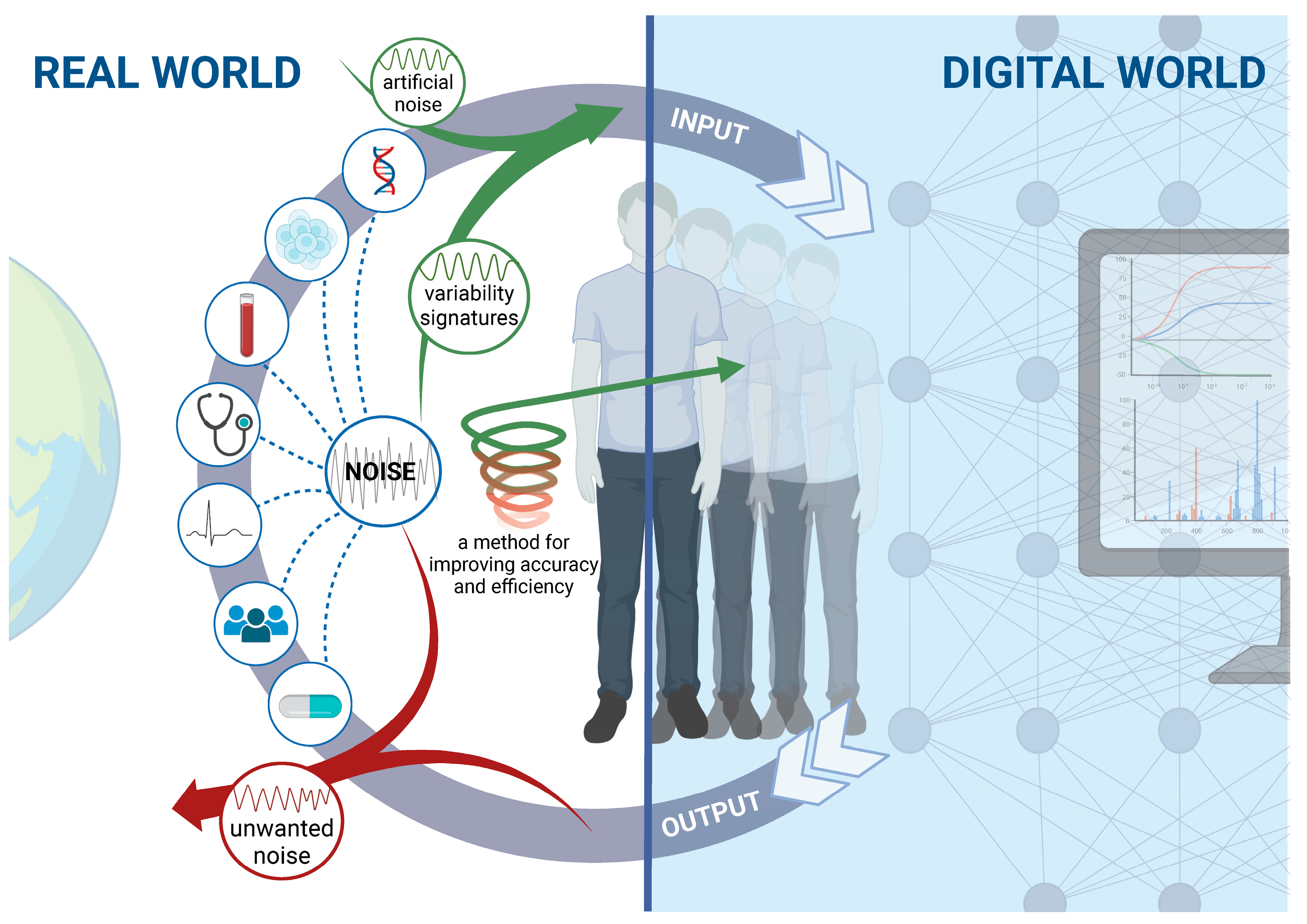
4. Digital Twins Use Real-World Data to Create Simulations
5. Using Digital Twin Systems in Biology
6. Applications of Digital Twins in Healthcare
7. The Need to Model Uncertainties and Noise in Complex Systems
8. Digital Twins’ Methods for Dealing with Uncertainties
- i.
- Complete Bayesian analysis is a component of probability statistics derived from the Bayesian theorem used for uncertainty quantification [41,87]. Bayesian inference estimates the probability of a hypothesis under updated knowledge (i.e., posterior probability). It uses prior probability (the probability of the hypothesis occurring irrespective of the updated knowledge), model evidence (the observation of experimental or simulated data), and likelihood (the probability of specific parameters being observed if the hypothesis is correct) [85,87]. Under the Bayesian principles, a prior distribution for the uncertain parameters is assumed based on expert knowledge. Using model evidence, the posterior distribution of these uncertain parameters is estimated via the formula, and a confidence interval reflecting the reliability of the result is extracted [38,68,85,87]. As more evidence accumulates in subsequent simulations, the parameters are updated, and the posterior distribution shows improved accuracy [41]. Combining the Bayesian approach with deep learning is helpful for uncertainty quantification, providing a framework for the training process, Bayesian deep learning [41,87].
- ii.
- The Markov Chain Monte Carlo (MCMC) method is used to estimate the posterior distribution, which is computationally intensive and sometimes cannot be calculated analytically [41,68]. MCMC addresses the sampling problem via probability distribution and approximation methods (e.g., Variational Inference and Monte Carlo dropouts) [68]. Monte Carlo (MC) simulations attempt to predict all the possible results of a system with random variables [41]. The algorithm runs multiple possible values within the known range of each input parameter, producing an output of a probability distribution that reflects every possible result and its likelihood [70]. The MCMC method enables the expression of the posterior probability of complex real-world processes by using computer simulations of random samplings from the probability distribution [87]. MCMC is generated within the space of all possible results. The progression from one possible value to the next is random, but using different algorithms, it is set up so that values derived from more plausible models appear more frequently [87]. This process approximates the most probable results and achieves more accurate results as more samples are obtained [70].
- iii.
- Variational inference (VI) for approximate Bayesian inference provides a computational approximation of the intractable posterior probability distribution by solving an optimization problem and finding a tractable distribution similar to the unknown one [68,70]. VI is faster than MCMC, and the convergence into a result is unequivocal [68]. However, it involves complex calculations, approximates the desired distribution rather than the theoretically optimal solution with considerably fewer samplings, and is applicable to large-scale datasets and complex models [68,70].
- iv.
