Three different optimization techniques are used to find protein conformations with the lowest amount of energy.
4.1. Monte Carlo Simulation
Monte Carlo (MC) simulation is a stochastic method that explores the conformational space through random sampling [
46]. It applies the Metropolis criterion [
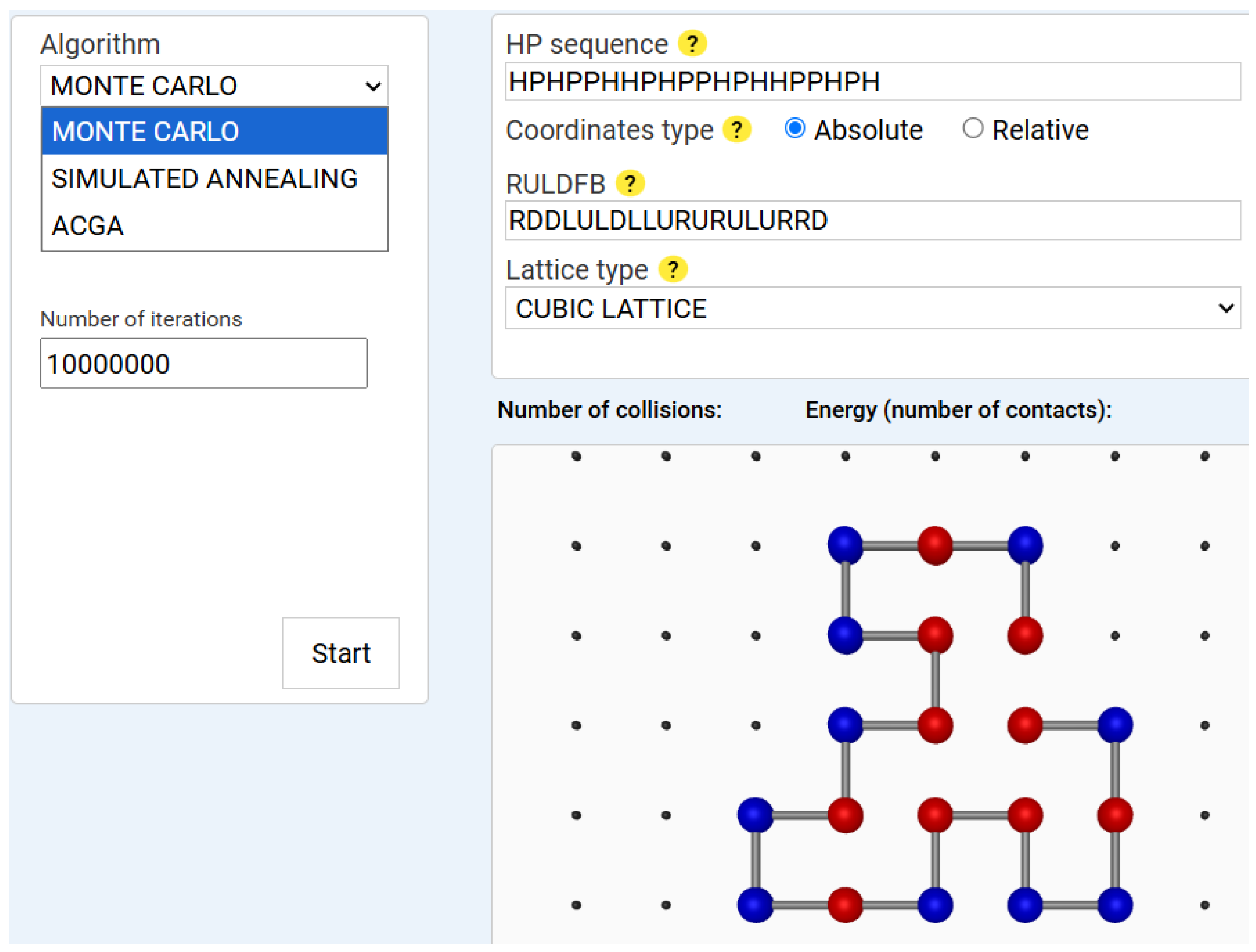
47] to decide whether to accept or reject a structural mutation based on energy change. Our implementation includes custom move sets, such as crankshaft motions, translational moves and pivot (rotational) moves. An example of a Monte Carlo execution result is displayed in
Figure 1.
The system was evaluated with 10,000,000 iterations and a constant temperature of 1.0, which probabilistically accepts the new conformation, according to:
where
(is the energy difference), and
T is the constant temperature.
