Hierarchical Structure of the Program Used by Filamentous Fungi to Navigate in Confining Microenvironments
Abstract
1. Introduction
2. Materials and Methods
3. Results and Discussion
3.1. Microfluidic Systems Testing the Space Navigation of Filamentous Fungi
3.2. Selecting Fungal Species for the Study of Space Navigation
3.3. Space Navigation at the Individual Hypha Level
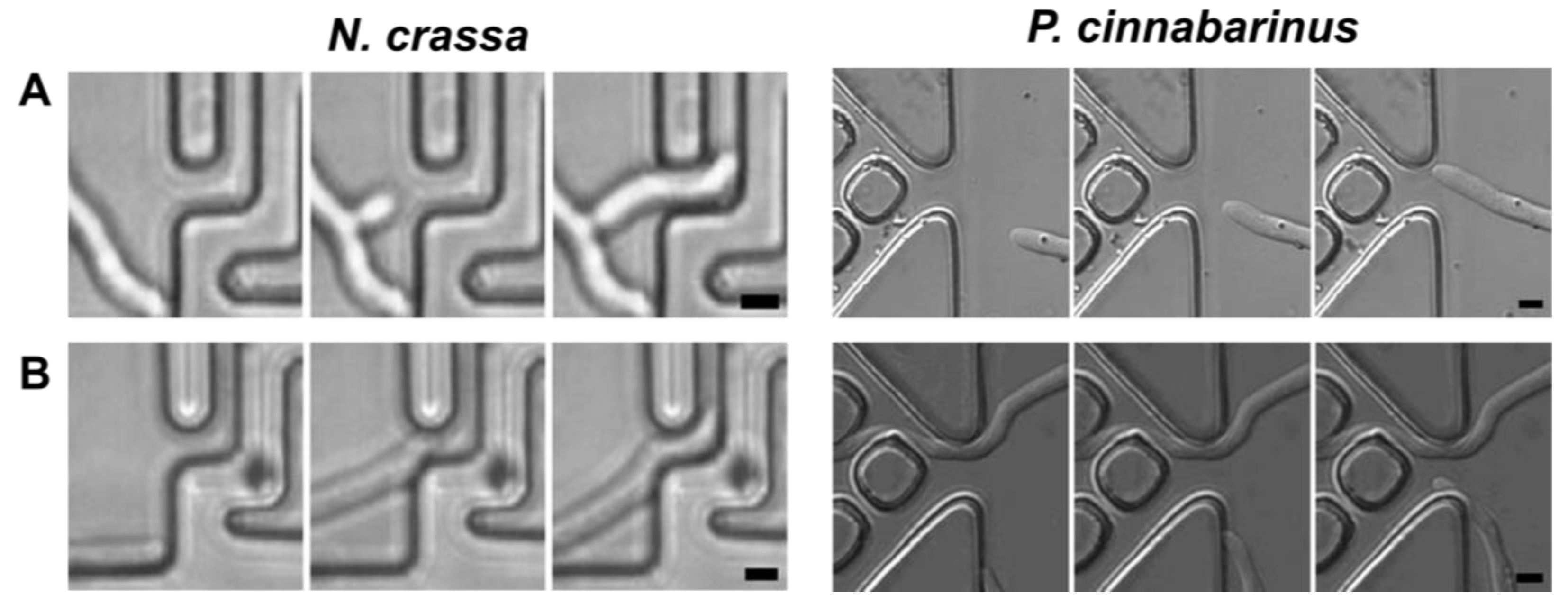
3.3.1. Sensing of Narrow Entries
- Remote sensing
- Contact-based sensing
3.3.2. Directional Memory
3.3.3. Branching
- Collision-induced branching
- Stochastic branching
3.4. Space Navigation at the Co-Located Hyphae Level
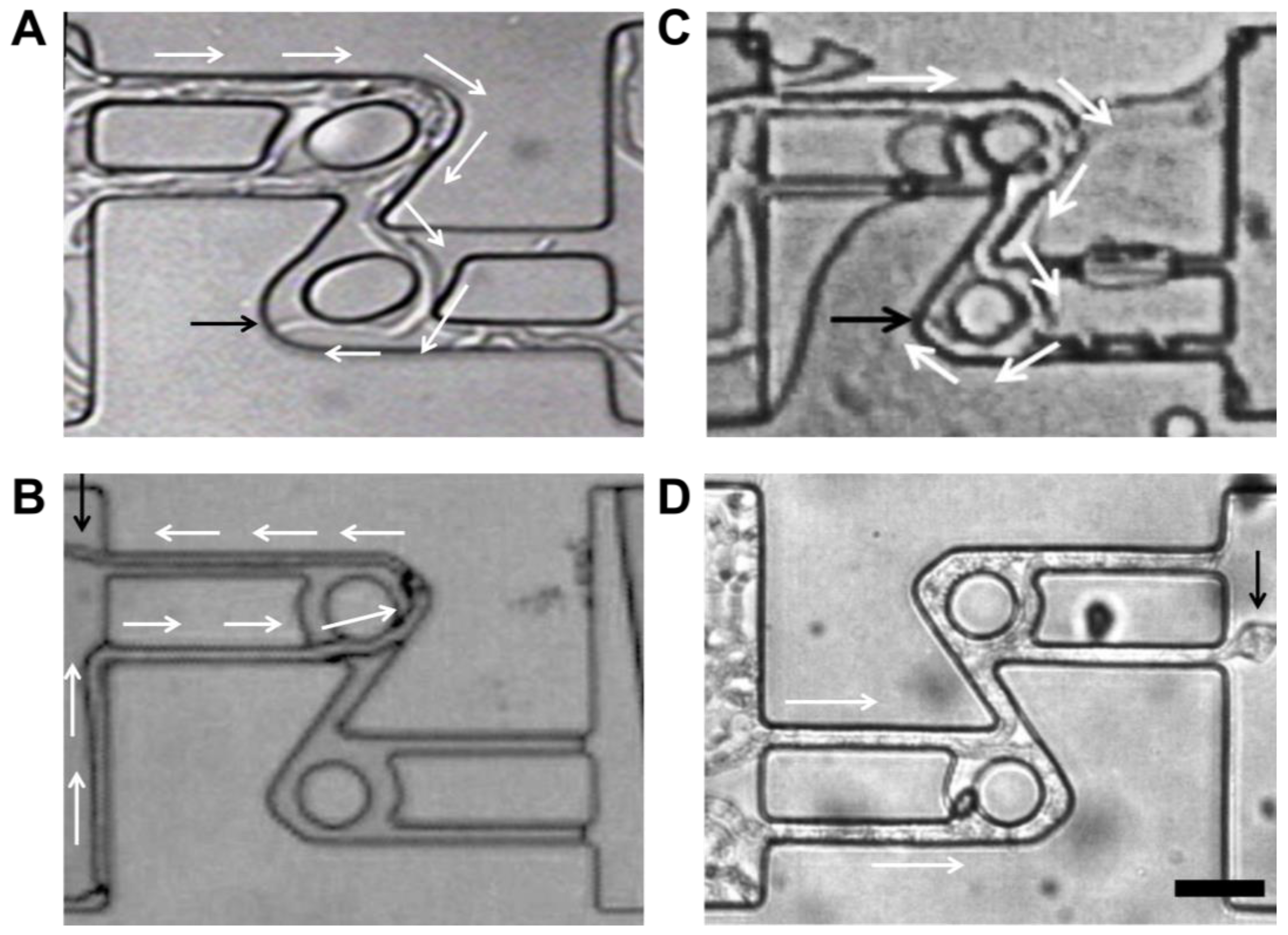
3.4.1. Negative Autotropism
- Side-by-side negative autotropism
- Head-on negative autotropism
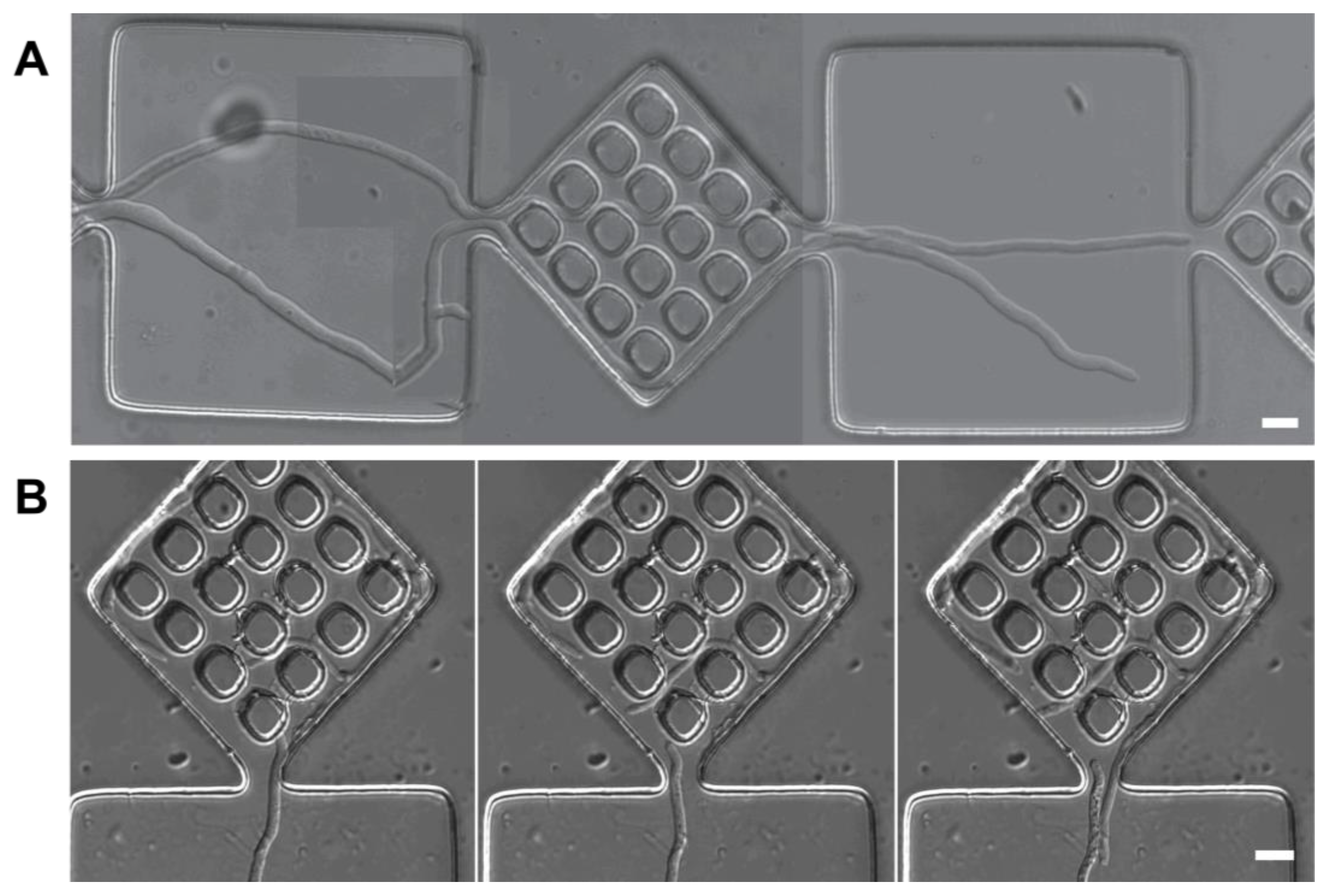
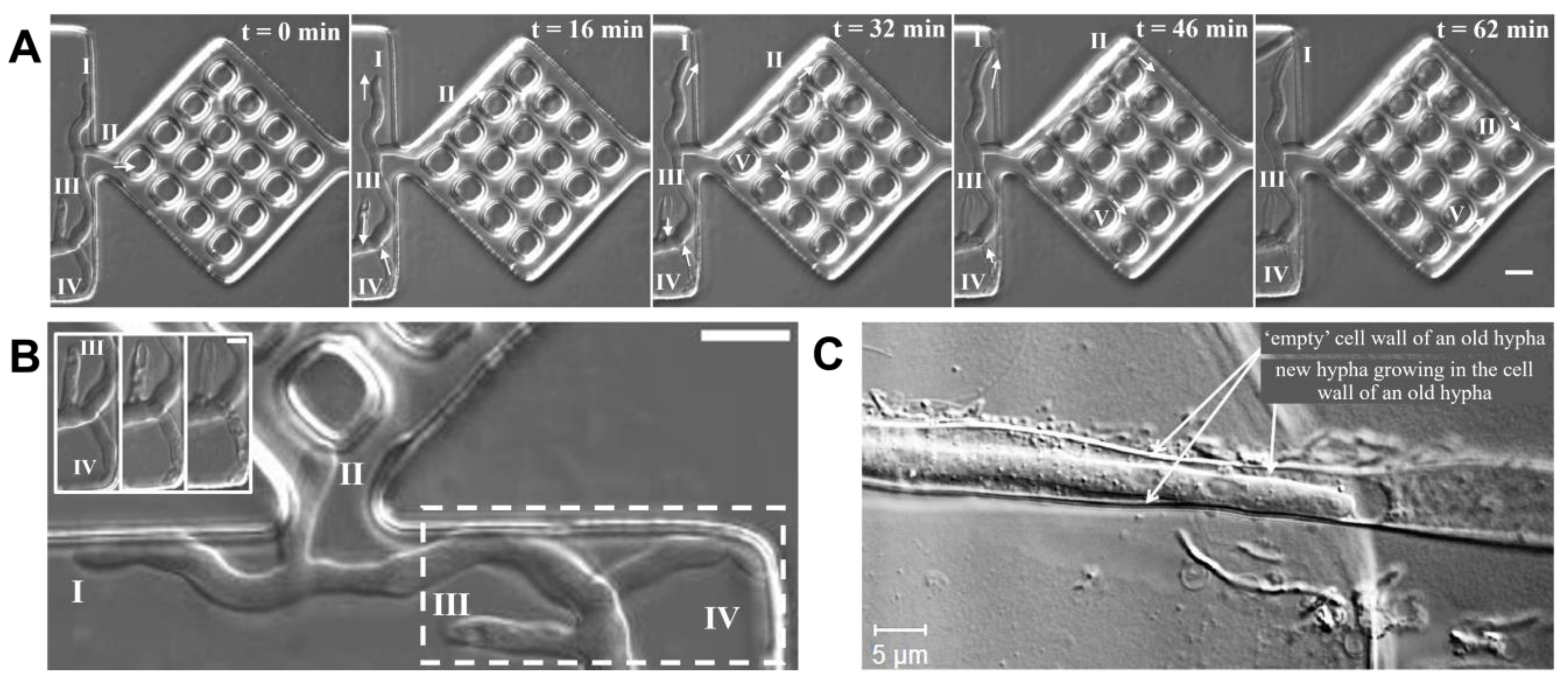
3.4.2. Cytoplasm Reallocation
- Anastomosis