- The Monte Carlo dropout method for approximate Bayesian inference prevents overfitting during the training of deep learning systems, improving generalization and prediction abilities from unseen data during the testing phase [68]. Some neurons within the hidden layers of a deep NN are randomly omitted, including their incoming and outgoing connections, resulting in diminished network complexity. As the neuron elimination is random, each training iteration is performed on a different edited network, resulting in multiple predictions generated from the same data. The output is a distribution of predictions produced by ensembles of smaller networks, reflecting the model’s uncertainty [38,70]. This improves the system’s performance by capturing randomness and quantifying uncertainties [38].
9. Improving Digital Twins for Biological Systems by Differentiating between Inherent Noise and Measurement-Related Unwanted Noise
10. Augmented Digital Twins Make Use of Noise to Improve the Performance of Biological Systems
11. Challenges Faced by Augmented Digital Twins in Medicine
Author Contributions
Funding
Institutional Review Board Statement
Conflicts of Interest
Abbreviations
References
- Nasirahmadi, A.; Hensel, O. Toward the Next Generation of Digitalization in Agriculture Based on Digital Twin Paradigm. Sensors 2022, 22, 498. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.; He, Z. Digital twin-driven intelligence disaster prevention and mitigation for infrastructure: Advances, challenges, and opportunities. Nat. Hazards 2022, 112, 1–36. [Google Scholar]
- Drobnyi, V.; Hu, Z.; Fathy, Y.; Brilakis, I. Construction and Maintenance of Building Geometric Digital Twins: State of the Art Review. Sensors 2023, 23, 4382. [Google Scholar] [CrossRef]
- Wang, J.; Qiao, L.; Lv, H.; Lv, Z. Deep Transfer Learning-based Multi-modal Digital Twins for Enhancement and Diagnostic Analysis of Brain MRI Image. IEEE/ACM Trans. Comput. Biol. Bioinform. 2022. [Google Scholar] [CrossRef]
- Kampczyk, A.; Dybel, K. The Fundamental Approach of the Digital Twin Application in Railway Turnouts with Innovative Monitoring of Weather Conditions. Sensors 2021, 21, 5757. [Google Scholar] [CrossRef] [PubMed]
- Finn, E.H.; Misteli, T. Molecular basis and biological function of variability in spatial genome organization. Science 2019, 365, eaaw9498. [Google Scholar] [CrossRef] [PubMed]
- Ilan, Y. Overcoming randomness does not rule out the importance of inherent randomness for functionality. J. Biosci. 2019, 44, 132. [Google Scholar] [CrossRef] [PubMed]
- Ilan, Y. Generating randomness: Making the most out of disordering a false order into a real one. J. Transl. Med. 2019, 17, 49. [Google Scholar] [CrossRef]
- Ilan, Y. Advanced Tailored Randomness: A Novel Approach for Improving the Efficacy of Biological Systems. J. Comput. Biol. 2020, 27, 20–29. [Google Scholar]
- Ilan, Y. Order Through Disorder: The Characteristic Variability of Systems. Front. Cell Dev. Biol. 2020, 8, 495391. [Google Scholar] [CrossRef]
- El-Haj, M.; Kanovitch, D.; Ilan, Y. Personalized inherent randomness of the immune system is manifested by an individualized response to immune triggers and immunomodulatory therapies: A novel platform for designing personalized immunotherapies. Immunol. Res. 2019, 67, 337–347. [Google Scholar] [CrossRef] [PubMed]
- Ilan, Y. Randomness in microtubule dynamics: An error that requires correction or an inherent plasticity required for normal cellular function? Cell Biol. Int. 2019, 43, 739–748. [Google Scholar]
- Ilan, Y. Microtubules: From understanding their dynamics to using them as potential therapeutic targets. J. Cell Physiol. 2019, 234, 7923–7937. [Google Scholar] [PubMed]
- Ilan-Ber, T.; Ilan, Y. The role of microtubules in the immune system and as potential targets for gut-based immunotherapy. Mol. Immunol. 2019, 111, 73–82. [Google Scholar] [CrossRef] [PubMed]
- Forkosh, E.; Kenig, A.; Ilan, Y. Introducing variability in targeting the microtubules: Review of current mechanisms and future directions in colchicine therapy. Pharmacol. Res. Perspect. 2020, 8, e00616. [Google Scholar] [CrossRef]
- Ilan, Y. beta-Glycosphingolipids as Mediators of Both Inflammation and Immune Tolerance: A Manifestation of Randomness in Biological Systems. Front. Immunol. 2019, 10, 1143. [Google Scholar] [CrossRef] [PubMed]
- Schutte, A.E.; Kollias, A.; Stergiou, G.S. Blood pressure and its variability: Classic and novel measurement techniques. Nat. Rev. Cardiol. 2022, 19, 643–654. [Google Scholar] [CrossRef] [PubMed]
- Chiera, M.; Cerritelli, F.; Casini, A.; Barsotti, N.; Boschiero, D.; Cavigioli, F.; Corti, G.C.; Manzotti, A. Heart Rate Variability in the Perinatal Period: A Critical and Conceptual Review. Front. Neurosci. 2020, 14, 561186. [Google Scholar] [CrossRef]
- Forte, G.; Favieri, F.; Casagrande, M. Heart Rate Variability and Cognitive Function: A Systematic Review. Front. Neurosci. 2019, 13, 710. [Google Scholar]
- Tian, Q.; Chastan, N.; Bair, W.N.; Resnick, S.M.; Ferrucci, L.; Studenski, S.A. The brain map of gait variability in aging, cognitive impairment and dementia-A systematic review. Neurosci. Biobehav. Rev. 2017, 74, 149–162. [Google Scholar] [CrossRef]
- Ilan, Y. Microtubules as a potential platform for energy transfer in biological systems: A target for implementing individualized, dynamic variability patterns to improve organ function. Mol. Cell. Biochem. 2022, 478, 375–392. [Google Scholar] [CrossRef] [PubMed]
- Ilan, Y. Constrained disorder principle-based variability is fundamental for biological processes: Beyond biological relativity and physiological regulatory networks. Prog. Biophys. Mol. Biol. 2023, 180–181, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Urban, E.A.; Johnston, R.J., Jr. Buffering and Amplifying Transcriptional Noise During Cell Fate Specification. Front. Genet. 2018, 9, 591. [Google Scholar] [CrossRef]
- Dar, R.D.; Weiss, R. Perspective: Engineering noise in biological systems towards predictive stochastic design. APL Bioeng. 2018, 2, 020901. [Google Scholar] [PubMed]
- Sejdić, E.; Lipsitz, L.A. Necessity of noise in physiology and medicine. Comput. Methods Programs Biomed. 2013, 111, 459–470. [Google Scholar] [CrossRef]
- Wu, F.; Tan, C. The Engineering of Artificial Cellular Systems using Synthetic Biology Approaches. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2014, 6, 369–383. [Google Scholar] [CrossRef] [PubMed]
- Ruppert, T.; Abonyi, J. Integration of real-time locating systems into digital twins. J. Ind. Inf. Integr. 2020, 20, 100174. [Google Scholar]
- Schluse, M.; Rossmann, J. From simulation to experimentable digital twins: Simulation-based development and operation of complex technical systems. In Proceedings of the 2016 IEEE International Symposium on Systems Engineering (ISSE), Edinburgh, UK, 3–5 October 2016. [Google Scholar]
- Shahzad, M.; Shafiq, M.T.; Douglas, D.; Kassem, M. Digital Twins in Built Environments: An Investigation of the Characteristics, Applications, and Challenges. Buildings 2022, 12, 120. [Google Scholar] [CrossRef]
- Bandiera, L.; Furini, S.; Giordano, E. Phenotypic Variability in Synthetic Biology Applications: Dealing with Noise in Microbial Gene Expression. Front. Microbiol. 2016, 7, 479. [Google Scholar] [PubMed]
- Tsimring, L.S. Noise in biology. Rep. Prog. Phys. 2014, 77, 026601. [Google Scholar] [CrossRef] [PubMed]
- Grieves, M. Origins of the Digital Twin Concept. 2016. Available online: https://www.researchgate.net/publication/307509727_Origins_of_the_Digital_Twin_Concept (accessed on 9 July 2017).