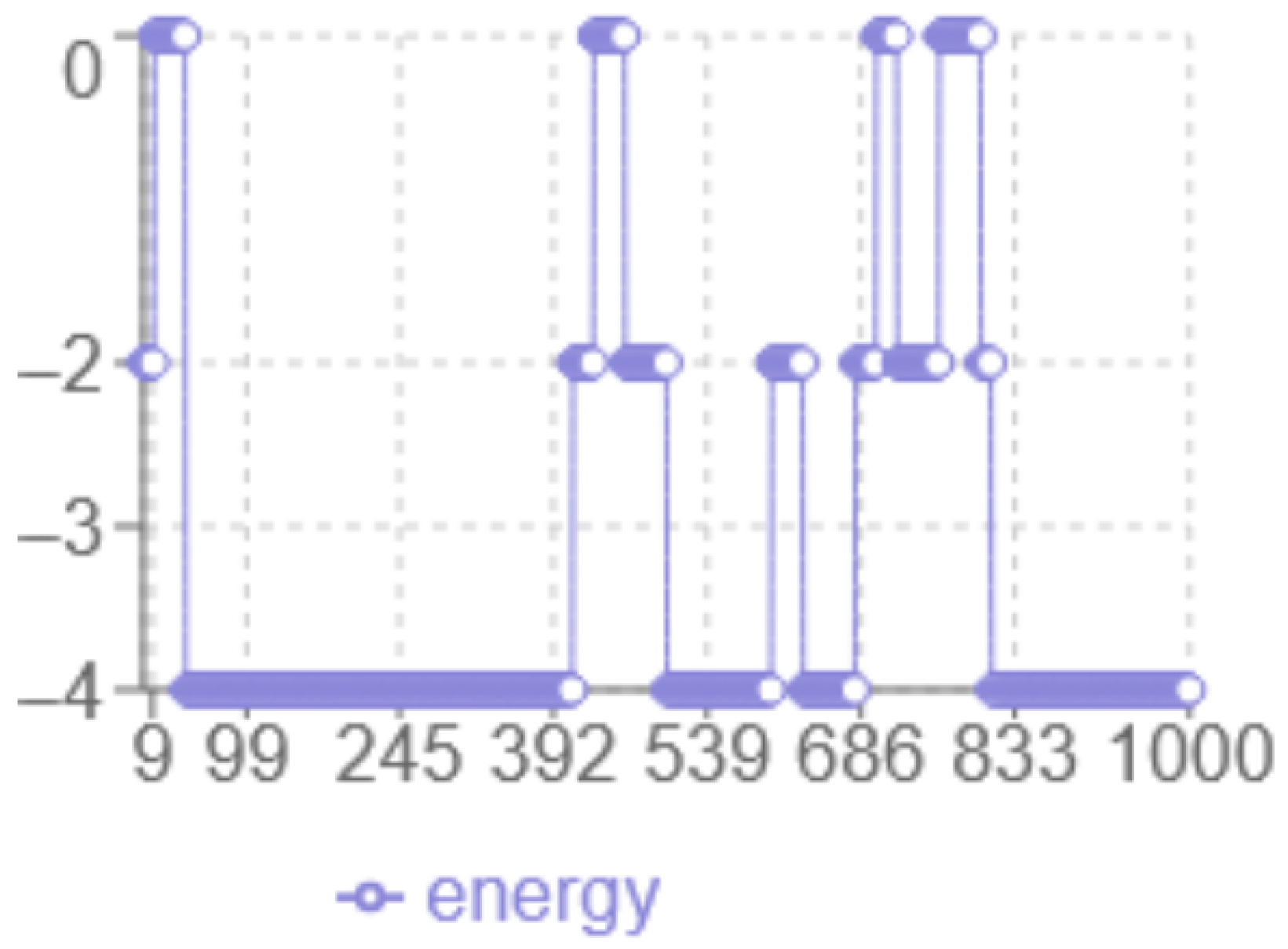
The analysis of the energy profile in
Figure 2 reveals the following characteristics: the
x-axis corresponds to the iteration number, while the
y-axis represents the energy level multiplied by two. The minimum energy achieved is
in this case. The algorithm displays a dynamic behaviour with significant fluctuations between 0 and −2, indicating more in-depth exploration of the solution space, with abrupt transitions that indicate the algorithm ability to escape local optima.
Algorithm 1 implements the Metropolis Monte Carlo (MC) strategy for exploring conformational space in the HP lattice protein folding model. The input consists of a binary HP sequence , where each monomer is either hydrophobic (H) or polar (P), along with the number of iterations and the lattice type (2D square or 3D cubic). The algorithm begins by initializing a valid conformation and computing its energy , which serves as the initial reference.