3.5. Space Navigation at the Mycelium Level
3.6. Multilayered Program Used by Filamentous Fungi for Space Navigation
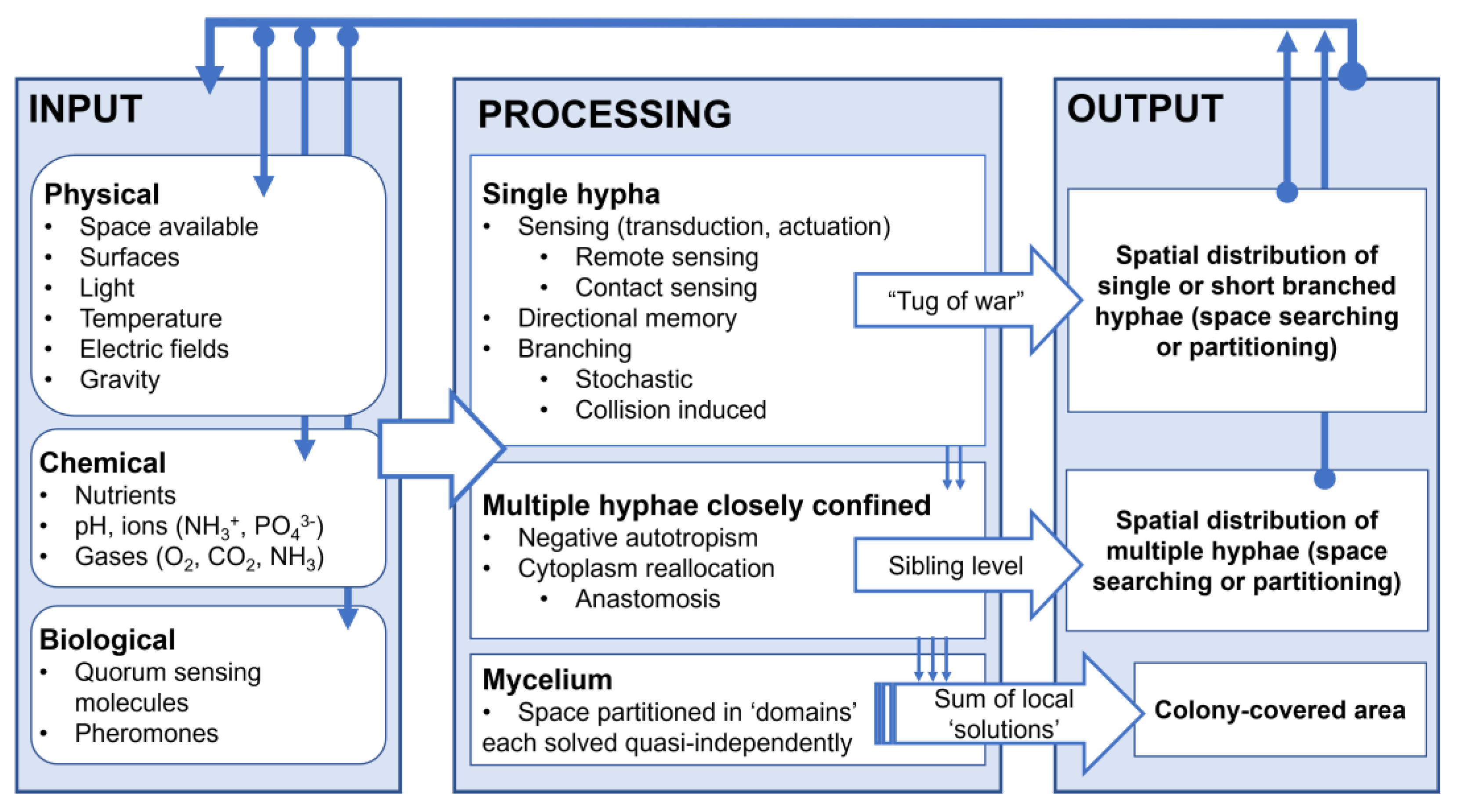
3.6.1. Information Processing in Spatial Navigation by Filamentous Fungi
3.6.2. The Structure of the Fungal Program for Space Navigation
- (i)
- At the single-hypha level, it appears that three programs for space navigation, some with their own subroutines, are in play:
- Remote sensing;
- Contact-based sensing.
- Collision-induced. Branching due to external sources, specifically collision-induced branching, is triggered when most or all physical avenues for growth are closed, e.g., corners, or when bluntly encountering obstacles. Because of the isotropic nature of turgor pressure, which appears to be its main driver, collision-induced branching presents a largely isotropic, opportunistic space search, especially in very small areas. Finally, collision-induced branching appears to have its own memory, manifesting as the gradual return to a lower branching frequency when encountering more open spaces.
- Stochastic. Fungi stochastically branch to enhance nutrient assimilation and interaction with their environment, partitioning the available space. This process is coordinated with the cell cycle through internal biochemical mechanisms.
- (ii)
- At the level of multiple, closely confined hyphae, space navigation comprises two programs, that is, one implemented through external communication, and the other implemented internally.
- (iii)
- At the mycelial level, the localized character of space searching is most evident, with the spatial distribution of fungal biomass being the result of a multitude of concatenated, but largely independent, local procedures for space searching, similar to scatter searching for optimization problems [98].
