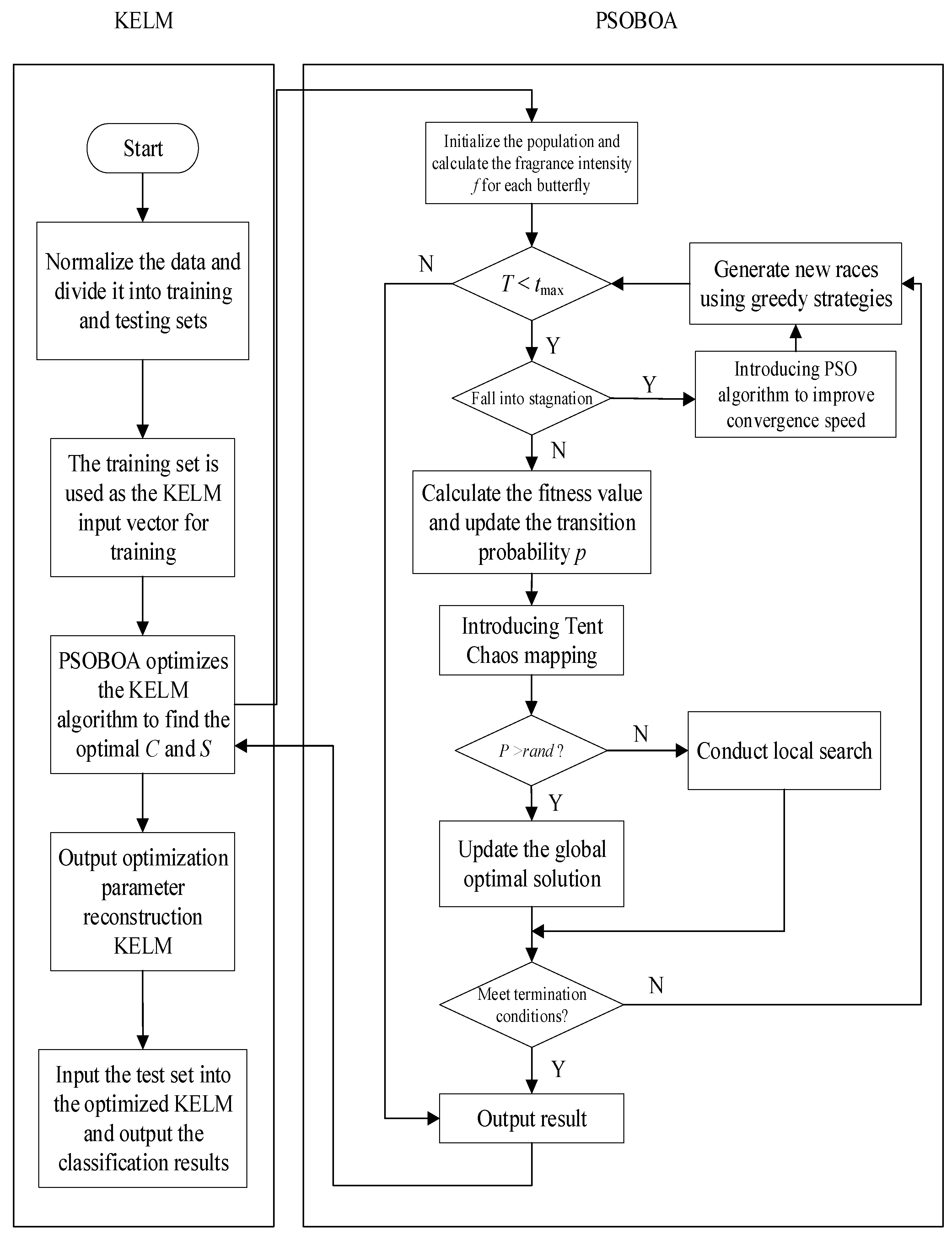
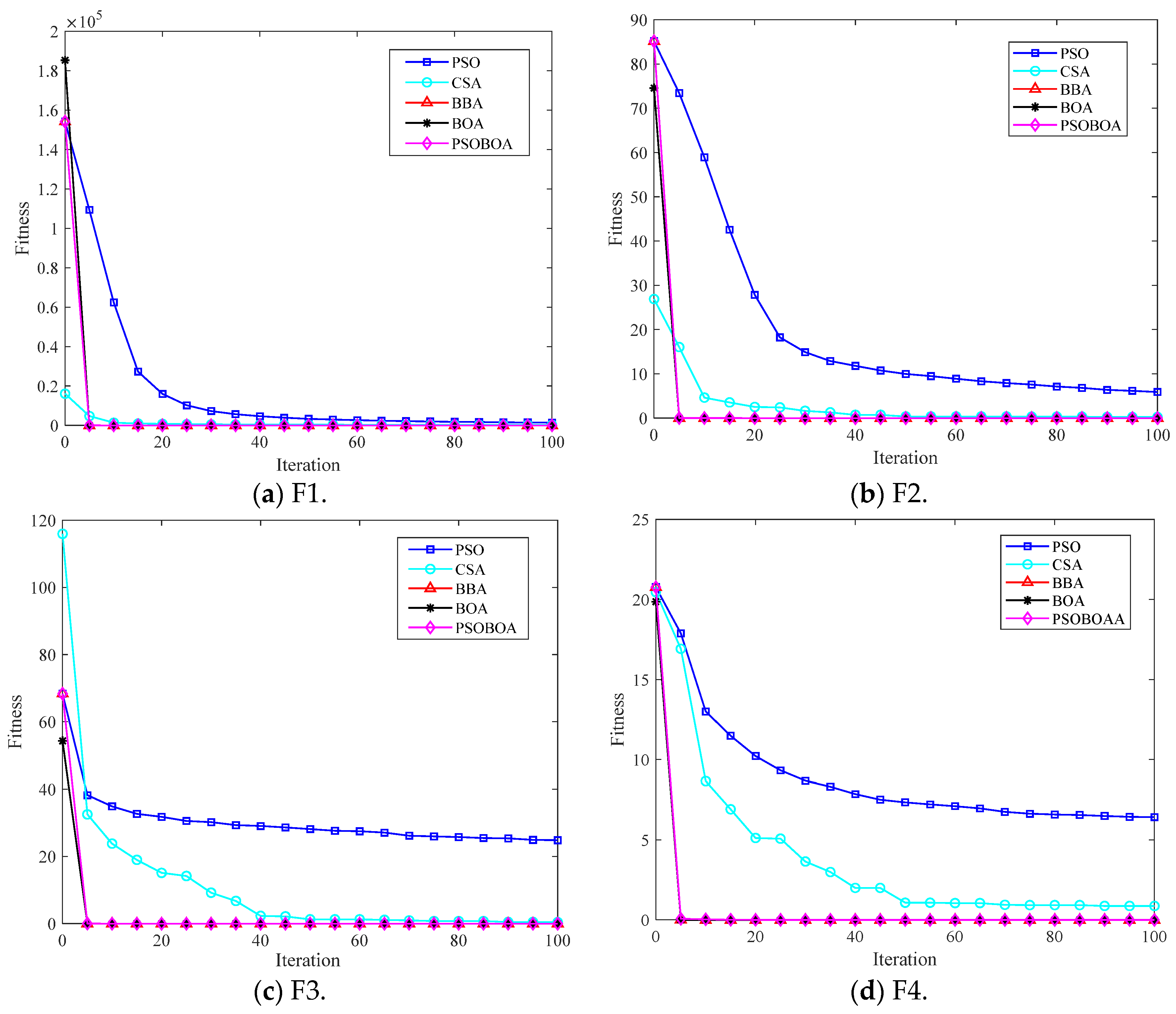
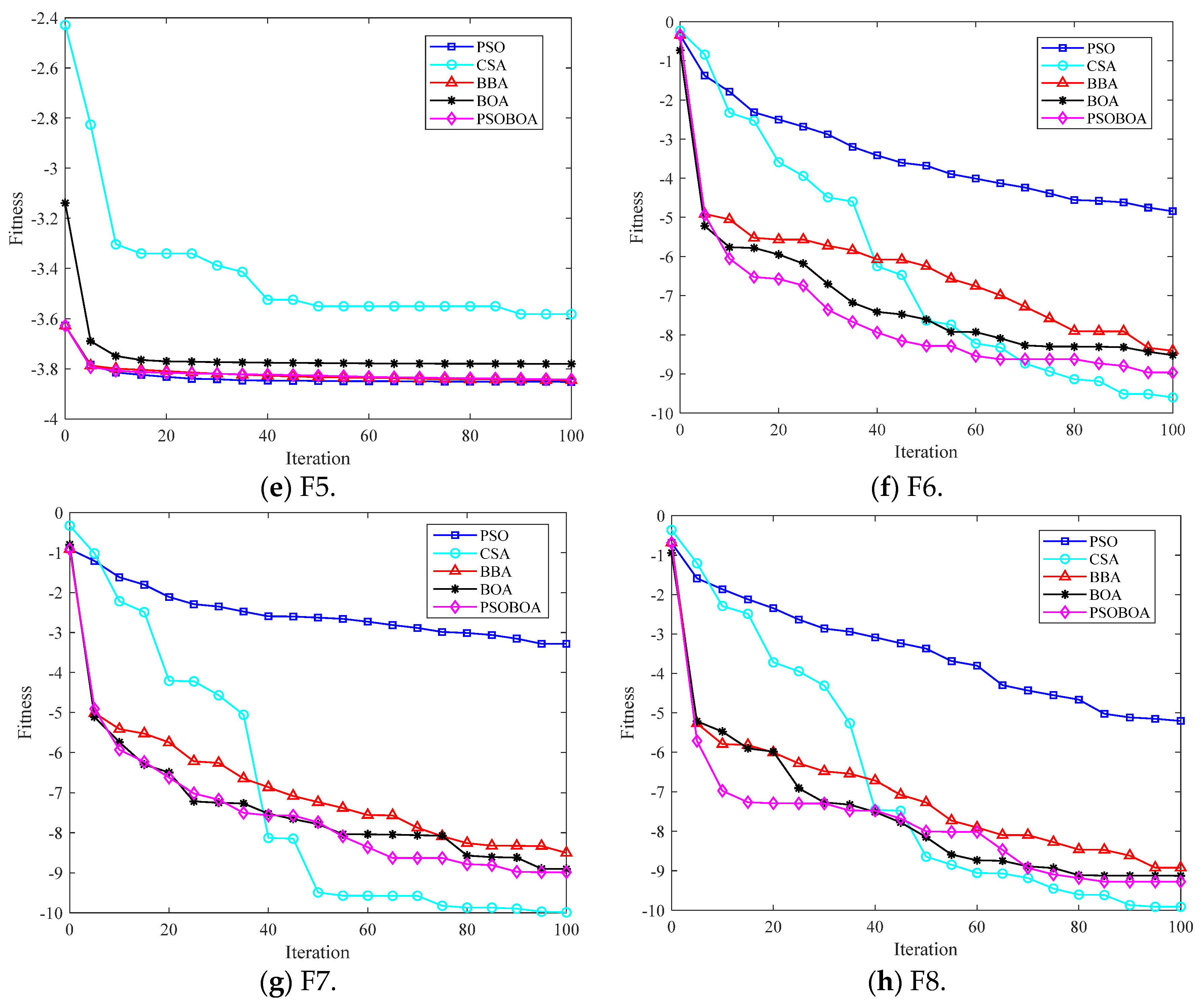
6.3. Algorithm Test Comparison and Result Analysis
In order to verify the effectiveness of the proposed method, this part experiments on the PSOBOA-KELM algorithm on seven classification data sets, which are BreastEW, CongressEW, Hepatitis, JPNdata, Parkinson, SpectEW, and Wdbc. The data sets are from the UCI Machine Learning Library (
http://archive.ics.uci.edu/mL/datasets, accessed on 1 October 2022). These data sets are mainly divided into binary classification problems, multi-classification problems, and regression fitting problems. The Breastcancer dataset has 699 data, including 9 features and two categories; the Parkinson dataset has 195 data, including 23 features and two categories; the BreastEW dataset has 569 data, including 30 features and two categories; and the Dermatology dataset has 358 data, including 35 features and six categories. The experiments selected seven real datasets widely used for multi-label classification. The learning factor (
c1 =
c2 = 2) in the important parameters of the particle swarm optimization algorithm was the inertia weight factor
w1 = 0.9 and
w2 = 0.4.