At each iteration, a new candidate conformation is generated via a mutation operator. If is valid (i.e., free of overlaps), its energy is computed and compared to the previous energy . If the energy difference is negative, the new conformation is accepted. Otherwise, it is accepted with a probability , where T is the system temperature. A random number determines acceptance based on p. If rejected, the previous conformation is retained.
The algorithm iteratively optimized the conformation, storing the best solution
encountered. This approach allows escape from local minima. The output is the optimal conformation string encoded in
RULDFB notation.
| Algorithm 1 Metropolis Monte Carlo Algorithm for HP lattice Model |
- 1:
Input: S, iterations, lattice_type ▹ HP sequence: with - 2:
Output: ▹ RULDFB string (Best conformation found) - 3:
Initialize a valid conformation - 4:
Compute energy of , - 5:
- 6:
for to do - 7:
▹ Generate a new conformation, - 8:
if is valid then ▹ Check for overlaps - 9:
Compute energy - 10:
- 11:
if then - 12:
Accept : , , - 13:
else - 14:
▹ Compute acceptance probability - 15:
Generate random number - 16:
if then - 17:
Accept : , - 18:
else - 19:
Reject : , - 20:
end if - 21:
end if - 22:
else - 23:
Reject : , - 24:
end if - 25:
end for - 26:
return;
|
4.2. Simulated Annealing
Simulated Annealing (SA) extends the Monte Carlo approach by progressively lowering the temperature to reduce the acceptance probability of suboptimal solutions [
48]. This mimics the annealing process in metallurgy. We use exponential cooling and adaptive temperature steps based on energy fluctuations.
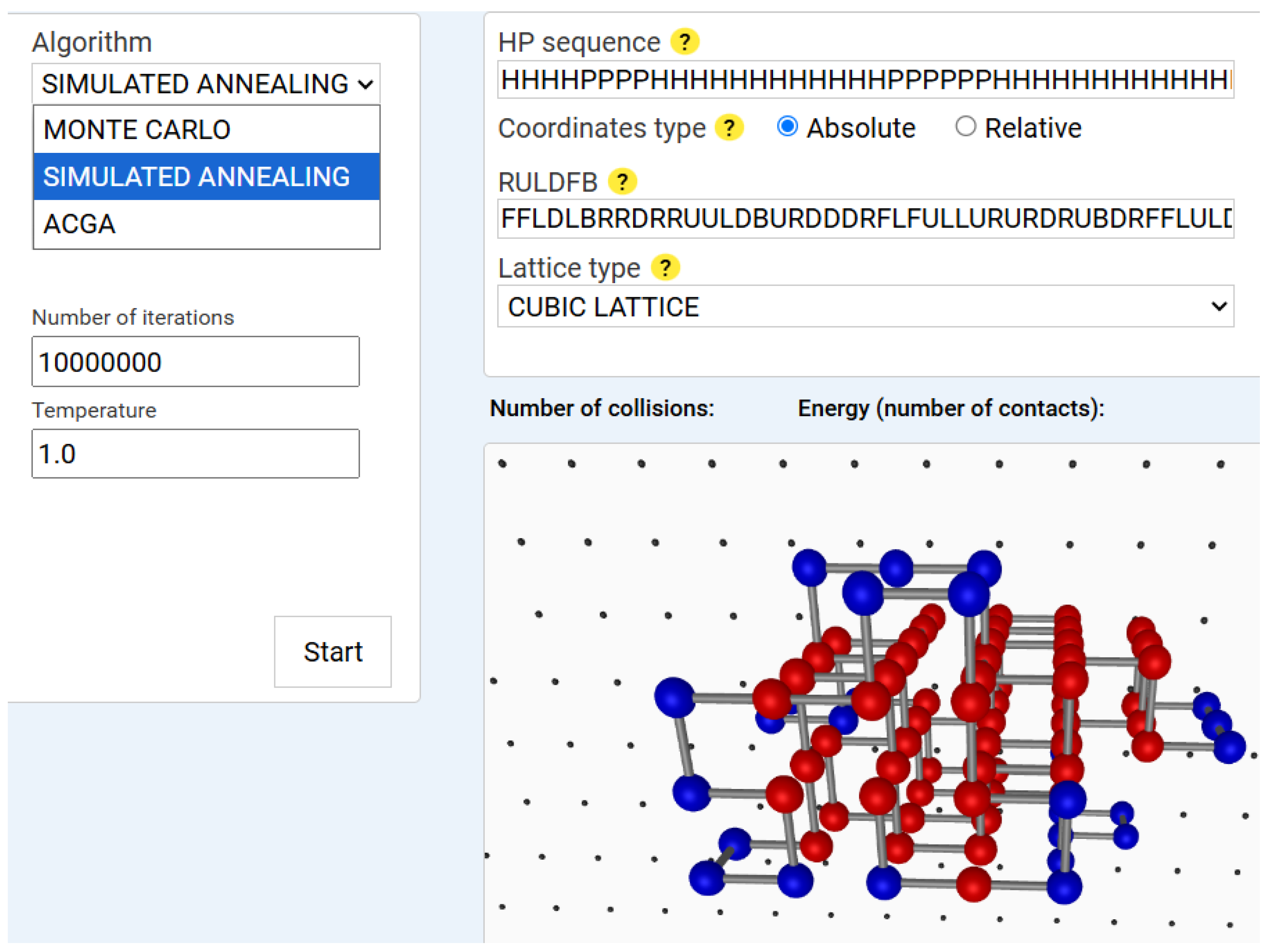
Figure 3 shows the results of a sample Simulated Annealing run.