- Various levels of confinement can blur the distinction between space partitioning, prevalent in open spaces, and space searching, prevalent in tightly confining spaces. Indeed, while the navigation of mazes requires space searching subroutines, e.g., directional memory and collision-induced branching, the growth of fungi in larger, quasi-open chambers exhibit partially space partitioning characters, e.g., negative autotropism and stochastic branching.
- The space navigation programs for single and multiple co-located hyphae are ‘vertically’ connected, e.g., cytoplasm reallocation is often triggered by collision-induced branching, which itself is also triggered by the remote sensing of close by narrow passage points. Furthermore, while a ‘tug-of-war’ between the different programs manifests at the single-hypha level, it can also manifest ‘vertically’, e.g., when negative autotropism takes precedence over remote sensing and directional memory.
- There is a large, species-specific variability in the usage of space navigation subroutines, with some being turned “off”. For instance, while both P. cinnabarinus and N. crassa exhibit the collision-induced branching subroutine, A. mellea has it ‘turned off’ most of the time, as well as its directional memory.
- The framework proposed above does not explore, in detail, the impact of other biological software and their associated hardware, e.g., sporulation, which do not immediately contribute to space navigation by filamentous fungi.
3.7. Significance and Future Work
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Peay, K.G.; Kennedy, P.G.; Talbot, J.M. Dimensions of biodiversity in the Earth mycobiome. Nat. Rev. Microbiol. 2016, 14, 434–447. [Google Scholar] [CrossRef] [PubMed]
- Větrovský, T.; Morais, D.; Kohout, P.; Lepinay, C.; Algora, C.; Awokunle Hollá, S.; Bahnmann, B.D.; Bílohnědá, K.; Brabcová, V.; D’Alò, F. GlobalFungi, a global database of fungal occurrences from high-throughput-sequencing metabarcoding studies. Sci. Data 2020, 7, 228. [Google Scholar] [CrossRef] [PubMed]
- Bahn, Y.-S.; Xue, C.; Idnurm, A.; Rutherford, J.C.; Heitman, J.; Cardenas, M.E. Sensing the environment: Lessons from fungi. Nat. Rev. Microbiol. 2007, 5, 57–69. [Google Scholar] [CrossRef] [PubMed]
- Naranjo-Ortiz, M.A.; Gabaldón, T. Fungal evolution: Diversity, taxonomy and phylogeny of the Fungi. Biol. Rev. 2019, 94, 2101–2137. [Google Scholar] [CrossRef]
- Riquelme, M.; Roberson, R.; Sánchez-León, E. 3 Hyphal tip growth in filamentous fungi. In Growth, Differentiation and Sexuality; Springer: Berlin/Heidelberg, Germany, 2016; pp. 47–66. [Google Scholar]
- Roberson, R. Subcellular structure and behaviour in fungal hyphae. J. Microsc. 2020, 280, 75–85. [Google Scholar] [CrossRef]
- Coudert, Y.; Harris, S.; Charrier, B. Design principles of branching morphogenesis in filamentous organisms. Curr. Biol. 2019, 29, R1149–R1162. [Google Scholar] [CrossRef]
- Fricker, M.D.; Heaton, L.L.; Jones, N.S.; Boddy, L. The mycelium as a network. Fungal Kingd. 2017, 335–367. [Google Scholar]
- Aleklett, K.; Ohlsson, P.; Bengtsson, M.; Hammer, E.C. Fungal foraging behaviour and hyphal space exploration in micro-structured Soil Chips. ISME J. 2021, 15, 1782–1793. [Google Scholar] [CrossRef]
- Fukuda, S.; Yamamoto, R.; Yanagisawa, N.; Takaya, N.; Sato, Y.; Riquelme, M.; Takeshita, N. Trade-off between plasticity and velocity in mycelial growth. MBio 2021, 12, e03120–e03196. [Google Scholar] [CrossRef]
- Smith, M.L.; Bruhn, J.N.; Anderson, J.B. The fungus Armillaria bulbosa is among the largest and oldest living organisms. Nature 1992, 356, 428–431. [Google Scholar] [CrossRef]
- Boswell, G.P.; Jacobs, H.; Davidson, F.A.; Gadd, G.M.; Ritz, K. Functional consequences of nutrient translocation in mycelial fungi. J. Theor. Biol. 2002, 217, 459–477. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, H.; Boswell, G.P.; Scrimgeour, C.M.; Davidson, F.A.; RITZ, K. Translocation of carbon by Rhizoctonia solani in nutritionally-heterogeneous microcosms. Mycol. Res. 2004, 108, 453–462. [Google Scholar] [CrossRef] [PubMed]
- Buffi, M.; Cailleau, G.; Kuhn, T.; Li Richter, X.-Y.; Stanley, C.E.; Wick, L.Y.; Chain, P.S.; Bindschedler, S.; Junier, P. Fungal drops: A novel approach for macro-and microscopic analyses of fungal mycelial growth. Microlife 2023, 4, uqad042. [Google Scholar] [CrossRef]
- Tegmark, M. Life 3.0: Being Human in the Age Of Artificial Intelligence; Knopf Doubleday Publishing Group: New York, NY, USA, 2018. [Google Scholar]
- Held, M.; Binz, M.; Edwards, C.; Nicolau, D.V. Dynamic behaviour of fungi in microfluidics: A comparative study. In Proceedings of the Imaging Manipulation, and Analysis of Biomolecules, Cells, and Tissues VII, San Jose, CA, USA, 12 February 2009; pp. 219–227. [Google Scholar]
- Held, M.; Edwards, C.; Nicolau, D.V. Probing the growth dynamics of Neurospora crassa with microfluidic structures. Fungal Biol. 2011, 115, 493–505. [Google Scholar] [CrossRef]
- Held, M.; Kašpar, O.; Edwards, C.; Nicolau, D.V. Intracellular mechanisms of fungal space searching in microenvironments. Proc. Natl. Acad. Sci. 2019, 116, 13543–13552. [Google Scholar] [CrossRef]
- Tokárová, V.; Perumal, A.S.; Nayak, M.; Shum, H.; Kašpar, O.; Rajendran, K.; Mohammadi, M.; Tremblay, C.; Gaffney, E.A.; Martel, S. Patterns of bacterial motility in microfluidics-confining environments. Proc. Natl. Acad. Sci. USA 2021, 118, e2013925118. [Google Scholar] [CrossRef]
