Current Advances and Future Perspectives of Liver-on-a-Chip Platforms Incorporating Dynamic Fluid Flow
Abstract
1. Introduction
2. Cell Culture Platforms for Hepatic Cells
2.1. Traditional Cell Culture Platforms
2.2. Organ-on-a-Chip Platform
3. Liver-on-a-Chip Platforms Utilizing Static Culture Techniques
4. Liver-on-a-Chip Platforms with Controlled Dynamic Fluidic Flow
4.1. Dynamic Fluid Flow Driven by Gravity Gradient
4.2. Unidirectional Medium Perfusion by Pump Systems
4.3. Medium Circulation Mediated by Pump Systems
5. Multi-Organ-on-a-Chip Systems Incorporating a Liver Compartment
6. Conclusions and Future Perspectives
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Rui, L. Energy metabolism in the liver. Compr. Physiol. 2014, 4, 177–197. [Google Scholar] [CrossRef] [PubMed]
- Petersen, M.C.; Vatner, D.F.; Shulman, G.I. Regulation of hepatic glucose metabolism in health and disease. Nat. Rev. Endocrinol. 2017, 13, 572–587. [Google Scholar] [CrossRef] [PubMed]
- Adeva-Andany, M.M.; Pérez-Felpete, N.; Fernández-Fernández, C.; Donapetry-García, C.; Pazos-García, C. Liver glucose metabolism in humans. Biosci. Rep. 2016, 36, e00416. [Google Scholar] [CrossRef] [PubMed]
- Gebhardt, R.; Matz-Soja, M. Liver zonation: Novel aspects of its regulation and its impact on homeostasis. World J. Gastroenterol. 2014, 20, 8491–8504. [Google Scholar] [CrossRef] [PubMed]
- Panwar, A.; Das, P.; Tan, L.P. 3D Hepatic Organoid-Based Advancements in LIVER Tissue Engineering. Bioengineering 2021, 8, 185. [Google Scholar] [CrossRef]
- Liu, J.; Du, Y.; Xiao, X.; Tan, D.; He, Y.; Qin, L. Construction of in vitro liver-on-a-chip models and application progress. Biomed. Eng. OnLine 2024, 23, 33. [Google Scholar] [CrossRef]
- Tonon, F.; Giobbe, G.G.; Zambon, A.; Luni, C.; Gagliano, O.; Floreani, A.; Grassi, G.; Elvassore, N. In vitro metabolic zonation through oxygen gradient on a chip. Sci. Rep. 2019, 9, 13557. [Google Scholar] [CrossRef]
- Cunningham, R.P.; Porat-Shliom, N. Liver Zonation—Revisiting Old Questions with New Technologies. Front. Physiol. 2021, 12, 732929. [Google Scholar] [CrossRef]
- Vekemans, K.; Braet, F. Structural and functional aspects of the liver and liver sinusoidal cells in relation to colon carcinoma metastasis. World J. Gastroenterol. 2005, 11, 5095–5102. [Google Scholar] [CrossRef]
- Messelmani, T.; Morisseau, L.; Sakai, Y.; Legallais, C.; Le Goff, A.; Leclerc, E.; Jellali, R. Liver organ-on-chip models for toxicity studies and risk assessment. Lab Chip 2022, 22, 2423–2450. [Google Scholar] [CrossRef]
- Buyl, K.; De Kock, J.; Bolleyn, J.; Rogiers, V.; Vanhaecke, T. Measurement of Albumin Secretion as Functionality Test in Primary Hepatocyte Cultures. Methods Mol. Biol. 2015, 1250, 303–308. [Google Scholar] [CrossRef] [PubMed]
- Bolleyn, J.; Rogiers, V.; Vanhaecke, T. Functionality Testing of Primary Hepatocytes in Culture by Measuring Urea Synthesis. Methods Mol. Biol. 2015, 1250, 317–321. [Google Scholar] [CrossRef] [PubMed]
- Gracia-Sancho, J.; Caparrós, E.; Fernández-Iglesias, A.; Francés, R. Role of liver sinusoidal endothelial cells in liver diseases. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 411–431. [Google Scholar] [CrossRef]
- Braet, F.; Wisse, E. Structural and functional aspects of liver sinusoidal endothelial cell fenestrae: A review. Comp. Hepatol. 2002, 1, 1. [Google Scholar] [CrossRef]