The SA algorithm also ran for 1,000,000 iterations, starting from an initial temperature
, which was gradually decreased using an exponential cooling schedule:
where
is the temperature at iteration
i;
is the initial temperature;
is the cooling rate, and
i is the iteration index.
According to the energy analysis in
Figure 4, the following observations were made: the
x-axis corresponds to the iteration number, while the
y-axis represents the energy level multiplied by two. The minimum energy achieved is
. The dynamic behaviour shows a transition from
to
without abrupt jumps, and a noticeable plateau around
, indicating prolonged exploration in that region. The final structure have increased linearity with fewer compact hydrophobic interactions, suggesting potential risks in a local optimum.
Algorithm 2 implements the SA optimisation strategy for exploring the conformational space of protein sequences modelled using the HP lattice framework. The input comprises an HP sequence , where each residue belongs to the set , alongside the initial temperature , cooling rate , number of iterations, and the lattice type.
The procedure begins by generating a valid initial conformation and computing its associated energy , which serves as the reference state. At each iteration, a new candidate conformation is produced via a random mutation applied to the current conformation. If is valid (i.e., free from overlaps and maintaining chain connectivity), its energy is evaluated and compared to the previous energy .
If the energy difference is non-positive, the candidate is accepted unconditionally. Otherwise, it is accepted probabilistically with , where represents the temperature at iteration i according to an exponential cooling schedule. A random number is drawn, and is accepted if ; otherwise, the previous conformation is retained.
Throughout the iterative process, the algorithm tracks the best conformation encountered. This stochastic approach enables efficient exploration of the solution space, balancing exploitation of low-energy states with the ability to escape local minima. The final output is the conformation with the lowest energy identified during the search.
As illustrated in Algorithm 2, the key difference between SA and MMC lies in the temperature schedule. While both methods start with an initial temperature set to 1, SA applies an exponential decay across iterations (see line 13 in the algorithm). This adjustment balances the rate between exploration and exploitation. Initially, exploration is encouraged by accepting solutions or conformations with higher energy than the parent conformation. Towards the end of the algorithm, the acceptance probability decreases significantly, thereby promoting exploitation of the combinatorial search space.
| Algorithm 2 Simulated Annealing for HP Lattice Model |
- 1:
Input: S, , , iterations, lattice_type ▹ S - HP String, - initial temperature - 2:
Initialize a valid conformation - 3:
Compute energy of , - 4:
- 5:
for to do - 6:
▹ Generate a new conformation, by random mutation - 7:
if is valid then ▹ Check for overlaps and chain connectivity - 8:
Compute energy - 9:
- 10:
if then - 11:
Accept : , , - 12:
else - 13:
- 14:
▹ Compute acceptance probability - 15:
Generate random number - 16:
if then - 17:
Accept : , - 18:
else - 19:
Reject : , - 20:
end if - 21:
end if - 22:
else - 23:
Reject : , - 24:
end if - 25:
end for - 26:
return
|
4.3. ACGA Algorithm
The proposed ACGA is a population-based method that evolves a set of protein conformations using selection, crossover, and mutation operations. The fitness function incorporates both energy minimisation and structural compactness metrics. ACGA involves the following steps:
- 1.