- Aufrecht, J.A.; Fowlkes, J.D.; Bible, A.N.; Morrell-Falvey, J.; Doktycz, M.J.; Retterer, S.T. Pore-scale hydrodynamics influence the spatial evolution of bacterial biofilms in a microfluidic porous network. PLoS ONE 2019, 14, e0218316. [Google Scholar] [CrossRef]
- Agudelo, C.; Sanati, A.; Ghanbari, M.; Packirisamy, M.; Geitmann, A. A microfluidic platform for the investigation of elongation growth in pollen tubes. J. Micromechanics Microengineering 2012, 22, 115009. [Google Scholar] [CrossRef]
- Stanley, C.E.; Shrivastava, J.; Brugman, R.; Heinzelmann, E.; van Swaay, D.; Grossmann, G. Dual-flow-RootChip reveals local adaptations of roots towards environmental asymmetry at the physiological and genetic levels. New Phytol. 2018, 217, 1357–1369. [Google Scholar] [CrossRef]
- Aufrecht, J.; Khalid, M.; Walton, C.L.; Tate, K.; Cahill, J.F.; Retterer, S.T. Hotspots of root-exuded amino acids are created within a rhizosphere-on-a-chip. Lab. A Chip 2022, 22, 954–963. [Google Scholar] [CrossRef]
- Johari, S.; Nock, V.; Alkaisi, M.M.; Wang, W. On-chip analysis of C. elegans muscular forces and locomotion patterns in microstructured environments. Lab. A Chip 2013, 13, 1699–1707. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Le, J.; Chen, Y.; Cai, Y.; Hong, Z.; Chai, Y. Recent advances in microfluidic devices for bacteria and fungus research. TrAC Trends Anal. Chem. 2019, 112, 175–195. [Google Scholar] [CrossRef]
- Bedekovic, T.; Brand, A.C. Microfabrication and its use in investigating fungal biology. Mol. Microbiol. 2022, 117, 569–577. [Google Scholar] [CrossRef] [PubMed]
- Richter, F.; Bindschedler, S.; Calonne-Salmon, M.; Declerck, S.; Junier, P.; Stanley, C.E. Fungi-on-a-Chip: Microfluidic platforms for single-cell studies on fungi. FEMS Microbiol. Rev. 2022, 46, fuac039. [Google Scholar] [CrossRef]
- Mafla-Endara, P.M.; Arellano-Caicedo, C.; Aleklett, K.; Pucetaite, M.; Ohlsson, P.; Hammer, E.C. Microfluidic chips provide visual access to in situ soil ecology. Commun. Biol. 2021, 4, 889. [Google Scholar] [CrossRef]
- Schmieder, S.S.; Stanley, C.E.; Rzepiela, A.; van Swaay, D.; Sabotič, J.; Nørrelykke, S.F.; deMello, A.J.; Aebi, M.; Künzler, M. Bidirectional propagation of signals and nutrients in fungal networks via specialized hyphae. Curr. Biol. 2019, 29, 217–228.e214. [Google Scholar] [CrossRef]
- Gimeno, A.; Stanley, C.E.; Ngamenie, Z.; Hsung, M.-H.; Walder, F.; Schmieder, S.S.; Bindschedler, S.; Junier, P.; Keller, B.; Vogelgsang, S. A versatile microfluidic platform measures hyphal interactions between Fusarium graminearum and Clonostachys rosea in real-time. Commun. Biol. 2021, 4, 262. [Google Scholar] [CrossRef]
- Bhushan, B. Biomimetics: Lessons from Nature - an overview. Philos. Trans. R. Soc. A: Math. Phys. Eng. Sci. 2009, 367, 1445–1486. [Google Scholar] [CrossRef]
- Barthelat, F. Biomimetics for next generation materials. Philos. Trans. R. Soc. A: Math. Phys. Eng. Sci. 2007, 365, 2907–2919. [Google Scholar] [CrossRef]
- Pasparakis, G.; Krasnogor, N.; Cronin, L.; Davis, B.G.; Alexander, C. Controlled polymer synthesis—from biomimicry towards synthetic biology. Chem. Soc. Rev. 2010, 39, 286–300. [Google Scholar] [CrossRef]
- Rajput, V.; Mulay, P.; Mahajan, C.M. Bio-inspired algorithms for feature engineering: Analysis, applications and future research directions. Inf. Discov. Deliv. 2025, 53, 56–71. [Google Scholar] [CrossRef]
- Vincent, J.F.V.; Bogatyreva, O.A.; Bogatyrev, N.R.; Bowyer, A.; Pahl, A.K. Biomimetics: Its practice and theory. J. R. Soc. Interface 2006, 3, 471–482. [Google Scholar] [CrossRef] [PubMed]
- Hanson, K.L.; Nicolau Jr, D.V.; Filipponi, L.; Wang, L.; Lee, A.P.; Nicolau, D.V. Fungi use efficient algorithms for the exploration of microfluidic networks. Small 2006, 2, 1212–1220. [Google Scholar] [CrossRef]
- Held, M.; Lee, A.P.; Edwards, C.; Nicolau, D.V. Microfluidics structures for probing the dynamic behaviour of filamentous fungi. Microelectron. Eng. 2010, 87, 786–789. [Google Scholar] [CrossRef]
- Anderson, J.R.; Chiu, D.T.; Jackman, R.J.; Cherniavskaya, O.; McDonald, J.C.; Wu, H.; Whitesides, S.H.; Whitesides, G.M. Fabrication of topologically complex three-dimensional microfluidic systems in PDMS by rapid prototyping. Anal. Chem. 2000, 72, 3158–3164. [Google Scholar] [CrossRef]
- Fister Jr, I.; Yang, X.-S.; Fister, I.; Brest, J.; Fister, D. A brief review of nature-inspired algorithms for optimization. arXiv preprint 2013, arXiv:1307.4186. [Google Scholar]
- Molina, D.; LaTorre, A.; Herrera, F. An insight into bio-inspired and evolutionary algorithms for global optimization: Review, analysis, and lessons learnt over a decade of competitions. Cogn. Comput. 2018, 10, 517–544. [Google Scholar] [CrossRef]
- Johnvictor, A.C.; Durgamahanthi, V.; Pariti Venkata, R.M.; Jethi, N. Critical review of bio-inspired optimization techniques. Wiley Interdiscip. Rev. Comput. Stat. 2022, 14, e1528. [Google Scholar] [CrossRef]
- Ni, J.; Wu, L.; Fan, X.; Yang, S.X. Bioinspired intelligent algorithm and its applications for mobile robot control: A survey. Comput. Intell. Neurosci. 2016, 2016, 3810903. [Google Scholar] [CrossRef]