- Singh, M.; Fuenmayor, E.; Hinchy, E.P.; Qiao, Y.; Murray, N.; Devine, D. Digital Twin: Origin to Future. Appl. Syst. Innov. 2021, 4, 36. [Google Scholar] [CrossRef]
- Thiong’o, G.M.; Rutka, J.T. Digital Twin Technology: The Future of Predicting Neurological Complications of Pediatric Cancers and Their Treatment. Front. Oncol. 2021, 11, 781499. [Google Scholar] [CrossRef]
- Botín-Sanabria, D.M.; Mihaita, A.-S.; Peimbert-García, R.E.; Ramírez-Moreno, M.A.; Ramírez-Mendoza, R.A.; Lozoya-Santos, J.d.J. Digital Twin Technology Challenges and Applications: A Comprehensive Review. Remote Sens. 2022, 14, 1335. [Google Scholar] [CrossRef]
- Meierhofer, J.; Schweiger, L.; Lu, J.; Züst, S.; West, S.; Stoll, O.; Kiritsis, D. Digital Twin-Enabled Decision Support Services in Industrial Ecosystems. Appl. Sci. 2021, 11, 11418. [Google Scholar] [CrossRef]
- Verdouw, C.; Tekinerdogan, B.; Beulens, A.; Wolfert, S. Digital twins in smart farming. Agric. Syst. 2021, 189, 103046. [Google Scholar] [CrossRef]
- Mozo, A.; Karamchandani, A.; Gómez-Canaval, S.; Sanz, M.; Moreno, J.I.; Pastor, A. B5GEMINI: AI-Driven Network Digital Twin. Sensors 2022, 22, 4106. [Google Scholar] [CrossRef]
- Kritzinger, W.; Karner, M.; Traar, G.; Henjes, J.; Sihn, W. Digital Twin in manufacturing: A categorical literature review and classification. IFAC-PapersOnline 2018, 51, 1016–1022. [Google Scholar] [CrossRef]
- Henrichs, E.; Noack, T.; Pinzon Piedrahita, A.M.; Salem, M.A.; Stolz, J.; Krupitzer, C. Can a Byte Improve Our Bite? An Analysis of Digital Twins in the Food Industry. Sensors 2021, 22, 115. [Google Scholar] [CrossRef] [PubMed]
- Moingeon, P.; Chenel, M.; Rousseau, C.; Voisin, E.; Guedj, M. Virtual patients, digital twins and causal disease models: Paving the ground for in silico clinical trials. Drug Discov. Today 2023, 28, 13596446. [Google Scholar] [CrossRef] [PubMed]
- Bado, M.F.; Tonelli, D.; Poli, F.; Zonta, D.; Casas, J.R. Digital Twin for Civil Engineering Systems: An Exploratory Review for Distributed Sensing Updating. Sensors 2022, 22, 3168. [Google Scholar] [CrossRef]
- Wagg, D.; Worden, K.; Barthorpe, R.; Gardner, P. Digital Twins: State-of-The-Art Future Directions for Modelling and Simulation in Engineering Dynamics Applications. ASCE-ASME J. Risk Uncertain. Eng. Syst. Part B Mech. Eng. 2020, 6, 030901. [Google Scholar] [CrossRef]
- Stavropoulos, P.; Mourtzis, D. Chapter 10—Digital twins in industry 4.0. In Design and Operation of Production Networks for Mass Personalization in the Era of Cloud Technology; Mourtzis, D., Ed.; Elsevier: Amsterdam, The Netherlands, 2022; pp. 277–316. [Google Scholar]
- Jones, D.; Snider, C.; Nassehi, A.; Yon, J.; Hicks, B. Characterising the Digital Twin: A systematic literature review. CIRP J. Manuf. Sci. Technol. 2020, 29, 36–52. [Google Scholar] [CrossRef]
- Qi, Q.; Tao, F.; Hu, T.; Anwer, N.; Liu, A.; Wei, Y.; Wang, L.; Nee, A.Y.C. Enabling technologies and tools for digital twin. J. Manuf. Syst. 2021, 58, 3–21. [Google Scholar] [CrossRef]
- Qiao, L.; Cheng, Y. Analysis of Logistics Linkage by Digital Twins Technology and Lightweight Deep Learning. Comput. Intell. Neurosci. 2022, 2022, 6602545. [Google Scholar] [CrossRef]
- Kosse, S.; Vogt, O.; Wolf, M.; König, M.; Gerhard, D. Digital Twin Framework for Enabling Serial Construction. Front. Built Environ. 2022, 8, 864722. [Google Scholar] [CrossRef]
- Wu, Y.; Li, Y. Digital Twin-Driven Performance Optimization for Hazardous Waste Landfill Systems. Math. Probl. Eng. 2022, 2022, 7778952. [Google Scholar] [CrossRef]
- Bertoni, M.; Bertoni, A. Designing solutions with the product-service systems digital twin: What is now and what is next? Comput. Ind. 2022, 138, 103629. [Google Scholar] [CrossRef]
- Liu, Z.; Meyendorf, N.; Mrad, N. The role of data fusion in predictive maintenance using digital twin. AIP Conf. Proc. 2018, 1949, 020023. [Google Scholar]
- Laubenbacher, R.; Sluka, J.P.; Glazier, J.A. Using digital twins in viral infection. Science 2021, 371, 1105–1106. [Google Scholar] [CrossRef]
- Bruynseels, K.; Santoni de Sio, F.; van den Hoven, J. Digital Twins in Health Care: Ethical Implications of an Emerging Engineering Paradigm. Front. Genet. 2018, 9, 31. [Google Scholar] [CrossRef]
- Rabah, S.; Zacharewicz, G.; Chapurlat, V. Digital Twin for Services (DT4S): Conceptual Strategy. IFAC-PapersOnline 2022, 55, 3256–3261. [Google Scholar] [CrossRef]
- Haleem, A.; Javaid, M.; Singh, R.P.; Suman, R. Medical 4.0 technologies for healthcare: Features, capabilities, and applications. Internet Things Cyber-Phys. Syst. 2022, 2, 12–30. [Google Scholar] [CrossRef]
- Guo, J.; Lv, Z. Application of Digital Twins in multiple fields. Multimed. Tools Appl. 2022, 81, 26941–26967. [Google Scholar] [CrossRef]
- Braun, M. Represent me: Please! Towards an ethics of digital twins in medicine. J. Med. Ethics 2021, 47, 394–400. [Google Scholar] [CrossRef]
- Gunasegaram, D.R.; Murphy, A.B.; Barnard, A.; DebRoy, T.; Matthews, M.J.; Ladani, L.; Gu, D. Towards developing multiscale-multiphysics models and their surrogates for digital twins of metal additive manufacturing. Addit. Manuf. 2021, 46, 102089. [Google Scholar] [CrossRef]