- Fujita, T.; Narumiya, S. Roles of hepatic stellate cells in liver inflammation: A new perspective. Inflamm. Regen. 2016, 36, 1. [Google Scholar] [CrossRef]
- Ju, C.; Tacke, F. Hepatic macrophages in homeostasis and liver diseases: From pathogenesis to novel therapeutic strategies. Cell. Mol. Immunol. 2016, 13, 316–327. [Google Scholar] [CrossRef] [PubMed]
- Kolios, G.; Valatas, V.; Kouroumalis, E. Role of Kupffer cells in the pathogenesis of liver disease. World J. Gastroenterol. 2006, 12, 7413–7420. [Google Scholar] [CrossRef]
- Dixon, L.J.; Barnes, M.; Tang, H.; Pritchard, M.T.; Nagy, L.E. Kupffer cells in the liver. Compr. Physiol. 2013, 3, 785–797. [Google Scholar] [CrossRef] [PubMed]
- Jang, M.; Kim, H.N. From Single- to Multi-organ-on-a-Chip System for Studying Metabolic Diseases. BioChip J. 2023, 17, 133–146. [Google Scholar] [CrossRef]
- Kaur, S.; Kidambi, S.; Ortega-Ribera, M.; Thuy, L.T.T.; Nieto, N.; Cogger, V.C.; Xie, W.F.; Tacke, F.; Gracia-Sancho, J. In Vitro Models for the Study of Liver Biology and Diseases: Advances and Limitations. Cell Mol. Gastroenterol. Hepatol. 2023, 15, 559–571. [Google Scholar] [CrossRef]
- Ramos, M.J.; Bandiera, L.; Menolascina, F.; Fallowfield, J.A. In vitro models for non-alcoholic fatty liver disease: Emerging platforms and their applications. iScience 2022, 25, 103549. [Google Scholar] [CrossRef]
- Montgomery, M.K.; Hallahan, N.L.; Brown, S.H.; Liu, M.; Mitchell, T.W.; Cooney, G.J.; Turner, N. Mouse strain-dependent variation in obesity and glucose homeostasis in response to high-fat feeding. Diabetologia 2013, 56, 1129–1139. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Zhang, W.; Qiu, H.; Xiao, C.; Shi, J.; Reid, L.M.; He, Z. Mouse Models of Liver Parenchyma Injuries and Regeneration. Front. Cell Dev. Biol. 2022, 10, 903740. [Google Scholar] [CrossRef] [PubMed]
- Van Herck, M.A.; Vonghia, L.; Francque, S.M. Animal Models of Nonalcoholic Fatty Liver Disease—A Starter’s Guide. Nutrients 2017, 9, 1072. [Google Scholar] [CrossRef] [PubMed]
- Seok, J.; Warren, H.S.; Cuenca, A.G.; Mindrinos, M.N.; Baker, H.V.; Xu, W.; Richards, D.R.; McDonald-Smith, G.P.; Gao, H.; Hennessy, L.; et al. Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc. Natl. Acad. Sci. USA 2013, 110, 3507–3512. [Google Scholar] [CrossRef]
- Ingber, D.E. Human organs-on-chips for disease modelling, drug development and personalized medicine. Nat. Rev. Genet. 2022, 23, 467–491. [Google Scholar] [CrossRef]
- Malinowska, J.M.; Palosaari, T.; Sund, J.; Carpi, D.; Bouhifd, M.; Weber, R.J.M.; Whelan, M.; Viant, M.R. Integrating in vitro metabolomics with a 96-well high-throughput screening platform. Metabolomics 2022, 18, 11. [Google Scholar] [CrossRef]
- Kim, N.-J.; Bang, J.-H.; Yi, H.; Ku, H.-O.; Kim, J.-S.; Kim, J.-Y.; Jeon, B.-S. Comparison of in vitro models for drug-induced liver injury assessment. J. Biomed. Transl. Res. 2024, 25, 53–67. [Google Scholar] [CrossRef]
- Dufva, M. A quantitative meta-analysis comparing cell models in perfused organ on a chip with static cell cultures. Sci. Rep. 2023, 13, 8233. [Google Scholar] [CrossRef]
- Zhao, J.; Zhou, L.; Zhang, Y.; Cheng, J.; Zeng, Y.; Li, X. TDF and TAF inhibit liver cancer cell migration, invasion via p7TP3. Sci. Rep. 2024, 14, 8161. [Google Scholar] [CrossRef]