Table 2 summarizes the data size, attribute dimension, number of tags, and cardinality of the seven datasets. The specific description information of the dataset is shown in
Table 2.
The data set had to be preprocessed before the experiment, and certain features were missing. These records were averaged in this experiment to guarantee the accuracy of the sample data. To reduce the gap between the eigenvalues and prevent the larger eigenvalues from adversely affecting the smaller eigenvalues, we normalized each eigenvalue to the [−1, 1] interval. The normalized calculation formula is:
where
x is the original value of the data,
is the normalized value,
is the maximum value in feature
a, and
is the minimum value in feature
a.
At the same time, in order to obtain an unbiased estimate of the algorithm’s generalization accuracy, k-fold CV is generally used to evaluate the classification accuracy. In this method, all test sets are independent, which can improve the reliability of the results. In this study, the k value was set to 10, that is, each experimental data set was divided into 10 subsets, one of which was taken as the test set each time, and the rest was used as the training set, and then the average value of 10 experiments was calculated as the result of the ten-fold crossover. Each of the above classification experiments was run independently 20 times to ensure the stability of the algorithm.
The parameter settings of the contrast swarm intelligent optimization algorithm involved in this paper are shown in
Table 3.
From the results of
Table 4 and
Table 5, it can be seen that the method proposed in this paper is accurate in accuracy, precision, F-measure, sensitivity, specificity, and MCC. The indicator performs significantly better than other comparative feature selection methods.
For the accuracy indicator, the PSOBOA-KELM feature selection method proposed in this paper has an accuracy rate of 96.49%, 96.56%, 87.87%, 83.96%, and 90% on BreastEW, CongressEW, hepatitisfulldata, JPNdata, Parkinson, SpectEW, and wdbc data sets, respectively. Compared with the PSO-KELM, BBA-KELM, and BOA-KELM feature selection methods, the method proposed in this paper has the highest accuracy rate. For example, in the BreastEW dataset, the accuracy of the method proposed in this paper is 0.91% higher than the PSO-KELM method, 0.91% higher than the BBA-KELM method, and 0.03% higher than the BOA-KELM method. For the precision indicator, on the BreastEW, CongressEW, hepatitisfulldata, JPNdata, Parkinson, SpectEW, and wdbc data sets, the accuracy of the PSOBOA-KELM feature selection method proposed in this paper is 95.98%, 100%, 100%, 78.89%, 92.86%, 87.5%, and 100%, respectively. Compared with the PSO-KELM, BBA-KELM, and BOA-KELM feature selection methods, the proposed method has the highest accuracy. For example, in the BreastEW dataset, the accuracy of the method proposed in this paper is 1.31% higher than the PSO-KELM method, 1.46% higher than the BBA-KELM method, and 1.31% higher than the BOA-KELM method. For the F-measure index, on the BreastEW, CongressEW, hepatitisfulldata, JPNdata, Parkinson, SpectEW, and wdbc data sets, the F-measure of the PSOBOA-KELM feature selection method proposed in this paper is 97.3%, 97.10%, 70.83%, 84.03%, 93.75%, 33.33%, and 96.4%, respectively. Compared with the PSO-KELM, BBA-KELM, and BOA-KELM feature selection methods, the proposed method’s F-measure works better. For example, in the CongressEW dataset, the F-measure value of the method proposed in this paper is 2.76% higher than the PSO-KELM method, 4.79% higher than the BBA-KELM method, and 0.95% higher than the BOA-KELM method.