- Corral-Acero, J.; Margara, F.; Marciniak, M.; Rodero, C.; Loncaric, F.; Feng, Y.; Gilber, A.; Fernandes, J.F.; Bukhari, H.A.; Wajdan, A.; et al. The ‘Digital Twin’ to enable the vision of precision cardiology. Eur. Heart J. 2020, 41, 4556–4564. [Google Scholar] [CrossRef] [PubMed]
- Voigt, I.; Inojosa, H.; Dillenseger, A.; Haase, R.; Akgün, K.; Ziemssen, T. Digital Twins for Multiple Sclerosis. Front. Immunol. 2021, 12, 669811. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.-Q.; Videiro, P.M.; Guedes Soares, C. Opportunities and Challenges to Develop Digital Twins for Subsea Pipelines. J. Mar. Sci. Eng. 2022, 10, 739. [Google Scholar] [CrossRef]
- Bjornsson, B.; Borrebaeck, C.; Elander, N.; Gasslander, T.; Gawel, D.R.; Gustafsson, M.; Jörnsten, R.; Lee, E.J.; Li, X.; Lilja, S.; et al. Digital twins to personalize medicine. Genome Med. 2019, 12, 4. [Google Scholar] [CrossRef]
- Kamel Boulos, M.N.; Zhang, P. Digital Twins: From Personalised Medicine to Precision Public Health. J. Pers. Med. 2021, 11, 745. [Google Scholar] [CrossRef] [PubMed]
- Masison, J.; Beezley, J.; Mei, Y.; Ribeiro, H.; Knapp, A.C.; Vieira, L.S.; Adhikari, B.; Scindia, Y.; Grauer, M.; Helba, B.; et al. A modular computational framework for medical digital twins. Proc. Natl. Acad. Sci. USA 2021, 118, e2024287118. [Google Scholar] [CrossRef] [PubMed]
- Douthwaite, J.A.; Lesage, B.; Gleirscher, M.; Calinescu, R.; Aitken, J.M.; Alexander, R.; Law, J. A Modular Digital Twinning Framework for Safety Assurance of Collaborative Robotics. Front. Robot. AI 2021, 8, 758099. [Google Scholar] [CrossRef] [PubMed]
- Erol, T.; Mendi, A.F.; Doğan, D. The digital twin revolution in healthcare. In Proceedings of the 2020 4th International Symposium on Multidisciplinary Studies and Innovative Technologies (ISMSIT), Istanbul, Turkey, 22–24 October 2020. [Google Scholar]
- Ghoshal, B.; Tucker, A.; Sanghera, B.; Wong, W.L. Estimating uncertainty in deep learning for reporting confidence to clinicians when segmenting nuclei image data. In Proceedings of the 2019 IEEE 32nd International Symposium on Computer-Based Medical Systems (CBMS), Cordoba, Spain, 5–7 June 2019. [Google Scholar]
- Barricelli, B.R.; Casiraghi, E.; Fogli, D. A survey on digital twin: Definitions, characteristics, applications, and design implications. IEEE Access 2019, 7, 167653–167671. [Google Scholar] [CrossRef]
- Gargalo, C.L.; de las Heras, S.C.; Jones, M.N.; Udugama, I.; Mansouri, S.S.; Krühne, U.; Gernaey, K.V. Towards the Development of Digital Twins for the Bio-manufacturing Industry. In Digital Twins: Tools and Concepts for Smart Biomanufacturing; Herwig, C., Pörtner, R., Möller, J., Eds.; Springer: Cham, Switzerland, 2021; pp. 1–34. [Google Scholar]
- Boughton, C.K.; Hovorka, R. The artificial pancreas. Curr. Opin. Organ. Transplant. 2020, 25, 336–342. [Google Scholar] [CrossRef]
- Steil, G.; Clark, B.; Kanderian, S.; Rebrin, K. Modeling Insulin Action for Development of a Closed-Loop Artificial Pancreas. Diabetes Technol. Ther. 2005, 7, 94–108. [Google Scholar] [CrossRef] [PubMed]
- Tyler, N.S.; Jacobs, P.G. Artificial Intelligence in Decision Support Systems for Type 1 Diabetes. Sensors 2020, 20, 3214. [Google Scholar] [CrossRef] [PubMed]
- Shamanna, P.; Joshi, S.; Shah, L.; Dharmalingam, M.; Saboo, B.; Mohammed, J.; Mohamed, M.; Poon, T.; Kleinman, N.; Thajudeen, M.; et al. Type 2 diabetes reversal with digital twin technology-enabled precision nutrition and staging of reversal: A retrospective cohort study. Clin. Diabetes Endocrinol. 2021, 7, 21. [Google Scholar] [CrossRef]
- Shamanna, P.; Saboo, B.; Damodharan, S.; Mohammed, J.; Mohamed, M.; Poon, T.; Kleinman, N.; Thajudeen, M. Reducing HbA1c in type 2 diabetes using digital twin technology-enabled precision nutrition: A retrospective analysis. Diabetes Ther. 2020, 11, 2703–2714. [Google Scholar] [CrossRef] [PubMed]
- Vaskovsky, A.M.; Chvanova, M.S.; Rebezov, M.B. Creation of digital twins of neural network technology of personalization of food products for diabetics. In Proceedings of the 2020 4th Scientific School on Dynamics of Complex Networks and their Application in Intellectual Robotics (DCNAIR), Innopolis, Russia, 7–9 September 2020. [Google Scholar]
- Azelton, K.R.; Crowley, A.P.; Vence, N.; Underwood, K.; Morris, G.; Kelly, J.; Landry, M.J. Digital Health Coaching for Type 2 Diabetes: Randomized Controlled Trial of Healthy at Home. Front. Digit. Health 2021, 3, 764735. [Google Scholar] [CrossRef] [PubMed]
- Popa, E.O.; van Hilten, M.; Oosterkamp, E.; Bogaardt, M.-J. The use of digital twins in healthcare: Socio-ethical benefits and socio-ethical risks. Life Sci. Soc. Policy 2021, 17, 6. [Google Scholar] [CrossRef]
- Vogenberg, F.R.; Isaacson Barash, C.; Pursel, M. Personalized medicine: Part 1: Evolution and development into theranostics. Pharm. Ther. 2010, 35, 560–576. [Google Scholar]