- Bitam, S.; Mellouk, A.; Zeadally, S. Bio-inspired routing algorithms survey for vehicular ad hoc networks. IEEE Commun. Surv. Tutor. 2014, 17, 843–867. [Google Scholar] [CrossRef]
- Abdel-Basset, M.; Mohamed, R.; Abouhawwash, M. Fungal growth optimizer: A novel nature-inspired metaheuristic algorithm for stochastic optimization. Comput. Methods Appl. Mech. Eng. 2025, 437, 117825. [Google Scholar] [CrossRef]
- Foudeh, A.M.; Didar, T.F.; Veres, T.; Tabrizian, M. Microfluidic designs and techniques using lab-on-a-chip devices for pathogen detection for point-of-care diagnostics. Lab A Chip 2012, 12, 3249–3266. [Google Scholar] [CrossRef] [PubMed]
- Du, G.; Fang, Q.; den Toonder, J.M. Microfluidics for cell-based high throughput screening platforms—A review. Anal. Chim. Acta 2016, 903, 36–50. [Google Scholar] [CrossRef] [PubMed]
- Stanley, C.E.; Grossmann, G.; i Solvas, X.C.; deMello, A.J. Soil-on-a-Chip: Microfluidic platforms for environmental organismal studies. Lab A Chip 2016, 16, 228–241. [Google Scholar] [CrossRef] [PubMed]
- Banik, S.; Uchil, A.; Kalsang, T.; Chakrabarty, S.; Ali, M.A.; Srisungsitthisunti, P.; Mahato, K.K.; Surdo, S.; Mazumder, N. The revolution of PDMS microfluidics in cellular biology. Crit. Rev. Biotechnol. 2023, 43, 465–483. [Google Scholar] [CrossRef]
- Coetzee, M.P.; Wingfield, B.D.; Harrington, T.C.; Steimel, J.; Coutinho, T.A.; Wingfield, M.J. The root rot fungus Armillaria mellea introduced into South Africa by early Dutch settlers. Mol. Ecol. 2001, 10, 387–396. [Google Scholar] [CrossRef]
- Eggert, C.; Temp, U.; Eriksson, K.-E. The ligninolytic system of the white rot fungus Pycnoporus cinnabarinus: Purification and characterization of the laccase. Appl. Environ. Microbiol. 1996, 62, 1151–1158. [Google Scholar] [CrossRef]
- Aramayo, R.; Selker, E.U. Neurospora crassa, a model system for epigenetics research. Cold Spring Harb. Perspect. Biol. 2013, 5, a017921. [Google Scholar] [CrossRef]
- Minke, P.F.; Lee, I.H.; Tinsley, J.H.; Bruno, K.S.; Plamann, M. Neurospora crassa ro-10 and ro-11 genes encode novel proteins required for nuclear distribution. Mol. Microbiol. 1999, 32, 1065–1076. [Google Scholar] [CrossRef]
- Singh, N.; Gaur, S. GRAS Fungi: A New Horizon in Safer Food Product. In Fungi in Sustainable Food Production; Dai, X., Sharma, M., Chen, J., Eds.; Springer International Publishing: Cham, Switzerland, 2021; pp. 27–37. [Google Scholar]
- Frisvad, J.C.; Møller, L.L.H.; Larsen, T.O.; Kumar, R.; Arnau, J. Safety of the fungal workhorses of industrial biotechnology: Update on the mycotoxin and secondary metabolite potential of Aspergillus niger, Aspergillus oryzae, and Trichoderma reesei. Appl. Microbiol. Biotechnol. 2018, 102, 9481–9515. [Google Scholar] [CrossRef]
- Albuquerque, P.; Casadevall, A. Quorum sensing in fungi–a review. Med. Mycol. 2012, 50, 337–345. [Google Scholar] [CrossRef]
- Wongsuk, T.; Pumeesat, P.; Luplertlop, N. Fungal quorum sensing molecules: Role in fungal morphogenesis and pathogenicity. J. Basic Microbiol. 2016, 56, 440–447. [Google Scholar] [CrossRef] [PubMed]
- Municio-Diaz, C.; Muller, E.; Drevensek, S.; Fruleux, A.; Lorenzetti, E.; Boudaoud, A.; Minc, N. Mechanobiology of the cell wall–insights from tip-growing plant and fungal cells. J. Cell Sci. 2022, 135, jcs259208. [Google Scholar] [CrossRef] [PubMed]
- Kumamoto, C.A. Molecular mechanisms of mechanosensing and their roles in fungal contact sensing. Nat. Rev. Microbiol. 2008, 6, 667–673. [Google Scholar] [CrossRef]
- Takeuchi, Y.; Schmid, J.; Caldwell, J.H.; Harold, F.M. Transcellular ion currents and extension of Neurospora crassa hyphae. J. Membr. Biol. 1988, 101, 33–41. [Google Scholar] [CrossRef]
- Xiong, B.-J.; Stanley, C.E.; Dusny, C.; Schlosser, D.; Harms, H.; Wick, L.Y. pH Distribution along Growing Fungal Hyphae at Microscale. J. Fungi 2022, 8, 599. [Google Scholar] [CrossRef]
- Lomascolo, A.; Uzan-Boukhris, E.; Herpoël-Gimbert, I.; Sigoillot, J.C.; Lesage-Meessen, L. Peculiarities of Pycnoporus species for applications in biotechnology. Appl. Microbiol. Biotechnol. 2011, 92, 1129–1149. [Google Scholar] [CrossRef]
- Bowen, A.D.; Davidson, F.A.; Keatch, R.; Gadd, G.M. Induction of contour sensing in Aspergillus niger by stress and its relevance to fungal growth mechanics and hyphal tip structure. Fungal Genet. Biol. 2007, 44, 484–491. [Google Scholar] [CrossRef]
- Kolattukudy, P.; Rogers, L.M.; Li, D.; Hwang, C.-S.; Flaishman, M.A. Surface signaling in pathogenesis. Proc. Natl. Acad. Sci. USA 1995, 92, 4080–4087. [Google Scholar] [CrossRef]
- Hoch, H.C.; Staples, R.C.; Whitehead, B.; Comeau, J.; Wolf, E.D. Signaling for growth orientation and cell differentiation by surface topography in Uromyces. Science 1987, 235, 1659–1662. [Google Scholar] [CrossRef]
- Sherwood, J.; Gow, N.; Gooday, G.; Gregory, D.; Marshall, D. Contact sensing in Candida albicans: A possible aid to epithelial penetration. J. Med. Vet. Mycol. 1992, 30, 461–469. [Google Scholar] [CrossRef] [PubMed]
- Brand, A.; Shanks, S.; Duncan, V.M.; Yang, M.; Mackenzie, K.; Gow, N.A. Hyphal orientation of Candida albicans is regulated by a calcium-dependent mechanism. Curr. Biol. 2007, 17, 347–352. [Google Scholar] [CrossRef] [PubMed]
- Hopke, A.; Mela, A.; Ellett, F.; Carter-House, D.; Peña, J.F.; Stajich, J.E.; Altamirano, S.; Lovett, B.; Egan, M.; Kale, S. Crowdsourced analysis of fungal growth and branching on microfluidic platforms. Plos ONE 2021, 16, e0257823. [Google Scholar] [CrossRef] [PubMed]