- Arora, M. Cell Culture Media: A Review. Mater. Methods 2013, 3, 175. [Google Scholar] [CrossRef]
- Vis, M.A.M.; Ito, K.; Hofmann, S. Impact of Culture Medium on Cellular Interactions in in vitro Co-culture Systems. Front. Bioeng. Biotechnol. 2020, 8, 911. [Google Scholar] [CrossRef]
- Natarajan, V.; Simoneau, C.R.; Erickson, A.L.; Meyers, N.L.; Baron, J.L.; Cooper, S.; McDevitt, T.C.; Ott, M. Modelling T-cell immunity against hepatitis C virus with liver organoids in a microfluidic coculture system. Open Biol. 2022, 12, 210320. [Google Scholar] [CrossRef] [PubMed]
- Vega, J.M.H.; Hong, H.J.; Loutherback, K.; Stybayeva, G.; Revzin, A. A Microfluidic Device for Long-Term Maintenance of Organotypic Liver Cultures. Adv. Mater. Technol. 2023, 8, 2201121. [Google Scholar] [CrossRef]
- Zhao, X.; Gao, W.; Yin, J.; Fan, W.; Wang, Z.; Hu, K.; Mai, Y.; Luan, A.; Xu, B.; Jin, Q. A high-precision thermometry microfluidic chip for real-time monitoring of the physiological process of live tumour cells. Talanta 2021, 226, 122101. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Huang, Z.; Gao, W.; Gao, W.; He, R.; Li, Y.; Crawford, R.; Zhou, Y.; Xiao, L.; Xiao, Y. Current Advances in 3D Dynamic Cell Culture Systems. Gels 2022, 8, 829. [Google Scholar] [CrossRef]
- Tsai, H.-H.; Yang, K.-C.; Wu, M.-H.; Chen, J.-C.; Tseng, C.-L. The Effects of Different Dynamic Culture Systems on Cell Proliferation and Osteogenic Differentiation in Human Mesenchymal Stem Cells. Int. J. Mol. Sci. 2019, 20, 4024. [Google Scholar] [CrossRef]
- Mohamadian Namaqi, M.; Moll, F.; Wiedemeier, S.; Grodrian, A.; Lemke, K. Dynamic cell culture modulates colon cancer cell migration in a novel 3D cell culture system. Sci. Rep. 2024, 14, 18851. [Google Scholar] [CrossRef]
- Mahdavi, R.; Hashemi-Najafabadi, S.; Ghiass, M.A.; Adiels, C.B. Microfluidic design for in-vitro liver zonation—A numerical analysis using COMSOL Multiphysics. Med. Biol. Eng. Comput. 2024, 62, 121–133. [Google Scholar] [CrossRef]
- Esch, M.B.; Prot, J.-M.; Wang, Y.I.; Miller, P.; Llamas-Vidales, J.R.; Naughton, B.A.; Applegate, D.R.; Shuler, M.L. Multi-cellular 3D human primary liver cell culture elevates metabolic activity under fluidic flow. Lab Chip 2015, 15, 2269–2277. [Google Scholar] [CrossRef]
- Deng, J.; Zhang, X.; Chen, Z.; Luo, Y.; Lu, Y.; Liu, T.; Wu, Z.; Jin, Y.; Zhao, W.; Lin, B. A cell lines derived microfluidic liver model for investigation of hepatotoxicity induced by drug-drug interaction. Biomicrofluidics 2019, 13, 024101. [Google Scholar] [CrossRef]
- Kwon, D.; Choi, G.; Park, S.-A.; Cho, S.; Cho, S.; Ko, S. Liver Acinus Dynamic Chip for Assessment of Drug-Induced Zonal Hepatotoxicity. Biosensors 2022, 12, 445. [Google Scholar] [CrossRef] [PubMed]
- Du, K.; Li, S.; Li, C.; Li, P.; Miao, C.; Luo, T.; Qiu, B.; Ding, W. Modeling nonalcoholic fatty liver disease on a liver lobule chip with dual blood supply. Acta Biomater. 2021, 134, 228–239. [Google Scholar] [CrossRef]
- Jiao, D.; Xie, L.; Xing, W. A pumpless liver-on-a-chip for drug hepatotoxicity analysis. Analyst 2024, 149, 4675–4686. [Google Scholar] [CrossRef]