- Huang, P.H.; Kim, K.H.; Schermer, M. Ethical Issues of Digital Twins for Personalized Health Care Service: Preliminary Mapping Study. J. Med. Internet Res. 2022, 24, e33081. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, S.M.; Wildenhaus, K.; Bucher, A.; Byrd, B. Digital Twins and the Emerging Science of Self: Implications for Digital Health Experience Design and “Small” Data. Front. Comput. Sci. 2020, 2, 31. [Google Scholar] [CrossRef]
- Luther, W.; Baloian, N.; Biella, D.; Sacher, D. Digital Twins and Enabling Technologies in Museums and Cultural Heritage: An Overview. Sensors. 2023, 23, 1583. [Google Scholar] [CrossRef] [PubMed]
- Hernigou, P.; Olejnik, R.; Safar, A.; Martinov, S.; Hernigou, J.; Ferre, B. Digital twins, artificial intelligence, and machine learning technology to identify a real personalized motion axis of the tibiotalar joint for robotics in total ankle arthroplasty. Int. Orthop. 2021, 45, 2209–2217. [Google Scholar] [CrossRef]
- Gillette, K.; Gsell, M.A.; Prassl, A.J.; Karabelas, E.; Reiter, U.; Reiter, G.; Grandits, T.; Payer, C.; Štern, D.; Urschler, M.; et al. A framework for the generation of digital twins of cardiac electrophysiology from clinical 12-leads ECGs. Med. Image Anal. 2021, 71, 102080. [Google Scholar] [CrossRef] [PubMed]
- Vodovotz, Y. Towards systems immunology of critical illness at scale: From single cell ’omics to digital twins. Trends Immunol. 2023, 44, 345–355. [Google Scholar] [CrossRef] [PubMed]
- Chakshu, N.K.; Carson, J.; Sazonov, I.; Nithiarasu, P. A semi-active human digital twin model for detecting severity of carotid stenoses from head vibration—A coupled computational mechanics and computer vision method. Int. J. Numer. Methods Biomed. Eng. 2019, 35, e3180. [Google Scholar] [CrossRef]
- HamlAbadi, K.G.; Vahdati, M.; Saghiri, A.M.; Forestiero, A. Digital Twins in cancer: State-of-the-art and open research. In Proceedings of the 2021 IEEE/ACM Conference on Connected Health: Applications, Systems and Engineering Technologies (CHASE), Washington, DC, USA, 16–17 December 2021. [Google Scholar]
- Hernandez-Boussard, T.; Macklin, P.; Greenspan, E.J.; Gryshuk, A.L.; Stahlberg, E.; Syeda-Mahmood, T.; Shmulevich, I. Digital twins for predictive oncology will be a paradigm shift for precision cancer care. Nat. Med. 2021, 27, 2065–2066. [Google Scholar] [CrossRef]
- Laubenbacher, R.; Niarakis, A.; Helikar, T.; An, G.; Shapiro, B.; Malik-Sheriff, R.S.; Sego, T.J.; Knapp, A.; Macklin, P.; Glazier, J.A. Building digital twins of the human immune system: Toward a roadmap. Npj Digit. Med. 2022, 5, 64. [Google Scholar] [CrossRef] [PubMed]
- Pinton, P. Computational models in inflammatory bowel disease. Clin. Transl. Sci. 2022, 15, 824–830. [Google Scholar] [CrossRef]
- Khan, A.; Milne-Ives, M.; Meinert, E.; Iyawa, G.E.; Jones, R.B.; Josephraj, A.N. A scoping review of digital twins in the context of the COVID-19 pandemic. Biomed. Eng. Comput. Biol. 2022, 13, 11795972221102115. [Google Scholar] [CrossRef] [PubMed]
- Ahmadian, H.; Mageswaran, P.; Walter, B.A.; Blakaj, D.M.; Bourekas, E.C.; Mendel, E.; Marras, W.S.; Soghrati, S. Toward an artificial intelligence-assisted framework for reconstructing the digital twin of vertebra and predicting its fracture response. Int. J. Numer. Methods Biomed. Eng. 2022, 38, e3601. [Google Scholar] [CrossRef]
- Bjelland, Ø.; Rasheed, B.; Schaathun, H.G.; Pedersen, M.D.; Steinert, M.; Hellevik, A.I.; Bye, A.R.T. Towards a Digital Twin for Arthroscopic Knee Surgery: A Systematic Review. IEEE Access 2022, 10, 45029–45052. [Google Scholar] [CrossRef]
- Gawlikowski, J.; Tassi, C.R.N.; Ali, M.; Lee, J.; Humt, M.; Feng, J.; Kruspe, A.; Triebel, R.; Jung, P.; Roscher, R. A survey of uncertainty in deep neural networks. arXiv 2021, arXiv:2107.03342. [Google Scholar] [CrossRef]
- Stoean, C.; Stoean, R.; Atencia, M.; Abdar, M.; Velázquez-Pérez, L.; Khosravi, A.; Nahavandi, S.; Acharya, U.R.; Joya, G. Automated detection of presymptomatic conditions in Spinocerebellar Ataxia type 2 using Monte Carlo dropout and deep neural network techniques with electrooculogram signals. Sensors 2020, 20, 3032. [Google Scholar] [CrossRef] [PubMed]
- Ivanchuk, P.; Ivanchuk, M. One example of using Markov Chain Monte Carlo Method for predicting in medicine. Cardiol. Cardiovasc. Res. 2017, 1, 113–116. [Google Scholar]
- Yin, M.; Yazdani, A.; Karniadakis, G.E. One-dimensional modeling of fractional flow reserve in coronary artery disease: Uncertainty quantification and Bayesian optimization. Comput. Methods Appl. Mech. Eng. 2019, 353, 66–85. [Google Scholar] [CrossRef]
- Vyas, S.; Gupta, S.; Bhargava, D.; Boddu, R. Fuzzy Logic System Implementation on the Performance Parameters of Health Data Management Frameworks. J. Healthc. Eng. 2022, 2022, 9382322. [Google Scholar] [CrossRef] [PubMed]
- Vlamou, E.; Papadopoulos, B. Fuzzy logic systems and medical applications. AIMS Neurosci. 2019, 6, 266–272. [Google Scholar] [PubMed]
- Mauseth, R.; Wang, Y.; Dassau, E.; Kircher, R.; Matheson, D.; Zisser, H.; Jovanoviĉ, L.; Doyle, F.J. Proposed clinical application for tuning fuzzy logic controller of artificial pancreas utilizing a personalization factor. J. Diabetes Sci. Technol. 2010, 4, 913–922. [Google Scholar] [CrossRef]