- Harris, S.D. Branching of fungal hyphae: Regulation, mechanisms and comparison with other branching systems. Mycologia 2008, 100, 823–832. [Google Scholar] [CrossRef]
- Boddy, L. Saprotrophic cord-forming fungi: Meeting the challenge of heterogeneous environments. Mycologia 1999, 91, 13–32. [Google Scholar] [CrossRef]
- Boddy, L.; Wells, J.M.; Culshaw, C.; Donnelly, D.P. Fractal analysis in studies of mycelium in soil. Geoderma 1999, 88, 301–328. [Google Scholar] [CrossRef]
- Papagianni, M. Quantification of the fractal nature of mycelial aggregation in Aspergillus niger submerged cultures. Microb. Cell Factories 2006, 5, 1–13. [Google Scholar] [CrossRef]
- Juge, C.; Champagne, A.; Coughlan, A.P.; Juge, N.; Parrott, L.; Piché, Y. Quantifying the growth of arbuscular mycorrhizal fungi: Usefulness of the fractal dimension. Botany 2009, 87, 387–400. [Google Scholar] [CrossRef]
- Ryoo, D. Fungal fractal morphology of pellet formation in Aspergillus niger. Biotechnol. Tech. 1999, 13, 33–36. [Google Scholar] [CrossRef]
- Potapova, T. Structural and functional organization of growing tips of Neurospora crassa hyphae. Biochemestry 2014, 79, 593–607. [Google Scholar] [CrossRef]
- Hood, M.A.; Robinson, P.M. Formation of aerial hyphae in Saprolegnia ferax. Mycol. Res. 1989, 93, 101–105. [Google Scholar] [CrossRef]
- Brand, A.; Gow, N.A. Mechanisms of hypha orientation of fungi. Curr. Opin. Microbiol. 2009, 12, 350–357. [Google Scholar] [CrossRef] [PubMed]
- Lindahl, B.D.; Olsson, S. Fungal translocation-creating and responding to environmental heterogeneity. Mycologist 2004, 18, 79–88. [Google Scholar] [CrossRef]
- Giovannetti, M.; Sbrana, C.; Logi, C. Microchambers and video-enhanced light microscopy for monitoring cellular events in living hyphae of arbuscular mycorrhizal fungi. Plant Soil 2000, 226, 153–159. [Google Scholar] [CrossRef]
- Falconer, R.E.; Bown, J.L.; White, N.A.; Crawford, J.W. Biomass recycling: A key to efficient foraging by fungal colonies. Oikos 2007, 116, 1558–1568. [Google Scholar] [CrossRef]
- Hickey, P.C.; Jacobson, D.J.; Read, N.D.; Glass, N.L. Live-cell imaging of vegetative hyphal fusion in Neurospora crassa. Fungal Genet. Biol. 2002, 37, 109–119. [Google Scholar] [CrossRef]
- Simonin, A.; Palma-Guerrero, J.; Fricker, M.; Glass, N.L. Physiological significance of network organization in fungi. Eukaryot. Cell 2012, 11, 1345–1352. [Google Scholar] [CrossRef]
- Francisco, C.S.; Zwyssig, M.M.; Palma-Guerrero, J. The role of vegetative cell fusions in the development and asexual reproduction of the wheat fungal pathogen Zymoseptoria tritici. BMC Biol. 2020, 18, 1–16. [Google Scholar] [CrossRef]
- Bebber, D.P.; Hynes, J.; Darrah, P.R.; Boddy, L.; Fricker, M.D. Biological solutions to transport network design. Proc. R. Soc. B Biol. Sci. 2007, 274, 2307–2315. [Google Scholar] [CrossRef]
- Walker, G.M.; White, N.A. Introduction to fungal physiology. Fungi Biol. Appl. 2017, 1–35. [Google Scholar]
- Steiner, M.; Linkov, I.; Yoshida, S. The role of fungi in the transfer and cycling of radionuclides in forest ecosystems. J. Environ. Radioact. 2002, 58, 217–241. [Google Scholar] [CrossRef] [PubMed]
- van’t Padje, A.; Oyarte Galvez, L.; Klein, M.; Hink, M.A.; Postma, M.; Shimizu, T.; Kiers, E.T. Temporal tracking of quantum-dot apatite across in vitro mycorrhizal networks shows how host demand can influence fungal nutrient transfer strategies. ISME J. 2021, 15, 435–449. [Google Scholar] [CrossRef] [PubMed]
- de Ulzurrun, G.V.-D.; Baetens, J.; Van den Bulcke, J.; De Baets, B. Modelling three-dimensional fungal growth in response to environmental stimuli. J. Theor. Biol. 2017, 414, 35–49. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Ayouz, M.; Lv, P.; Perré, P. A lattice-based system for modeling fungal mycelial growth in complex environments. Phys. A Stat. Mech. Its Appl. 2018, 511, 191–206. [Google Scholar] [CrossRef]
- Moore, D.; Meskauskas, A. The Algorithmic Fungus; CreateSpace Independent Publishing Platform: Charleston, SC, USA, 2017. [Google Scholar]
- Adamatzky, A.; Ayres, P.; Beasley, A.E.; Roberts, N.; Wösten, H.A.B. Logics in Fungal Mycelium Networks. Log. Universalis 2022, 16, 655–669. [Google Scholar] [CrossRef]
- Fukasawa, Y.; Savoury, M.; Boddy, L. Ecological memory and relocation decisions in fungal mycelial networks: Responses to quantity and location of new resources. ISME J. 2020, 14, 380–388. [Google Scholar] [CrossRef]
- Heaton, L.; Obara, B.; Grau, V.; Jones, N.; Nakagaki, T.; Boddy, L.; Fricker, M.D. Analysis of fungal networks. Fungal Biol. Rev. 2012, 26, 12–29. [Google Scholar] [CrossRef]
- Whiteside, M.D.; Werner, G.D.; Caldas, V.E.; van’t Padje, A.; Dupin, S.E.; Elbers, B.; Bakker, M.; Wyatt, G.A.; Klein, M.; Hink, M.A. Mycorrhizal fungi respond to resource inequality by moving phosphorus from rich to poor patches across networks. Curr. Biol. 2019, 29, 2043–2050.e2048. [Google Scholar] [CrossRef]
- Trewavas, A. What is plant behaviour? Plant Cell Environ. 2009, 32, 606–616. [Google Scholar] [CrossRef]
- Asenova, E.; Lin, H.-Y.; Fu, E.; Nicolau, D.V. Optimal fungal space searching algorithms. IEEE Trans. Nanobioscience 2016, 15, 613–618. [Google Scholar] [CrossRef]