- Ferrari, E.; Monti, E.; Cerutti, C.; Visone, R.; Occhetta, P.; Griffith, L.G.; Rasponi, M. A method to generate perfusable physiologic-like vascular channels within a liver-on-chip model. Biomicrofluidics 2023, 17, 064103. [Google Scholar] [CrossRef] [PubMed]
- Freag, M.S.; Namgung, B.; Reyna Fernandez, M.E.; Gherardi, E.; Sengupta, S.; Jang, H.L. Human Nonalcoholic Steatohepatitis on a Chip. Hepatol. Commun. 2021, 5, 217–233. [Google Scholar] [CrossRef] [PubMed]
- Bircsak, K.M.; DeBiasio, R.; Miedel, M.; Alsebahi, A.; Reddinger, R.; Saleh, A.; Shun, T.; Vernetti, L.A.; Gough, A. A 3D microfluidic liver model for high throughput compound toxicity screening in the OrganoPlate®. Toxicology 2021, 450, 152667. [Google Scholar] [CrossRef]
- Bonanini, F.; Kurek, D.; Previdi, S.; Nicolas, A.; Hendriks, D.; de Ruiter, S.; Meyer, M.; Clapés Cabrer, M.; Dinkelberg, R.; García, S.B.; et al. In vitro grafting of hepatic spheroids and organoids on a microfluidic vascular bed. Angiogenesis 2022, 25, 455–470. [Google Scholar] [CrossRef]
- Bonanini, F.; Dinkelberg, R.; Torregrosa, M.C.; Kortekaas, N.; Hagens, T.M.S.; Treillard, S.; Kurek, D.; van Duinen, V.; Vulto, P.; Bircsak, K. A microvascularizedin vitroliver model for disease modeling and drug discovery. Biofabrication 2024, 17, 015007. [Google Scholar] [CrossRef]
- Busek, M.; Aizenshtadt, A.; Koch, T.; Frank, A.; Delon, L.; Martinez, M.A.; Golovin, A.; Dumas, C.; Stokowiec, J.; Gruenzner, S.; et al. Pump-less, recirculating organ-on-a-chip (rOoC) platform. Lab Chip 2023, 23, 591–608. [Google Scholar] [CrossRef]
- Liu, J.; Feng, C.; Zhang, M.; Song, F.; Liu, H. Design and Fabrication of a Liver-on-a-chip Reconstructing Tissue-tissue Interfaces. Front. Oncol. 2022, 12, 959299. [Google Scholar] [CrossRef]
- Boul, M.; Benzoubir, N.; Messina, A.; Ghasemi, R.; Mosbah, I.B.; Duclos-Vallée, J.-C.; Dubart-Kupperschmitt, A.; Le Pioufle, B. A versatile microfluidic tool for the 3D culture of HepaRG cells seeded at various stages of differentiation. Sci. Rep. 2021, 11, 14075. [Google Scholar] [CrossRef]
- Duivenvoorde, L.P.M.; Louisse, J.; Pinckaers, N.E.T.; Nguyen, T.; van der Zande, M. Comparison of gene expression and biotransformation activity of HepaRG cells under static and dynamic culture conditions. Sci. Rep. 2021, 11, 10327. [Google Scholar] [CrossRef]
- Zhu, L.; Shao, C.; Chen, H.; Chen, Z.; Zhao, Y. Hierarchical Hydrogels with Ordered Micro-Nano Structures for Cancer-on-a-Chip Construction. Research 2021, 2021, 9845679. [Google Scholar] [CrossRef]
- Tian, T.; Ho, Y.; Chen, C.; Sun, H.; Hui, J.; Yang, P.; Ge, Y.; Liu, T.; Yang, J.; Mao, H. A 3D bio-printed spheroids based perfusion in vitro liver on chip for drug toxicity assays. Chin. Chem. Lett. 2022, 33, 3167–3171. [Google Scholar] [CrossRef]
- Asif, A.; Park, S.H.; Manzoor Soomro, A.; Khalid, M.A.U.; Salih, A.R.C.; Kang, B.; Ahmed, F.; Kim, K.H.; Choi, K.H. Microphysiological system with continuous analysis of albumin for hepatotoxicity modeling and drug screening. J. Ind. Eng. Chem. 2021, 98, 318–326. [Google Scholar] [CrossRef]
- Etxeberria, L.; Messelmani, T.; Badiola, J.H.; Llobera, A.; Fernandez, L.; Vilas-Vilela, J.L.; Leclerc, E.; Legallais, C.; Jellali, R.; Zaldua, A.M. Validation of HepG2/C3A Cell Cultures in Cyclic Olefin Copolymer Based Microfluidic Bioreactors. Polymers 2022, 14, 4478. [Google Scholar] [CrossRef] [PubMed]