Population Initialization: Firstly, a population of potential solutions is created. Each solution, referred to as a chromosome, is randomly generated, and it has an associated objective function called fitness.
- 2.
Exploration Stage: The mutation and crossover operators are applied to a certain percentage of the chromosomes in the population, which are chosen randomly. These operators ensure the dispersion of the population in the space of possible solutions, promoting exploration of the solution space.
- 3.
Through the selection operation, from a percentage of the individuals of the population, those with the best fitness are selected. In this way, a new population is created, usually statistically better, and this represents the next generation.
- 4.
Exploitation Stage: Through the selection operation, a certain percentage of individuals with the best fitness are chosen from the population. This process creates a new population, which is usually statistically better and represents the next generation of potential solutions. The selection operation helps exploit the combinatorial space by favoring the fitter individuals for reproduction.
Steps (2) and (3) are iterated for a number of generations, allowing the population to evolve and improve over time. The mutation and crossover operators contribute to the exploration stage, while the selection operator contributes to the exploitation stage of the genetic algorithm.
Chromosomes encoding. We consider that the conformation of the modeled proteins can be encoded using either relative or absolute directions. We have chosen to use both in our approach to exploit the advantages of both encodings. The benefits of using relative encoding are as follows: (a) smaller combinatorial space compared to absolute encoding (relative—; absolute—); (b) implicit avoidance of returning the current AA to the previous AA position in the walk, during the creation of the initial population. (c) Mutation and crossover operators do not require modifications of letters that specify the next positions. On the other hand, absolute coding allows easy conversion into Cartesian coordinates.
Therefore, we have employed both the absolute and relative encodings, resulting in the following corresponding representations (strings):
HP string
RULD string—absolute 2D square
SRL string—relative 2D square
RULDFB string—absolute 3D cubic
SRLFB string—relative 3D cubic
HP string is a sequence string. We will generically call RULD string, SRL string, RULDFB string and SRLFB string as conformation strings.
For computational efficiency reasons, the exploration is performed using the relative encoding, which reduces the conformation space from to . However, the corresponding absolute encoding is also stored to enable easy and fast computation of the Cartesian coordinates. The size of the sequence string is equal to n, and the size of the conformation strings is equal to , with each letter representing the relative successive direction in the conformation, where n is number of AAs of the sequence.
Generation of the initial population. The sequence string represents the input data, and the conformation string is the output of the ACGA algorithm. The primary structure of the protein is represented by the HP string sequence string) of n letters corresponding to the n AAs of a sequence.
Below are the steps for building a conformation (chromosome) using SRL string representation in the population initialization stage:
- 1.
Set i = 1. Initialize SRL[i] = ‘S’.
- 2.
i = i + 1. if continue with the next step. Otherwise, the conformation is completely generated in the string.
- 3.
Choose a random direction ’d’ from the {S,R,L }.
For the 3D case, a similar construction is used based on the string. Then, the string is converted to the string. The first letter, which is S, is always converted to the R letter. This fact reduces the 4-exponential combinatorial space by four times. After that, the string is converted to an array of Cartesian coordinates. Based on these Cartesian coordinates, the number of collisions, the number of contacts, and the fitness are computed. The math formulas used for finding an H-H contact in the 2D square and 3D cubic lattices are as follows.
For the 2D square lattice:
If , where , are the Cartesian coordinates of the i-th AA, and , are the Cartesian coordinates of the j-th AA, then there is a contact between the two AAs at positions i and j, .
For the 3D cubic:
If , where , , are the Cartesian coordinates of the i-th AA, and , , are the Cartesian coordinates of the j-th AA, then there is a contact between the two AAs at positions i and j, .
For finding a collision (two AAs in the same place), we use the following equations, where the terms have the same understanding as above:
For 2D square lattice, if : ;
For 3D cubic lattice, if :
If these equations are satisfied, it indicates that there is a collision between the AAs at positions i and j in the conformation.