- Hirschfeld, L.; Swanson, K.; Yang, K.; Barzilay, R.; Coley, C.W. Uncertainty Quantification Using Neural Networks for Molecular Property Prediction. J. Chem. Inf. Model. 2020, 60, 3770–3780. [Google Scholar] [CrossRef] [PubMed]
- Mae, Y.; Kumagai, W.; Kanamori, T. Uncertainty propagation for dropout-based Bayesian neural networks. Neural Netw. 2021, 144, 394–406. [Google Scholar] [CrossRef] [PubMed]
- Oszkinat, C.; Luczak, S.E.; Rosen, I.G. Uncertainty Quantification in Estimating Blood Alcohol Concentration From Transdermal Alcohol Level With Physics-Informed Neural Networks. IEEE Trans. Neural Netw. Learn. Syst. 2022. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Jiang, D.; Tao, B.; Jiang, G.; Sun, Y.; Kong, J.; Tong, X.; Zhao, G.; Chen, B. Genetic Algorithm-Based Trajectory Optimization for Digital Twin Robots. Front. Bioeng. Biotechnol. 2022, 9, 793782. [Google Scholar] [CrossRef] [PubMed]
- Held, T.; Klemmer, D.; Lässig, M. Survival of the simplest in microbial evolution. Nat. Commun. 2019, 10, 2472. [Google Scholar] [CrossRef]
- Katoch, S.; Chauhan, S.S.; Kumar, V. A review on genetic algorithm: Past, present, and future. Multimed. Tools Appl. 2021, 80, 8091–8126. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Yuan, H.; Matthew, P.; Ray, J.; Bagdasar, O.; Trovati, M. GORTS: Genetic algorithm based on one-by-one revision of two sides for dynamic travelling salesman problems. Soft Comput. 2020, 24, 7197–7210. [Google Scholar] [CrossRef]
- Fung, I.W.H.; Huang, C.; Tam, C.M. Application of GA optimization for solving precedent relationship problem in project scheduling. Math. Comput. Model 2013, 57, 2067–2081. [Google Scholar] [CrossRef]
- McCall, J. Genetic algorithms for modelling and optimisation. J. Comput. Appl. Math. 2005, 184, 205–222. [Google Scholar] [CrossRef]
- Hou, S.; Wen, H.; Feng, S.; Wang, H.; Li, Z. Application of Layered Coding Genetic Algorithm in Optimization of Unequal Area Production Facilities Layout. Comput. Intell. Neurosci. 2019, 2019, 3650923. [Google Scholar] [CrossRef]
- Liu, Y.; Zhu, N.; Li, M. Solving Many-Objective Optimization Problems by a Pareto-Based Evolutionary Algorithm With Preprocessing and a Penalty Mechanism. IEEE Trans. Cybern. 2021, 51, 5585–5594. [Google Scholar] [CrossRef] [PubMed]
- Lemmon, A.R.; Milinkovitch, M.C. The metapopulation genetic algorithm: An efficient solution for the problem of large phylogeny estimation. Proc. Natl. Acad. Sci. USA 2002, 99, 10516–10521. [Google Scholar] [CrossRef] [PubMed]
- Belavkin, R.V.; Channon, A.; Aston, E.; Aston, J.; Krašovec, R.; Knight, C.G. Monotonicity of fitness landscapes and mutation rate control. J. Math. Biol. 2016, 73, 1491–1524. [Google Scholar] [CrossRef] [PubMed]
- Ilan, Y. Second-Generation Digital Health Platforms: Placing the Patient at the Center and Focusing on Clinical Outcomes. Front. Digit. Health 2020, 2, 569178. [Google Scholar] [CrossRef] [PubMed]
- Speelman, C.P.; McGann, M. How Mean. is the Mean? Front. Psychol. 2013, 4, 451. [Google Scholar] [CrossRef]
- Ilan, Y. Making use of noise in biological systems. Prog. Biophys. Mol. Biol. 2023, 178, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Potruch, A.; Schwartz, A.; Ilan, Y. The role of bacterial translocation in sepsis: A new target for therapy. Therap Adv. Gastroenterol. 2022, 15, 17562848221094214. [Google Scholar] [CrossRef] [PubMed]
- Aubert, K.; Germaneau, A.; Rochette, M.; Ye, W.; Severyns, M.; Billot, M.; Rigoard, P.; Vendeuvre, T. Development of Digital Twins to Optimize Trauma Surgery and Postoperative Management. A Case Study Focusing on Tibial Plateau Fracture. Front. Bioeng. Biotechnol. 2021, 9, 722275. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, H.; Devoto, L. The potential of a digital twin in surgery. Surg. Innov. 2021, 28, 509–510. [Google Scholar] [CrossRef]
- Golse, N.; Joly, F.; Combari, P.; Lewin, M.; Nicolas, Q.; Audebert, C.; Samuel, D.; Allard, M.-A.; Cunha, A.S.; Castaing, D. Predicting the risk of post-hepatectomy portal hypertension using a digital twin: A clinical proof of concept. J. Hepatol. 2021, 74, 661–669. [Google Scholar] [CrossRef]
- Croatti, A.; Gabellini, M.; Montagna, S.; Ricci, A. On the Integration of Agents and Digital Twins in Healthcare. J. Med. Syst. 2020, 44, 161. [Google Scholar] [CrossRef]
- Abdullah, A.A.; Hassan, M.M.; Mustafa, Y.T. A review on bayesian deep learning in healthcare: Applications and challenges. IEEE Access 2022, 10, 36538–36562. [Google Scholar]
- Alizadehsani, R.; Roshanzamir, M.; Hussain, S.; Khsoravi, A.; Koohestani, A.; Zangooei, M.H.; Abdar, M.; Beykikhoshk, A.; Shoeibi, A.; Zare, A.; et al. Handling of uncertainty in medical data using machine learning and probability theory techniques: A review of 30 years (1991–2020). Ann. Oper. Res. 2021, 1–42. [Google Scholar] [CrossRef]
- Grieves, M.; Vickers, J. Digital Twin: Mitigating Unpredictable, Undesirable Emergent Behavior in Complex Systems. In Transdisciplinary Perspectives on Complex Systems: New Findings and Approaches; Kahlen, F.-J., Flumerfelt, S., Alves, A., Eds.; Springer: Cham, Switzerland, 2017; pp. 85–113. [Google Scholar]