- Ortega-Ribera, M.; Gibert-Ramos, A.; Abad-Jordà, L.; Magaz, M.; Téllez, L.; Paule, L.; Castillo, E.; Pastó, R.; de Souza Basso, B.; Olivas, P.; et al. Increased sinusoidal pressure impairs liver endothelial mechanosensing, uncovering novel biomarkers of portal hypertension. JHEP Rep. 2023, 5, 100722. [Google Scholar] [CrossRef]
- Chethikkattuveli Salih, A.R.; Hyun, K.; Asif, A.; Soomro, A.M.; Farooqi, H.M.U.; Kim, Y.S.; Kim, K.H.; Lee, J.W.; Huh, D.; Choi, K.H. Extracellular Matrix Optimization for Enhanced Physiological Relevance in Hepatic Tissue-Chips. Polymers 2021, 13, 3016. [Google Scholar] [CrossRef]
- Xie, X.; Maharjan, S.; Kelly, C.; Liu, T.; Lang, R.J.; Alperin, R.; Sebastian, S.; Bonilla, D.; Gandolfo, S.; Boukataya, Y.; et al. Customizable Microfluidic Origami Liver-on-a-Chip (oLOC). Adv. Mater. Technol. 2022, 7, 2100677. [Google Scholar] [CrossRef]
- Chang, M.; Fu, A.; Liu, B.; Wang, Y.; Zeng, H. Dynamic Liver Chip Based on Well-Coupled Microfluidics: An Accurate NASH Model for Drug Evaluation. Anal. Chem. 2024, 96, 16280–16288. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Ren, H.; Liu, Y.; Sun, L.; Zhang, Z.; Zhao, Y.; Shi, X. Bioinspired Artificial Liver System with hiPSC-Derived Hepatocytes for Acute Liver Failure Treatment. Adv. Healthc. Mater. 2021, 10, e2101580. [Google Scholar] [CrossRef] [PubMed]
- Messelmani, T.; Le Goff, A.; Souguir, Z.; Maes, V.; Roudaut, M.; Vandenhaute, E.; Maubon, N.; Legallais, C.; Leclerc, E.; Jellali, R. Development of Liver-on-Chip Integrating a Hydroscaffold Mimicking the Liver’s Extracellular Matrix. Bioengineering 2022, 9, 443. [Google Scholar] [CrossRef] [PubMed]
- Grebenyuk, S.; Abdel Fattah, A.R.; Kumar, M.; Toprakhisar, B.; Rustandi, G.; Vananroye, A.; Salmon, I.; Verfaillie, C.; Grillo, M.; Ranga, A. Large-scale perfused tissues via synthetic 3D soft microfluidics. Nat. Commun. 2023, 14, 193. [Google Scholar] [CrossRef]
- Sugiura, S.; Satoh, T.; Shin, K.; Onuki-Nagasaki, R.; Kanamori, T. Perfusion culture of multi-layered HepG2 hepatocellular carcinoma cells in a pressure-driven microphysiological system. J. Biosci. Bioeng. 2022, 134, 348–355. [Google Scholar] [CrossRef]
- Mazhar, N.; Islam, M.S.; Raza, M.Z.; Mahin, S.K.H.; Islam, M.R.; Chowdhury, M.E.H.; Al-Ali, A.; Agouni, A.; Yalcin, H.C. Comparative Analysis of In Vitro Pumps Used in Cardiovascular Investigations: Focus on Flow Generation Principles and Characteristics of Generated Flows. Bioengineering 2024, 11, 1116. [Google Scholar] [CrossRef]
- McCarty, W.J.; Usta, O.B.; Yarmush, M.L. A Microfabricated Platform for Generating Physiologically-Relevant Hepatocyte Zonation. Sci. Rep. 2016, 6, 26868. [Google Scholar] [CrossRef]
- Tao, T.; Deng, P.; Wang, Y.; Zhang, X.; Guo, Y.; Chen, W.; Qin, J. Microengineered Multi-Organoid System from hiPSCs to Recapitulate Human Liver-Islet Axis in Normal and Type 2 Diabetes. Adv. Sci. 2022, 9, 2103495. [Google Scholar] [CrossRef]
- Zandi Shafagh, R.; Youhanna, S.; Keulen, J.; Shen, J.X.; Taebnia, N.; Preiss, L.C.; Klein, K.; Büttner, F.A.; Bergqvist, M.; van der Wijngaart, W.; et al. Bioengineered Pancreas–Liver Crosstalk in a Microfluidic Coculture Chip Identifies Human Metabolic Response Signatures in Prediabetic Hyperglycemia. Adv. Sci. 2022, 9, 2203368. [Google Scholar] [CrossRef]
