1. Introduction
Colorectal cancer (CRC) remains a leading cause of preventable cancer death worldwide [
1]. The classical model of CRC tumorigenesis is from sporadic colorectal polyps, which progress over a period of several years before developing malignant potential. This, therefore, allows a window of opportunity for the detection and resection of these premalignant polyps, which has been shown to reduce the incidence of and mortality from CRC [
2]. In common with any operator-dependent procedure, colonoscopy and polyp detection has certain limitations. It is understood that small or subtle polyps may be missed at colonoscopy, with an operator-dependent miss-rate for adenomas as high as 26% [
3]. In recognition of this, the adenoma detection rate (ADR) has been introduced as a surrogate marker for a thorough endoscopic examination and is one of the key performance indicators for endoscopists. It has been demonstrated that an improved ADR is inversely proportional to post-colonoscopy risk of CRC, with each 1% increase in ADR providing a 3% decrease in the subsequent risk of cancer [
4]. There is certainly incentive to maximize polyp detection at the time of endoscopy.
A proportion of the polyps missed during colonoscopy could potentially have been identified if the operator possessed optimal examination skills and polyp recognition capabilities. Evidence for this is shown by studies suggesting that ADR can be increased by improving behavioral and technical skills of the endoscopist [
5]. Training programs consisting of hands-on teaching and regular feedback showed positive results in increasing ADR in clinical trials [
5,
6]. However, the increased ADR in these studies was small and the ability of endoscopists to detect very small, subtle, or flat lesions remains a limiting factor.
Recently, there has been an explosion in the application of artificial intelligence (AI), and its branch deep learning (DL), to provide computer-aided diagnosis (CAD) technologies in medical and health-care diagnostics [
7]. The ability of AI, and specifically DL and computer vision approaches, to locate, differentiate, and characterize distinct pathologies can enhance and surpass traditional hand-crafted CAD techniques [
8]. Current endoscopic equipment now provides exceptionally high-quality images, with even 1080p HD images or 2K imaging technology, but the presence on non-informative frames due to blurry frames, water frames, and bubble frames is still high [
9], which makes it more difficult to distinguish tissues.
Comparison of polyp detection and localization methods is a necessary yet complex process as (1) performance results are highly sensitive to design choices, (2) common metrics do not reflect clinical needs, and (3) comparison across datasets can be misleading [
10]. In regard to the latter, it must also be recognized that most datasets present clearly visible polyps, well-centered in the image, although this is not representative of the polyp detection and localization in the clinical setting where lesions often first appear at the edges of the image. This would also be related to point two, as technical metrics do not consider clinical indicators. Thus, clinical needs and casuistry should not be left aside when validating DL methods in a laboratory environment.
Beside comparison between methods, it is also necessary to compare the DL models versus clinicians, to demonstrate the added value of these types of CAD systems in the clinical practice. Many efforts have been made recently in this regard. Li et al. [
11] analyzed the performance of AI models for polyp detection and classification in comparison to clinicians, using different imaging modalities. In general, AI systems obtained high sensitivity and moderate specificity for both tasks, similarly to human experts. Nevertheless, none of the works for polyp detection considered endoscopic images and experts, so comparison in this situation is not included. Similarly, Xu et al. [
12] do not include any work that compares the DL detection method with clinicians (neither experts nor non-experts) in their meta-analysis. In any case, the ideal validation for any DL model or CAD system would be a randomized control trial (RCT), to prove the actual added value of such systems [
13,
14,
15]. Hassan et al. [
16] just included five RCTs in their systematic review, which showed the significant increase in detection when AI systems were used. A small number of RCTs might be due to the limitations for such trials, which might be difficult to overcome for some research groups with limited possibilities, having to face common barriers such as lack of funding, complex regulatory requirements, or inadequate infrastructures [
17]. It would be useful to establish a preliminary test that could be used as a prior step to a fully clinical validation and that would allow for a comparison between the DL model and gastroenterologists in a laboratory environment.
The aim of this study is to present the ClinExpPICCOLO dataset, a dataset for the clinical benchmark of DL methods for localization of colorectal polyps when viewing unedited endoscopic images, and to establish an expert clinical performance baseline to use as a comparator when analysing DL methods against clinicians.