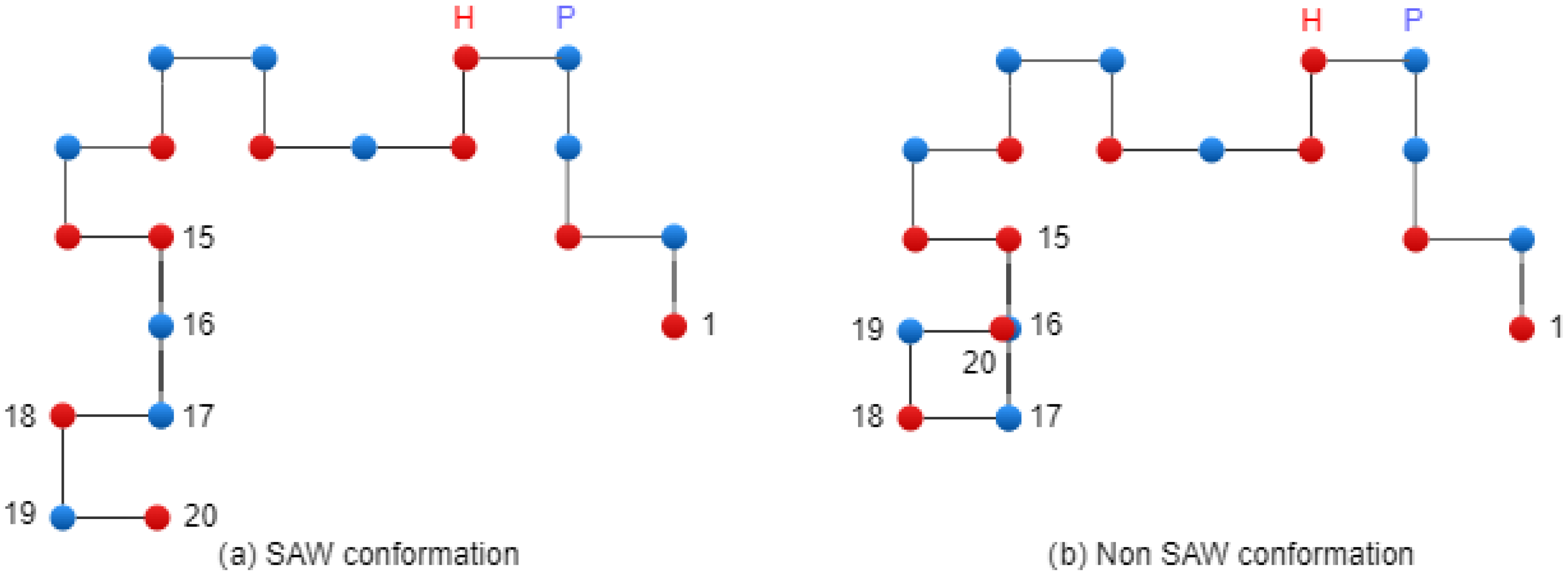
Figure 5 presents two conformations: on the left side, there is a SAW conformation, and on the right side, there is a conformation that has one collision (non-SAW conformation).
Fitness Evaluation Strategy. The fitness function evaluates how close a given chromosome is to the optimum solution. It determines how fit a chromosome is. We have used a fitness function (see Equation (
5)) inspired by the code of Alican Toprak. (
https://github.com/alican/GeneticAlgorithm accessed on 6 November 2025).
where
c is the conformation,
is the number of contacts, and the
is the number of collisions of the conformation. This is computed by checking the topological neighborhood of all AAs on the lattice, according to the number of contacts (Equation (
2)) and the number of collisions (
). There are two exceptions: if
then the formula becomes
and if
then
is replaced with 2. Thus, the fitness increases with the number of contacts and is strongly penalized by the number of collisions.
Adapted tournament selection. We proposed an adapted variant of tournament selection, which increases the probability of individuals with low energy values entering the next generation, while implicitly conserving the best individual. Specifically, the selection is applied to the previous population by choosing pairs of chromosomes at random, and after comparing them, the best one is copied into the position of the worst one. This way, the best chromosome is preserved through the generations.
Crossover. In addition to the rotational crossover used in our previous work [
49], we apply the translational crossover. For both types of crossover operators, the best chromosome (
) from the current population is protected. The crossover operation is performed using the following formula:
where
is the best chromosome from the current population, and
and
are parent chromosomes. The reason for introducing
, as parameter into the crossover operator is to protect this chromosome.