- Jeon, J.W.; Lee, S.H.; Kim, D.; Sung, J.H. In vitro hepatic steatosis model based on gut-liver-on-a-chip. Biotechnol. Prog. 2021, 37, e3121. [Google Scholar] [CrossRef]
- Yang, J.; Hirai, Y.; Iida, K.; Ito, S.; Trumm, M.; Terada, S.; Sakai, R.; Tsuchiya, T.; Tabata, O.; Kamei, K.-i. Integrated-gut-liver-on-a-chip platform as an in vitro human model of non-alcoholic fatty liver disease. Commun. Biol. 2023, 6, 310. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Priya Rajan, S.A.; Song, Q.; Zhao, Y.; Wan, M.; Aleman, J.; Skardal, A.; Bishop, C.; Atala, A.; Lu, B. 3D scaffold-free microlivers with drug metabolic function generated by lineage-reprogrammed hepatocytes from human fibroblasts. Biomaterials 2021, 269, 120668. [Google Scholar] [CrossRef]
- Shinohara, M.; Arakawa, H.; Oda, Y.; Shiraki, N.; Sugiura, S.; Nishiuchi, T.; Satoh, T.; Iino, K.; Leo, S.; Kato, Y.; et al. Coculture with hiPS-derived intestinal cells enhanced human hepatocyte functions in a pneumatic-pressure-driven two-organ microphysiological system. Sci. Rep. 2021, 11, 5437. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Wang, B.; Sun, Y.; Dai, B.; Fu, Y.; Zhang, Y.; Wang, Y.; Yang, Z.; Sun, Z.; Zhuang, S.; et al. An Oxygen-Concentration-Controllable Multiorgan Microfluidic Platform for Studying Hypoxia-Induced Lung Cancer-Liver Metastasis and Screening Drugs. ACS Sens. 2021, 6, 823–832. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, V.V.T.; Ye, S.; Gkouzioti, V.; van Wolferen, M.E.; Yengej, F.Y.; Melkert, D.; Siti, S.; de Jong, B.; Besseling, P.J.; Spee, B.; et al. A human kidney and liver organoid-based multi-organ-on-a-chip model to study the therapeutic effects and biodistribution of mesenchymal stromal cell-derived extracellular vesicles. J. Extracell. Vesicles 2022, 11, e12280. [Google Scholar] [CrossRef]
- Tao, T.P.; Brandmair, K.; Gerlach, S.; Przibilla, J.; Géniès, C.; Jacques-Jamin, C.; Schepky, A.; Marx, U.; Hewitt, N.J.; Maschmeyer, I.; et al. Demonstration of the first-pass metabolism in the skin of the hair dye, 4-amino-2-hydroxytoluene, using the Chip2 skin-liver microphysiological model. J. Appl. Toxicol. 2021, 41, 1553–1567. [Google Scholar] [CrossRef]
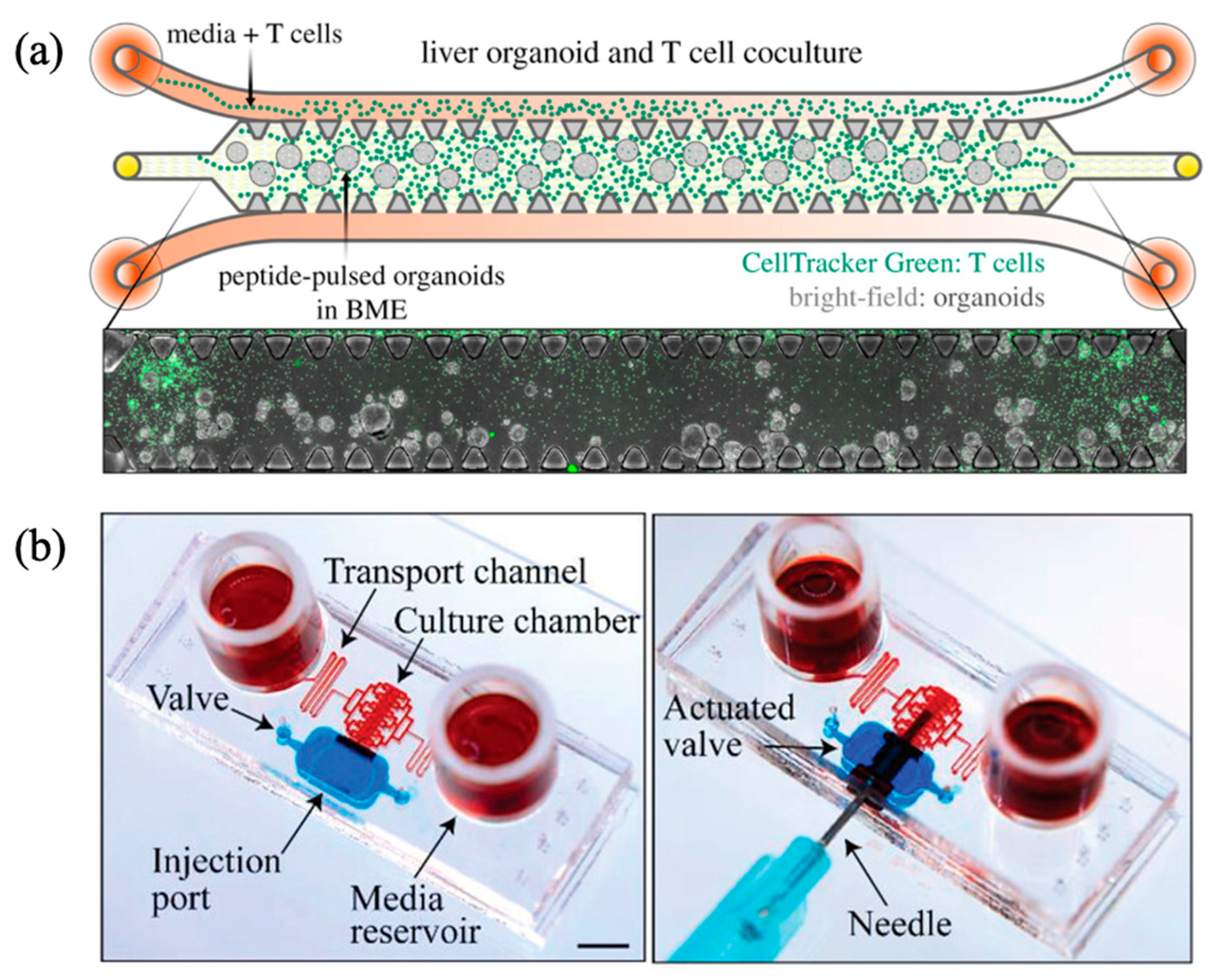
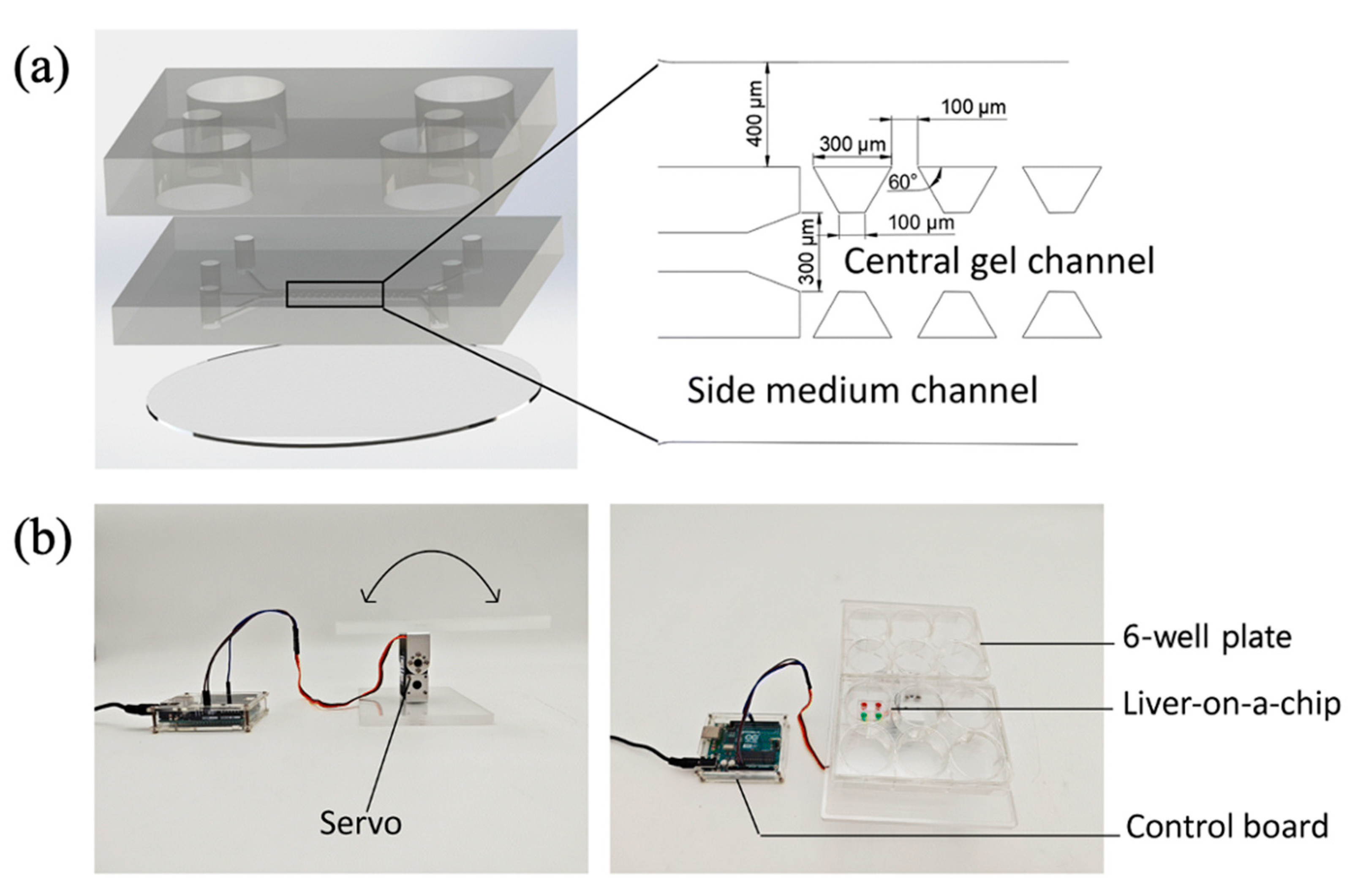
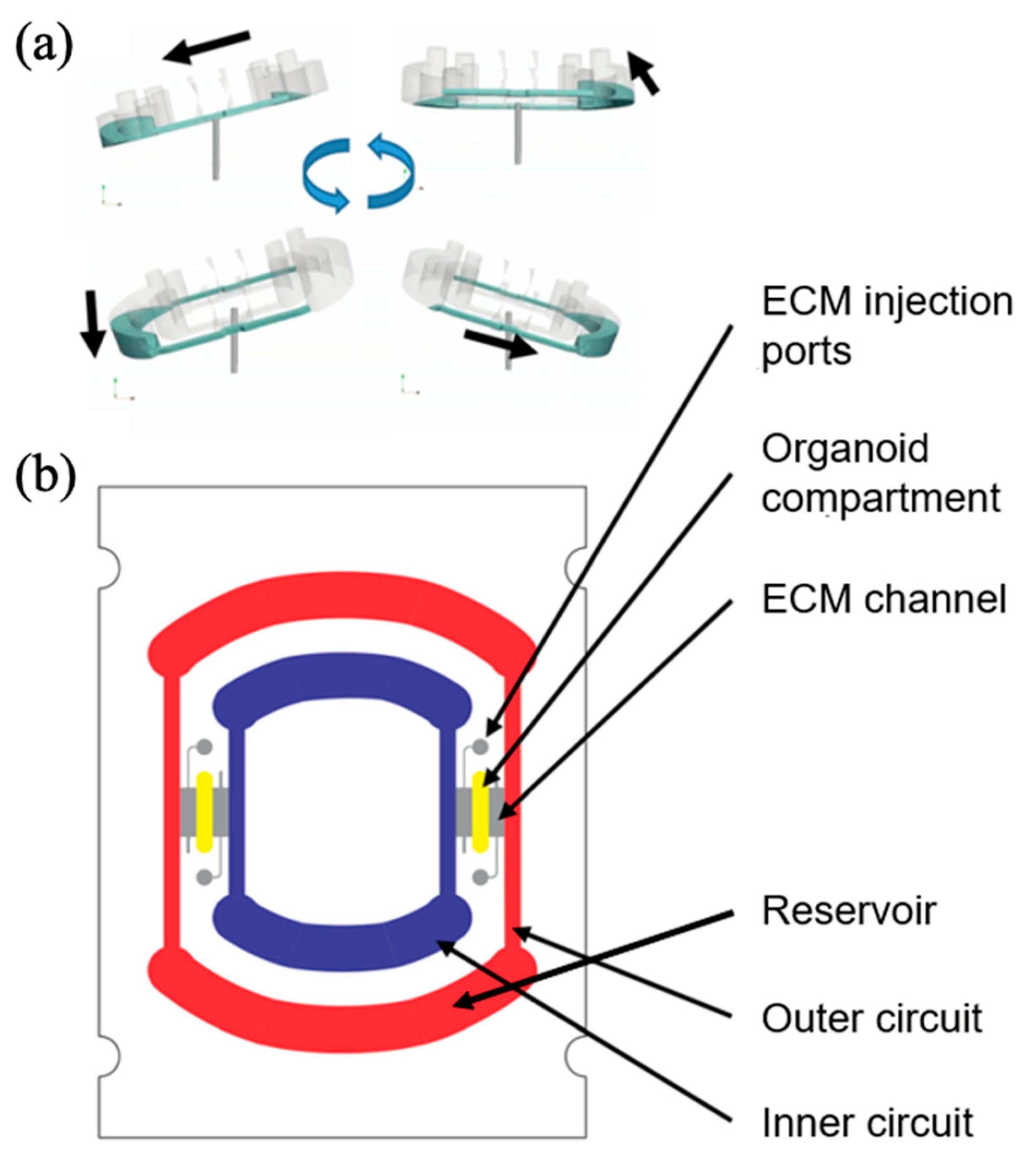
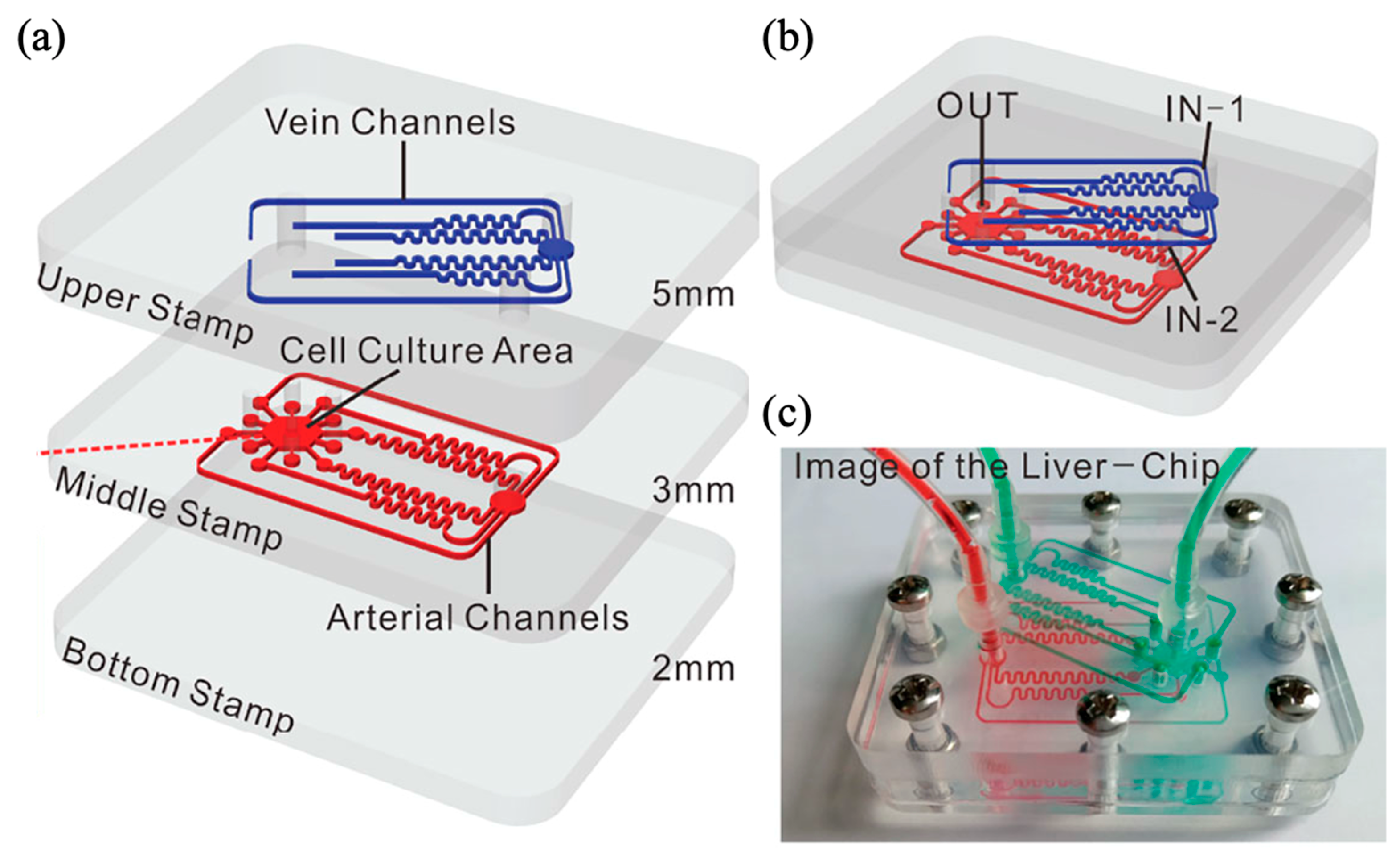
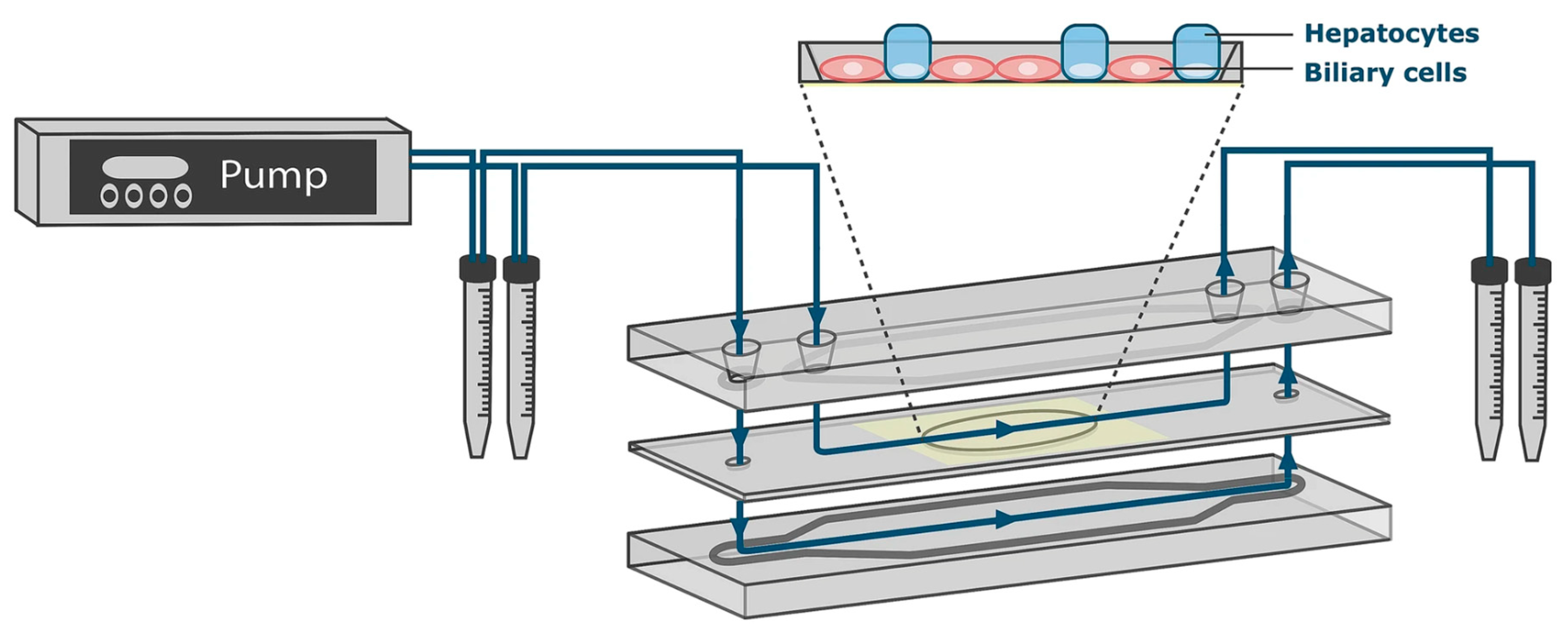
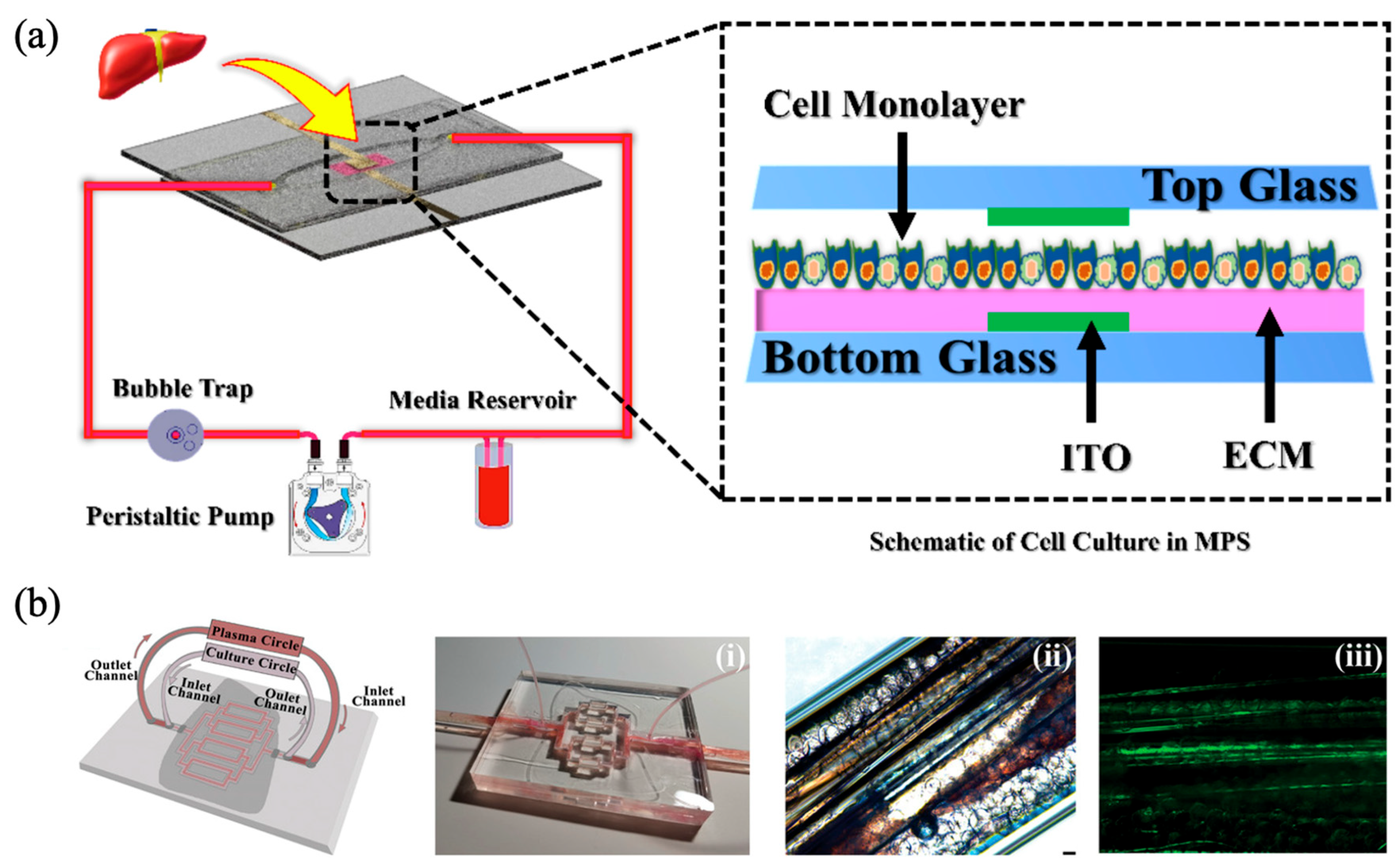
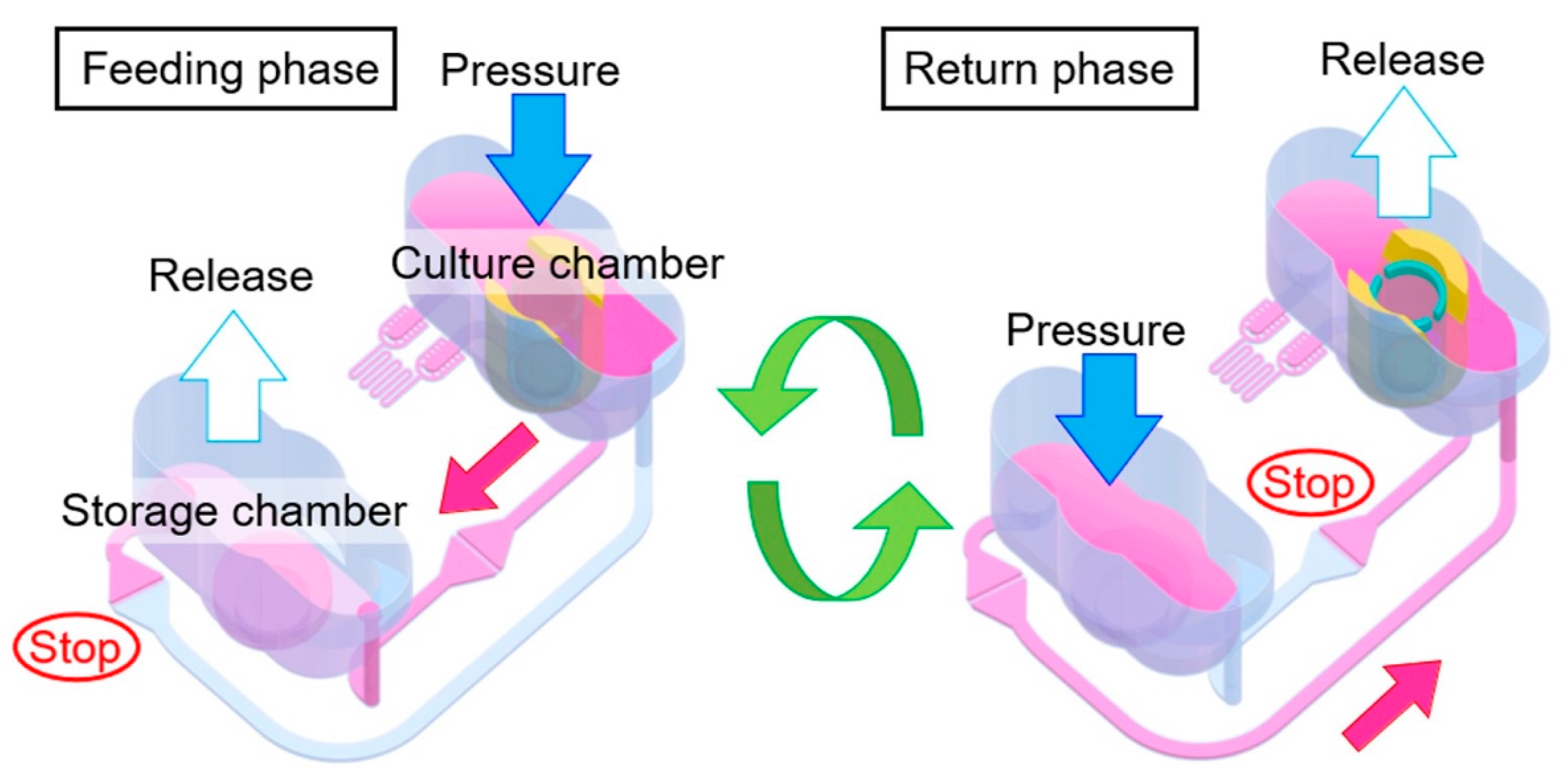
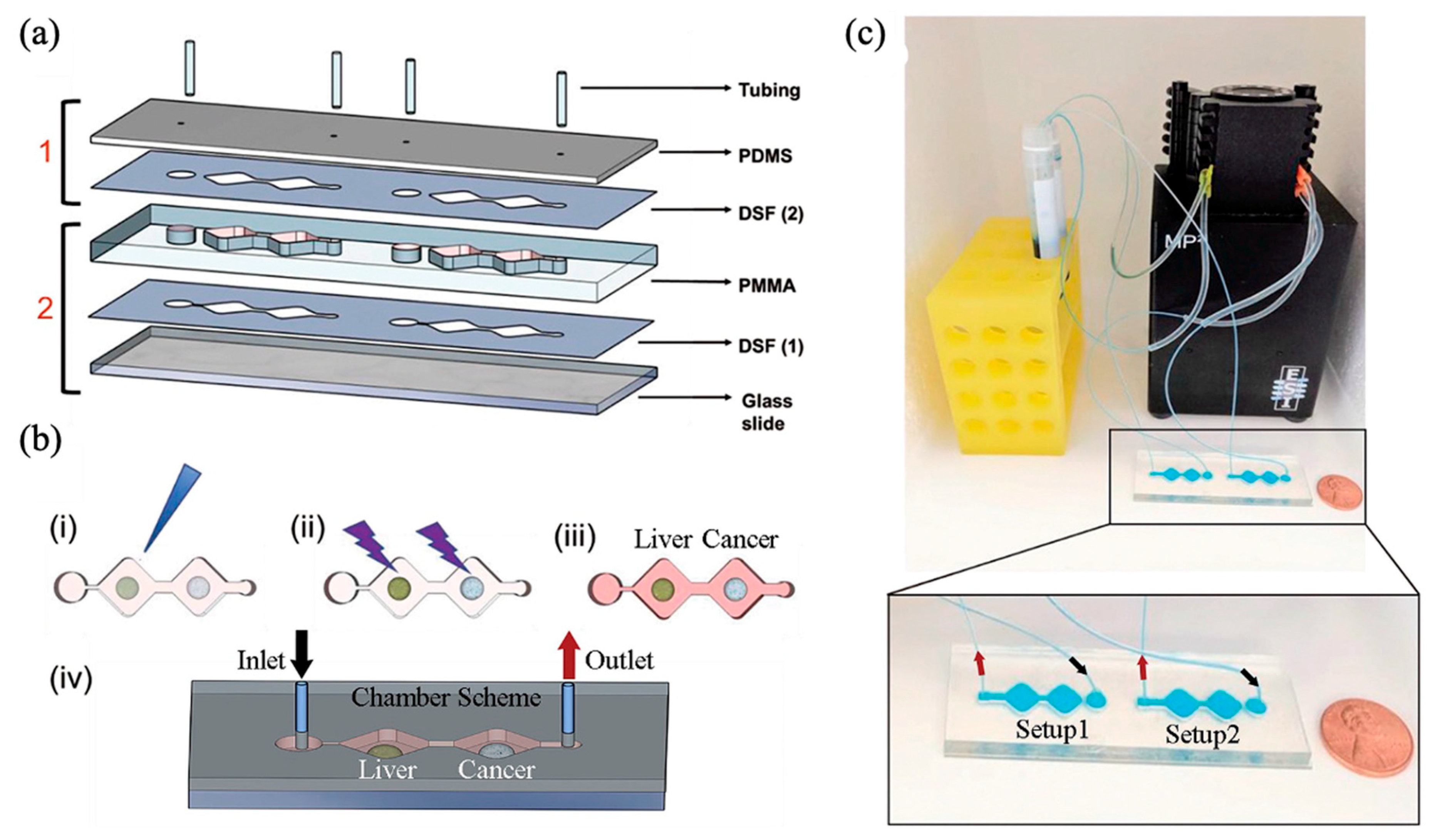
| Flow Control Strategy | Cell Type | Description | Ref. | ||
|---|---|---|---|---|---|
| Gravity gradient (Pump-less) | Height difference between medium reservoirs | HepG2, human umbilical vein endothelial cell (HUVEC) | 3D | A volume difference of 150 μL was given between the two reservoirs | [42] |
| Reproducing hepatic zonation, assessing APAP toxicity | |||||
| HepaRG, LX2, human hepatic sinusoidal endothelial cell (HHSEC) | 3D | A height difference of 15–18 mm was given between storage containers | [43] | ||
| Modeling NAFLD, reproducing dual hepatic lobular perfusion | |||||
| Rocker | HepG2, LX2 | 3D | Tilting ± 7°, every 7 min | [44] | |
| Assessing the effects of aristolochic acid I (AA) and aristololactam AII (AL) | |||||
| Primary human hepatocytes (PHH), HUVEC | 3D | Tilting ± 15°, 1 rpm velocity | [45] | ||