- Riquelme, M.; Yarden, O.; Bartnicki-Garcia, S.; Bowman, B.; Castro-Longoria, E.; Free, S.J.; Fleissner, A.; Freitag, M.; Lew, R.R.; Mouriño-Pérez, R. Architecture and development of the Neurospora crassa hypha–a model cell for polarized growth. Fungal Biol. 2011, 115, 446–474. [Google Scholar] [CrossRef] [PubMed]
- Lew, R.R. How does a hypha grow? The biophysics of pressurized growth in fungi. Nat. Rev. Microbiol. 2011, 9, 509–518. [Google Scholar] [CrossRef] [PubMed]
- Martí, R.; Laguna, M.; Glover, F. Principles of scatter search. Eur. J. Oper. Res. 2006, 169, 359–372. [Google Scholar] [CrossRef]
- Ibrahim, D. An overview of soft computing. Procedia Comput. Sci. 2016, 102, 34–38. [Google Scholar] [CrossRef]
- Gambhir, S.; Malik, S.K.; Kumar, Y. Role of soft computing approaches in healthcare domain: A mini review. J. Med. Syst. 2016, 40, 1–20. [Google Scholar] [CrossRef]
- Versaci, M.; Laganà, F.; Manin, L.; Angiulli, G. Soft computing and eddy currents to estimate and classify delaminations in biomedical device CFRP plates. J. Electr. Eng. 2025, 76, 72–79. [Google Scholar] [CrossRef]
- Huang, Y.; Lan, Y.; Thomson, S.J.; Fang, A.; Hoffmann, W.C.; Lacey, R.E. Development of soft computing and applications in agricultural and biological engineering. Comput. Electron. Agric. 2010, 71, 107–127. [Google Scholar] [CrossRef]
- Kliebenstein, D.J. Synthetic biology of metabolism: Using natural variation to reverse engineer systems. Curr. Opin. Plant Biol. 2014, 19, 20–26. [Google Scholar] [CrossRef]
- Xi, W.; Xiao, P.; Huang, H.; Hu, Y.; Huang, X. A minireview of fluorescent probes for the dual detection of viscosity and pH: Design and biological applications. Dye. Pigment. 2024, 112412. [Google Scholar] [CrossRef]
- Pullano, S.A.; Greco, M.; Oliva, G.; Laganà, F.; Islam, S.K.; Fiorillo, A.S. Thermoelectrical Analysis of Cell Lines with a Pyroelectric Sensor; Springer Nature: Cham, Switzerland, 2025; pp. 245–249. [Google Scholar]
- Yoshie, H.; Koushki, N.; Kaviani, R.; Tabatabaei, M.; Rajendran, K.; Dang, Q.; Husain, A.; Yao, S.; Li, C.; Sullivan, J.K.; et al. Traction Force Screening Enabled by Compliant PDMS Elastomers. Biophys. J. 2018, 114, 2194–2199. [Google Scholar] [CrossRef]
- Mehta, G.; Mehta, K.; Sud, D.; Song, J.W.; Bersano-Begey, T.; Futai, N.; Heo, Y.S.; Mycek, M.A.; Linderman, J.J.; Takayama, S. Quantitative measurement and control of oxygen levels in microfluidic poly(dimethylsiloxane) bioreactors during cell culture. Biomed. Microdevices 2007, 9, 123–134. [Google Scholar] [CrossRef] [PubMed]



| Organism | Pycnoporus cinnabarinus [36] | Armillaria mellea [16] | Neurospora crassa [16,17] | Neurospora crassa ro-1 [17] |
|---|---|---|---|---|
| Phylum | Basidiomycota | Basidiomycota | Ascomycota | Ascomycota |
| Ecological niche | decomposer | plant pathogen | decomposer | dynein mutants, exhibit pleiotropic phenotypes |
| Leading hypha width (μm) | 5.22 ± 0.74 | 3.86 ± 0.32 | 5.75 ± 0.50 | 5.30 ± 1.02 |
| Apical extension velocity (μm/min) | 1.63 ± 0.4 | 0.6 ± 0.2 | 0.8 ± 0.5 | 1.2 ± 0.7 |
| Apical splitting | no | yes | yes | no |
| Branching distance CIB (μm) | 81.4 ± 39.9 | - | 23.5 ± 15.0 | 17.7 ± 9.0 |
| Lateral branching angle (°) | 70.1 ± 14.6 | 54.6 ± 15.0 | 93.3 ± 15.4 | 89.4 ± 9.0 |
| Angle before CIB (°) | <90 | <70 | <50 | NA |
| Maze exit vs entrance rate | NA | 0.56 | 0. 90 | 0.73 |
| Fungal Level | Individual Hyphae | Co-Located Hyphae | ||||||
|---|---|---|---|---|---|---|---|---|
| Software Species | Sensing Entries | Branching | Directional Memory | Cytoplasm Reallocation | Negative Autotropism | |||
| Contact | Remote | Collision-Induced | Stochastic | Side-by-Side | Head-on | |||
| N. crassa | very strong | strong | strong, backwards | strong | strong | weak | strong | not observed |
| P. cinnabarinus | strong | very strong | strong, at the tip | strong | strong | strong | very strong | very strong |
| A. mellea | strong | weak | none | very weak | weak | very weak | weak | none |
| N. crassa ro-1 | strong | none | none | very strong | none | strong | none | none |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Montiel-Rubies, G.; Held, M.; Hanson, K.L.; Nicolau, D.V., Jr.; Mocanasu, R.C.; van Delft, F.C.M.J.M.; Nicolau, D.V. Hierarchical Structure of the Program Used by Filamentous Fungi to Navigate in Confining Microenvironments. Biomimetics 2025, 10, 287. https://doi.org/10.3390/biomimetics10050287
Montiel-Rubies G, Held M, Hanson KL, Nicolau DV Jr., Mocanasu RC, van Delft FCMJM, Nicolau DV. Hierarchical Structure of the Program Used by Filamentous Fungi to Navigate in Confining Microenvironments. Biomimetics. 2025; 10(5):287. https://doi.org/10.3390/biomimetics10050287
Chicago/Turabian StyleMontiel-Rubies, Gala, Marie Held, Kristi L. Hanson, Dan V. Nicolau, Jr., Radu C. Mocanasu, Falco C. M. J. M. van Delft, and Dan V. Nicolau. 2025. "Hierarchical Structure of the Program Used by Filamentous Fungi to Navigate in Confining Microenvironments" Biomimetics 10, no. 5: 287. https://doi.org/10.3390/biomimetics10050287
APA StyleMontiel-Rubies, G., Held, M., Hanson, K. L., Nicolau, D. V., Jr., Mocanasu, R. C., van Delft, F. C. M. J. M., & Nicolau, D. V. (2025). Hierarchical Structure of the Program Used by Filamentous Fungi to Navigate in Confining Microenvironments. Biomimetics, 10(5), 287. https://doi.org/10.3390/biomimetics10050287