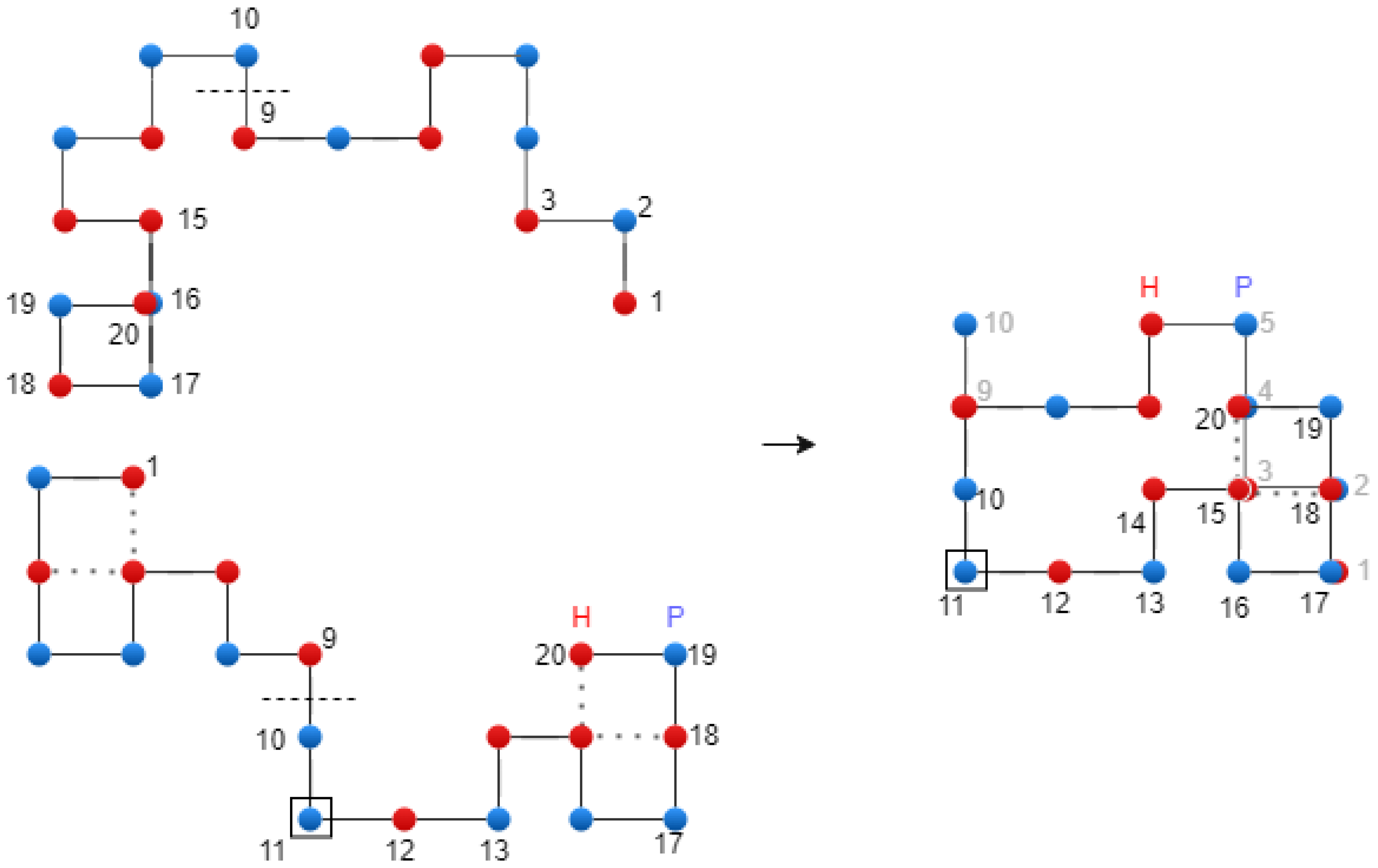
Figure 6 shows the translational crossover.
Mutation. We employ translational, rotational, and diagonal mutations. Given a chromosome , where for 2D (or for 3D), it is mutated to a new chromosome, C’. To achieve this, a position g (1 ≤ g ≤ n), known as the mutation point, is randomly chosen for each conformation. The letter at position g is then replaced by one letter sampled uniformly from the set of possible directions.
For the rotational mutation, the modification is applied to the
string, and the next letter after the
g point remains unchanged. Then, the
string is converted to the
string. This modification produces a rotation of the second part of the chain by
,
, or
, respectively. In the case of translational mutation, the modification is applied to the
string, and the next letter after the
g point remains unchanged.