| Recapitulating vascularized liver structure | |||||
| Primary human HC, HSC, KC, LSEC | 3D | Modeling the progression of non-alcoholic steatohepatitis (NASH), assessing the toxicity of elafibranor | [46] | ||
| iCell 2.0 Hepatocytes, THP-1, HMEC-1 | 3D | Tilting to a maximum angle of 15°, every 4 min | [47] | ||
| Metabolic analysis of phenacetin, coumarin, diclofenac, terfenadine, phenolphthalein | |||||
| Upcyte® Human Hepatocytes, HUVEC | 3D | Tilting ± 14°, every 8 min | [48] | ||
| Simulating veno-occlusive disease | |||||
| iCell® Hepatocytes 2.0, primary liver-derived endothelial cell (LDEC), primary HSC, HUVEC | 3D | Tilting ± 14°, every 8 min | [49] | ||
| Metabolic dysfunction-associated steatohepatitis (MASH) modeling, Analysis of the effects of SB-431542 and firsocostat | |||||
| Rotator | Human embryonic stem cell, human-induced pluripotent stem cell (hiPSC), HUVEC | 3D | Tilting 10–17°, 1–5 rpm speed | [50] | |
| Suggesting the possibility of circulating immune cells in the device | |||||
| Perfusion | Syringe pump | Murine primary HCs, LSECs LX2 | 3D | 2 mL/h flow rate in channel containing cells | [51] |
| Designed triple vessels in the liver and analyzed the effects of glucose concentration gradients | |||||
| HepaRG | 3D | 375 nl/min flow rate | [52] | ||
| Reproduction of the liver microstructure | |||||
| Air-pressurized pump | HepaRG, PHH | 2D | 100 µL/h flow rate | [53] | |
| Demonstration of enhanced CYP metabolic activity of hepatocytes, characterization of HepaRG cells | |||||
| Microfluidic pump | HepG2 | 3D | The inlet velocity is optimized to 10−5 m/s | [54] | |
| Replicating hepatic lobule structure | |||||
| Peristaltic Pump | HepG2, HUVEC | 2D & 3D | 99 μL/min flow rate | [55] | |
| Reproducing acute liver failure, assessing APAP toxicity | |||||
| Circulation | Peristaltic flow | HepG2, CCD-986sk | 2D | 50 μL/min flow rate | [56] |
| Integrated an electrochemical albumin measuring sensor and modeled diabetes to evaluate metformin | |||||
| HepG2/C3A | 2D | 25 µL/min flow rate | [57] | ||
| Evaluating dynamic cell culture on cyclic olefin copolymer (COC) biochips | |||||
| Primary rat hepatocytes, HSCs, LSECs, KCs | 2D | 1.5 mL/min flow rate | [58] | ||
| Modeled advanced chronic liver disease (ACLD), inducing dysregulation of hepatic endothelium-related pathways | |||||
| HepG2 | 2D | 60 μL/min flow rate | [59] | ||
| Assessing extracellular matrix (ECM) suitable for cell adhesion and the microenvironment reproduction | |||||
| PHH, HUVEC | 2D | 14 µL/min flow rate | [60] | ||
| Reproduced the structure, microenvironment, and function of the liver | |||||
| HepG2, HUVEC | 2D | 300 μL/min flow rate | [61] | ||
| Modeling NASH progression, evaluating antihyperlipidemic drugs | |||||
| hiPSC-Heps, PHH | 3D | 0.5, 1, and 2 mL/min flow rate | [62] | ||
| Developed a bioartificial liver (BAL) system with validated plasma filtration capabilities | |||||
| HepG2/C3A | 3D | 10 µL/min flow rate | [63] | ||
| Confirmed the functionality of 3D liver chips on hydroscaffolded biochips | |||||
| H3CX | 3D | Fabrication of microvascular networks | [64] | ||
| Pneumatic flow | HepG2 | 3D | 12–14, 3.4 μL/min flow rate in feed channel and bypass channel, 350–800 μL/min flow rate in return channel | [65] | |