- Jung, A.; Gsell, M.A.; Augustin, C.M.; Plank, G. An Integrated Workflow for Building Digital Twins of Cardiac Electromechanics-A Multi-Fidelity Approach for Personalising Active Mechanics. Mathematics 2022, 10, 823. [Google Scholar] [CrossRef]
- Kochunas, B.; Huan, X. Digital Twin Concepts with Uncertainty for Nuclear Power Applications. Energies 2021, 14, 4235. [Google Scholar] [CrossRef]
- Lin, L.; Bao, H.; Dinh, N. Uncertainty quantification and software risk analysis for digital twins in the nearly autonomous management and control systems: A review. Ann. Nucl. Energy 2021, 160, 108362. [Google Scholar] [CrossRef]
- Akkoyun, E.; Kwon, S.T.; Acar, A.C.; Lee, W.; Baek, S. Predicting abdominal aortic aneurysm growth using patient-oriented growth models with two-step Bayesian inference. Comput. Biol. Med. 2020, 117, 103620. [Google Scholar] [CrossRef] [PubMed]
- Yitmen, I.; Alizadehsalehi, S.; Akıner, İ.; Akıner, M.E. An Adapted Model of Cognitive Digital Twins for Building Lifecycle Management. Appl. Sci. 2021, 11, 4276. [Google Scholar] [CrossRef]
- Bykov, K.; Höhne, M.; Creosteanu, A.; Müller, K.-R.; Klauschen, F.; Nakajima, S.; Kloft, M. Explaining Bayesian Neural Networks. arXiv 2021, arXiv:2108.10346. [Google Scholar]
- Ilan, Y. Overcoming Compensatory Mechanisms toward Chronic Drug Administration to Ensure Long-Term, Sustainable Beneficial Effects. Mol. Ther. Methods Clin. Dev. 2020, 18, 335–344. [Google Scholar] [CrossRef]
- Ilan, Y. Improving Global Healthcare and Reducing Costs Using Second-Generation Artificial Intelligence-Based Digital Pills: A Market Disruptor. Int. J. Environ. Res. Public. Health 2021, 18, 811. [Google Scholar]
- Ilan, Y. Next-Generation Personalized Medicine: Implementation of Variability Patterns for Overcoming Drug Resistance in Chronic Diseases. J. Pers. Med. 2022, 12, 1303. [Google Scholar] [PubMed]
- Ching, T.; Himmelstein, D.S.; Beaulieu-Jones, B.K.; Kalinin, A.A.; Do, B.T.; Way, G.P.; Ferrero, E.; Agapow, P.-M.; Zietz, M.; Hoffman, M.; et al. Opportunities and obstacles for deep learning in biology and medicine. J. R. Soc. Interface 2018, 15, 20170387. [Google Scholar] [CrossRef]
- Hodas, N.O.; Stinis, P. Doing the Impossible: Why Neural Networks Can Be Trained at All. Front. Psychol. 2018, 9, 1185. [Google Scholar] [CrossRef] [PubMed]
- Parisi, G.I.; Kemker, R.; Part, J.L.; Kanan, C.; Wermter, S. Continual lifelong learning with neural networks: A review. Neural Netw. 2019, 113, 54–71. [Google Scholar] [CrossRef]
- Chen, X.; Liu, C.; Zhao, Y.; Jia, Z.; Jin, G. Improving adversarial robustness of Bayesian neural networks via multi-task adversarial training. Inf. Sci. 2022, 592, 156–173. [Google Scholar] [CrossRef]
- Gelman, R.; Berg, M.; Ilan, Y. A Subject-Tailored Variability-Based Platform for Overcoming the Plateau Effect in Sports Training: A Narrative Review. Int. J. Environ. Res. Public. Health 2022, 19, 1722. [Google Scholar]
- Kessler, A.; Weksler-Zangen, S.; Ilan, Y. Role of the Immune System and the Circadian Rhythm in the Pathogenesis of Chronic Pancreatitis: Establishing a Personalized Signature for Improving the Effect of Immunotherapies for Chronic Pancreatitis. Pancreas 2020, 49, 1024–1032. [Google Scholar] [CrossRef] [PubMed]
- Ishay, Y.; Kolben, Y.; Kessler, A.; Ilan, Y. Role of circadian rhythm and autonomic nervous system in liver function: A hypothetical basis for improving the management of hepatic encephalopathy. Am. J. Physiol.-Gastrointest. Liver Physiol. 2021, 321, G400–G412. [Google Scholar] [PubMed]
- Kolben, Y.; Weksler-Zangen, S.; Ilan, Y. Adropin as a potential mediator of the metabolic system-autonomic nervous system-chronobiology axis: Implementing a personalized signature-based platform for chronotherapy. Obes. Rev. 2021, 22, e13108. [Google Scholar]
- Kenig, A.; Kolben, Y.; Asleh, R.; Amir, O.; Ilan, Y. Improving Diuretic Response in Heart Failure by Implementing a Patient-Tailored Variability and Chronotherapy-Guided Algorithm. Front. Cardiovasc. Med. 2021, 8, 695547. [Google Scholar] [CrossRef] [PubMed]
- Azmanov, H.; Ross, E.L.; Ilan, Y. Establishment of an Individualized Chronotherapy, Autonomic Nervous System, and Variability-Based Dynamic Platform for Overcoming the Loss of Response to Analgesics. Pain. Physician 2021, 24, 243–252. [Google Scholar] [PubMed]
- Potruch, A.; Khoury, S.T.; Ilan, Y. The role of chronobiology in drug-resistance epilepsy: The potential use of a variability and chronotherapy-based individualized platform for improving the response to anti-seizure drugs. Seizure 2020, 80, 201–211. [Google Scholar] [PubMed]
- Isahy, Y.; Ilan, Y. Improving the long-term response to antidepressants by establishing an individualized platform based on variability and chronotherapy. Int. J. Clin. Pharmacol. Ther. 2021, 59, 768–774. [Google Scholar] [CrossRef] [PubMed]