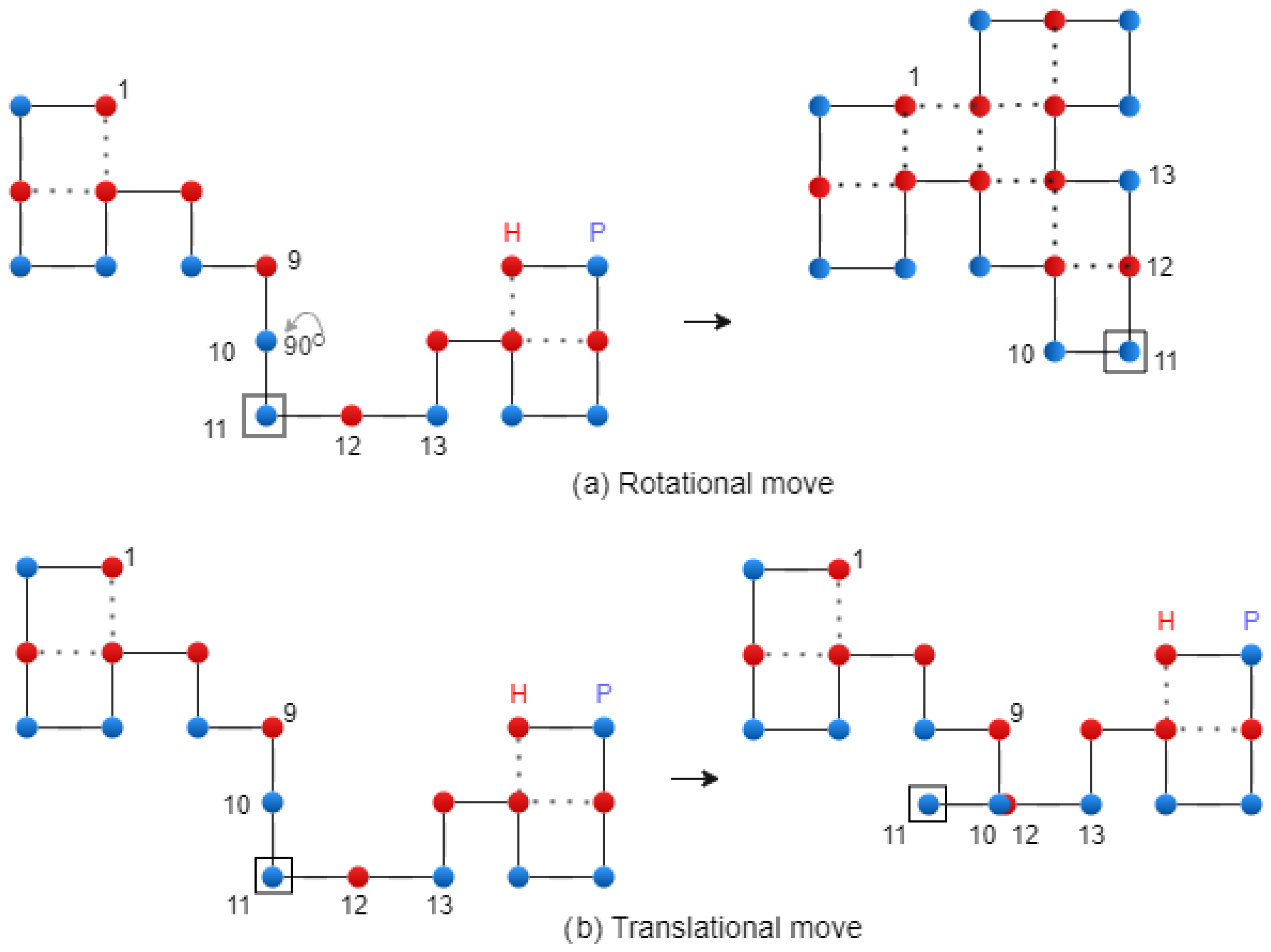
Figure 7 shows the two types of mutation. A diagonal move is executed on the two letters of the
string that form a corner. Finally, the mutation operation is performed using the following formula:
where
is the best chromosome from the current population and
t represents the iteration number (time). The reason for introducing
, as parameter into the mutation operator is to protect it.
The algorithm. For every generation, the next operations are executed: (a) rotational crossover, (b) translational crossover, (c) translational mutation, (d) rotational mutation, (e) diagonal mutation and (f) tournament selection. After iterating all generations, the algorithm returns the best conformation obtained.
Pseudocode of the ACGA algorithm skeleton is given in Algorithm 3. As can be seen, the stopping criterion of the algorithm consists of reaching the number of generations, given as an input parameter.
| Algorithm 3 All Conformations Genetic Algorithm (ACGA) |
- 1:
Input: - 2:
Output: - 3:
Initialization of the population - 4:
Compute fitness of each conformation conf Equation ( 5) - 5:
Adapted tournament selection - 6:
the best conformation - 7:
- 8:
while do - 9:
for (every chosen conformation) do - 10:
- 11:
- 12:
end for - 13:
Compute the fitness of modified chromosome - 14:
- 15:
the best conformation - 16:
- 17:
end while - 18:
return C*
|