For the sensitivity index, on the BreastEW, CongressEW, hepatitisfulldata, JPNdata, Parkinson, SpectEW, and wdbc data sets, the sensitivity values of the PSOBOA-KELM feature selection method proposed in this paper are 100%, 94.37%, 58.33%, 93.75%, 100%, 20%, and 93.07%, respectively. Compared with the PSO-KELM, BBA-KELM, and BOA-KELM feature selection methods, the proposed method has a higher sensitivity value. For example, in the CongressEW dataset, the sensitivity value of the method proposed in this paper is 1.92% higher than the PSO-KELM method, 1.78% higher than the BBA-KELM method, and 1.78% higher than the BOA-KELM method. Compared with the PSO-KELM, BBA-KELM, and BOA-KELM feature selection methods, the method proposed in this paper has a higher specificity value. For example, in the BreastEW dataset, the specificity value of the method proposed in this paper is 1.96% higher than the PSO-KELM method, 2.38% higher than the BBA-KELM method, and 2.38% higher than the BOA-KELM method. For the MCC index, the PSOBOA-KELM feature selection method proposed in this paper has the MCC values of 92.58%, 93.16%, 66.39%, 68.1%, and 72.81% on the BreastEW, CongressEW, hepatitisfulldata, JPNdata, Parkinson, SpectEW, and wdbc data sets, respectively. Compared with the PSO-KELM, BBA-KELM, and BOA-KELM feature selection methods, the method proposed in this paper has a higher MCC value. For example, in the wdbc data set, the MCC value of the method proposed in this paper is 3.64% higher than the PSO-KELM method, 5.61% higher than the BBA-KELM method, and 1.93% higher than the BOA-KELM method.
In addition, in order to compare the performance of these four algorithms more intuitively, as shown in
Figure 3, the performance evaluation indicators of these four methods are compared in detail. At the same time, the calculation and simulation time consumption of the four algorithms in the seven data sets are also presented, as shown in
Figure 4.
According to the experimental findings, the PSOBOA-KELM technique has an acceptable performance in terms of classification, and the calculation and simulation times are not too long. It may choose an acceptable and constrained feature subset, and its classification performance is noticeably better than that of comparable approaches. The algorithm also performs well when it comes to the challenge of classifying various data sets. It takes a fair amount of time and produces an excellent classification accuracy. The comparison of eight datasets is shown in
Table 6 and
Table 7. In addition, in order to compare the performance of these four algorithms more intuitively, the performance evaluation indicators of these four methods are compared in detail, as shown in
Figure 5. At the same time, the calculation and simulation time consumption of the four algorithms in the seven data sets are also presented, as shown in
Figure 6.
The four algorithms were tested in the Australian, Breastcancer, Dermatology, HeartEW, Diabetes, Glass, Heart, and Vote8 data sets regarding accuracy, precision, F-measure, sensitivity, specificity, MCC, and other six indicators, and achieved a good classification performance. The calculation and simulation time were also relatively short for the PSOBOA-KELM method. It may choose an acceptable and constrained feature subset, and its classification performance is noticeably better than that of comparable approaches. In addition to achieving an improved classification accuracy, the algorithm also performs well while classifying data from various data sets.
In addition, the simulation experiment comparison of the Sinc function was conducted. The four algorithms were compared by fitting the Sinc function. The expression of the Sinc function is as follows:
We set to generate 2000 [−10, 10] uniformly distributed data sets
x, calculated 2000 data sets
, and then generated 2000 [−0.2, 0.2] uniformly distributed noise
ε. Let the training set be
and then generate another set of 2000 data sets
as the test set. In addition, the root-mean-square error (
RMSE), mean absolute error (
MAE), and relative standard deviation (
RSD) were used as the evaluation indicators for error analysis. The calculation formulas of the three indicators are as follows:
where parameter
represents the measured value,
represents the predicted value, parameter
N is the number of samples, parameter
=
is the absolute error, and the numerator and denominator of RSD are in the form of standard deviation. The comparison of Sinc function fitting results is shown in
Table 8.
It can be seen from
Table 8 that, calculated by the PSO-KELM algorithm, the index values of RMSE and MAE are the largest, and the index value of RSD is closer to the smallest, and the performance of the test results is poor. The index value is even smaller, and the performance of the test result is average. The RMSE and MAE index values of the BOA-KELM algorithm are smaller, the RSD index values are closer to larger, and the test results have a better performance. The PSOBOA-KELM algorithm has the smallest RMSE and MAE index values, the RSD index value is closer to the largest, and the test results have the best performance. It shows that the error of the PSOBOA-KELM model is relatively smaller, and the prediction accuracy is better than that of the PSO-KELM, BBA-KELM, and BOA-KELM algorithms. At the same time, this can also be known from the data change trend in
Table 2, which indicates that the PSOBOA-KELM algorithm has the best performance, and optimizing the KELM regularization parameter
C and kernel function
S can improve the prediction accuracy of the KELM model.