- Khoury, T.; Ilan, Y. Introducing Patterns of Variability for Overcoming Compensatory Adaptation of the Immune System to Immunomodulatory Agents: A Novel Method for Improving Clinical Response to Anti-TNF Therapies. Front. Immunol. 2019, 10, 2726. [Google Scholar] [PubMed]
- Khoury, T.; Ilan, Y. Platform introducing individually tailored variability in nerve stimulations and dietary regimen to prevent weight regain following weight loss in patients with obesity. Obes. Res. Clin. Pract. 2021, 15, 114–123. [Google Scholar] [CrossRef]
- Kenig, A.; Ilan, Y. A Personalized Signature and Chronotherapy-Based Platform for Improving the Efficacy of Sepsis Treatment. Front. Physiol. 2019, 10, 1542. [Google Scholar] [CrossRef]
- Ilan, Y. Why targeting the microbiome is not so successful: Can randomness overcome the adaptation that occurs following gut manipulation? Clin. Exp. Gastroenterol. 2019, 12, 209–217. [Google Scholar]
- Gelman, R.; Bayatra, A.; Kessler, A.; Schwartz, A.; Ilan, Y. Targeting SARS-CoV-2 receptors as a means for reducing infectivity and improving antiviral and immune response: An algorithm-based method for overcoming resistance to antiviral agents. Emerg. Microbes Infect. 2020, 9, 1397–1406. [Google Scholar]
- Ishay, Y.; Potruch, A.; Schwartz, A.; Berg, M.; Jamil, K.; Agus, S.; Ilan, Y. A digital health platform for assisting the diagnosis and monitoring of COVID-19 progression: An adjuvant approach for augmenting the antiviral response and mitigating the immune-mediated target organ damage. Biomed. Pharmacother. 2021, 143, 112228. [Google Scholar] [PubMed]
- Ilan, Y.; Spigelman, Z. Establishing patient-tailored variability-based paradigms for anti-cancer therapy: Using the inherent trajectories which underlie cancer for overcoming drug resistance. Cancer Treat. Res. Commun. 2020, 25, 100240. [Google Scholar]
- Hurvitz, N.; Azmanov, H.; Kesler, A.; Ilan, Y. Establishing a second-generation artificial intelligence-based system for improving diagnosis, treatment, and monitoring of patients with rare diseases. Eur. J. Hum. Genet. 2021, 29, 1485–1490. [Google Scholar] [CrossRef]
- Ilan, Y. Digital Medical Cannabis as Market Differentiator: Second-Generation Artificial Intelligence Systems to Improve Response. Front. Med. 2021, 8, 788777. [Google Scholar]
- Azmanov, H.; Bayatra, A.; Ilan, Y. Digital Analgesic Comprising a Second-Generation Digital Health System: Increasing Effectiveness by Optimizing the Dosing and Minimizing Side Effects. J. Pain. Res. 2022, 15, 1051–1060. [Google Scholar]
- Hurvitz, N.; Elkhateeb, N.; Sigawi, T.; Rinsky-Halivni, L.; Ilan, Y. Improving the effectiveness of anti-aging modalities by using the constrained disorder principle-based management algorithms. Front. Aging 2022, 3, 1044038. [Google Scholar] [CrossRef] [PubMed]
- Gelman, R.; Hurvitz, N.; Nesserat, R.; Kolben, Y.; Nachman, D.; Jamin, K.; Agus, S.; Asleh, R.; Amir, O.; Berg, M.; et al. A second-generation artificial intelligence-based therapeutic regimen improves diuretic resistance in heart failure: Results of a feasibility open-labeled clinical trial. Biomed. Pharmacother. 2023, 161, 114334. [Google Scholar]
- Kolben, Y.; Azmanov, H.; Gelman, R.; Dror, D.; Ilan, Y. Using chronobiology-based second-generation artificial intelligence digital system for overcoming antimicrobial drug resistance in chronic infections. Ann. Med. 2023, 55, 311–318. [Google Scholar]
- Sigawi, T.; Lehmann, H.; Hurvitz, N.; Ilan, Y. Constrained disorder principle-based second-generation algorithms implement quantified variability signatures to improve the function of complex systems. J. Bioinform. Syst. Biol. 2023, 6, 82–89. [Google Scholar]
- Jospin, L.V.; Laga, H.; Boussaid, F.; Buntine, W.; Bennamound, M. Hands-on Bayesian neural networks—A tutorial for deep learning users. IEEE Comput. Intell. Mag. 2022, 17, 29–48. [Google Scholar] [CrossRef]
- Lipková, J.; Angelikopoulos, P.; Wu, S.; Alberts, E.; Wiestler, B.; Diehl, C.; Preibisch, C.; Pyka, T.; Combs, S.E.; Hadjidoukas, P.; et al. Personalized radiotherapy design for glioblastoma: Integrating mathematical tumor models, multimodal scans, and bayesian inference. IEEE Trans. Med. Imaging 2019, 38, 1875–1884. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sigawi, T.; Ilan, Y. Using Constrained-Disorder Principle-Based Systems to Improve the Performance of Digital Twins in Biological Systems. Biomimetics 2023, 8, 359. https://doi.org/10.3390/biomimetics8040359
Sigawi T, Ilan Y. Using Constrained-Disorder Principle-Based Systems to Improve the Performance of Digital Twins in Biological Systems. Biomimetics. 2023; 8(4):359. https://doi.org/10.3390/biomimetics8040359
Chicago/Turabian StyleSigawi, Tal, and Yaron Ilan. 2023. "Using Constrained-Disorder Principle-Based Systems to Improve the Performance of Digital Twins in Biological Systems" Biomimetics 8, no. 4: 359. https://doi.org/10.3390/biomimetics8040359
APA StyleSigawi, T., & Ilan, Y. (2023). Using Constrained-Disorder Principle-Based Systems to Improve the Performance of Digital Twins in Biological Systems. Biomimetics, 8(4), 359. https://doi.org/10.3390/biomimetics8040359