| Evaluation of the functionality of HepG2 in a 3D stacked system | |||||
| Flow Control Strategy | Cell Type | Description | Ref. | ||
|---|---|---|---|---|---|
| Circulation | Peristaltic flow | hiPSC (liver and islet) | 3D | 100 µL/h flow rate | [68] |
| Evaluating the effects of high glucose on glucose–insulin-associated function and the effect of metformin | |||||
| Pneumatic Flow | primary human cell (liver and islet) | 3D | Various flow rated from 0 to 4000 µL/min | [69] | |
| Mimicking high glucose environment, observing changes in insulin sensitivity and glucose responsive genes | |||||
| Gut (Intestine) | Rocker | HepG2 (liver), Caco-2 (intestine) | 2D | Tilting ± 10° every 5 min, 240 μL/h and 80 μL/h flow rates in gut and liver channels | [70] |
| Modeling hepatic steatosis, assessing the impact of gut-liver interactions on the anti-steatotic effects of XL-335 and metformin | |||||
| Peristaltic flow | HepG2 (liver), Caco-2 (intestine) | 2D | Flow rate controlled within 0 to 20 nL/min using pump on/off frequency | [71] | |
| Modeling NAFLD using FFA | |||||
| human induced hepatocyte-like cells, HLKC, HLECP2 (liver), HCT-116 (colon cancer) | 3D | 4 μL/min flow rate | [72] | ||
| Evaluating the decreased survival of tumor when capecitabine is metabolized in the liver | |||||
| Pneumatic flow | Human hepatocytes from PXB mouse (liver), hiPS-derived intestinal cell (intestine) | 2D | 100 μL/min flow rate | [73] | |
| Suggesting that the liver function is enhanced through gut–liver interactions | |||||
| Lung | syringe pump | L02 (liver), A549 and HFL-1 (lung) | 3D | Various unidirectional flow rates of 1, 10, 20, 30, and 40 μL/min | [74] |
| Analyzing the mechanisms of lung cancer metastasis to the liver and evaluating the cancer treatment efficacy of tirapazamine | |||||
| Kidney | peristaltic flow | primary human cell (liver and kidney) | 3D | Pump operation at 0.5 Hz at 500 mbar pressure and 500 mbar vacuum | [75] |
| Evaluating the renal therapeutic effect and distribution of mesenchymal stromal cell-derived small extracellular vesicles (EVs) to liver compartments | |||||
| Skin | peristaltic flow | HPR116, HHSteC (liver), EpiDerm™ (skin) | 3D | Pump operation at 0.5 Hz at 350 mbar pressure and 300 mbar vacuum | [76] |
| Metabolomic analysis of the hair dye 4-amino-2-hydroxytoluene | |||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yun, J.; Jeon, T.-J.; Kim, S.M. Current Advances and Future Perspectives of Liver-on-a-Chip Platforms Incorporating Dynamic Fluid Flow. Biomimetics 2025, 10, 443. https://doi.org/10.3390/biomimetics10070443
Yun J, Jeon T-J, Kim SM. Current Advances and Future Perspectives of Liver-on-a-Chip Platforms Incorporating Dynamic Fluid Flow. Biomimetics. 2025; 10(7):443. https://doi.org/10.3390/biomimetics10070443
Chicago/Turabian StyleYun, Jingyeong, Tae-Joon Jeon, and Sun Min Kim. 2025. "Current Advances and Future Perspectives of Liver-on-a-Chip Platforms Incorporating Dynamic Fluid Flow" Biomimetics 10, no. 7: 443. https://doi.org/10.3390/biomimetics10070443
APA StyleYun, J., Jeon, T.-J., & Kim, S. M. (2025). Current Advances and Future Perspectives of Liver-on-a-Chip Platforms Incorporating Dynamic Fluid Flow. Biomimetics, 10(7), 443. https://doi.org/10.3390/biomimetics10070443