2. Materials and Methods
2.1. ClinExpPICCOLO Dataset
The polyp image library for detection and localization of colorectal polyps was created from the endoscopic videos originating the ‘test’ subset of the PICCOLO dataset [
18] to assure subset independence in the event of DL methods trained with this dataset. The PICCOLO dataset in its entirety contains 3433 manually segmented images containing endoscopic images of polyps, captured under WLE and NBI during routine colonoscopy. The ‘test’ set includes 333 images, each of which contains one or more colorectal polyps. These were reviewed individually, and over 100 were selected for inclusion in this study, showing polyps at a variety of sizes, orientations, and distances. For specific details of the acquisition and annotation protocols, the reader is referred to [
18].
To supplement these images, representative images that did not contain polyps or showed one or more polyps not centered in the image were selected from the original endoscopic videos. A review group of endoscopists (BG, SN) selected an initial batch of 200 endoscopic images to be used in the study.
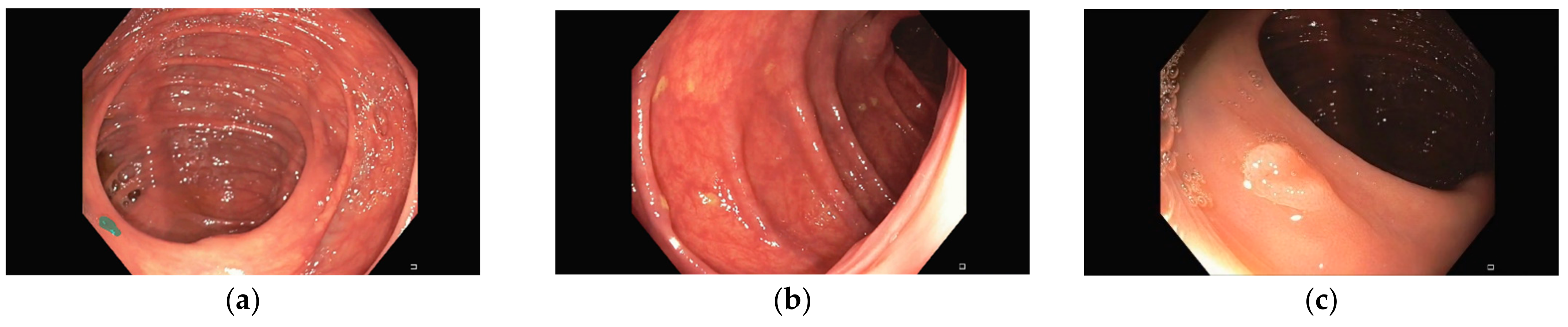
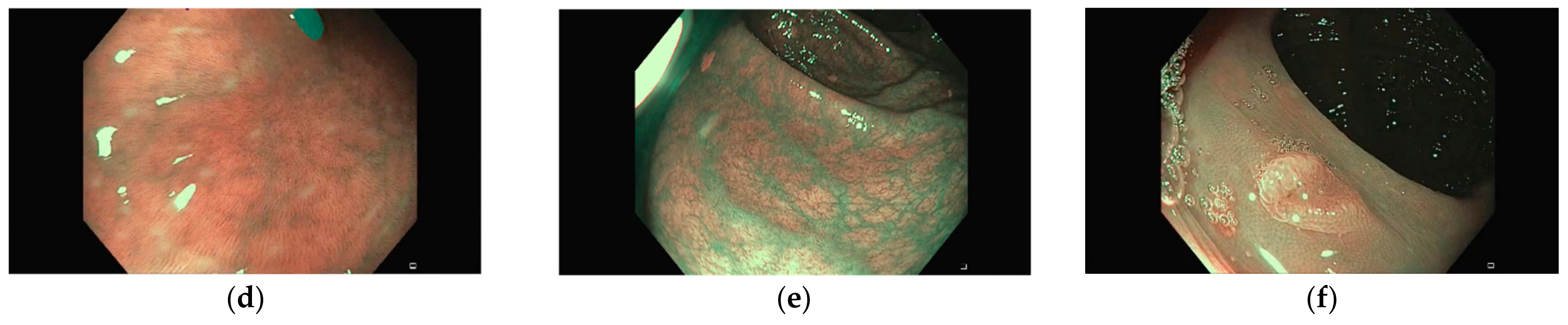
To give a comparable benchmark to the ADR of a skilled endoscopist, it was decided that approximately one third of the images should contain a polyp, with the remainder showing no abnormality. The second batch of images were combined with the polyp-containing images from the test set of PICCOLO dataset and reviewed again by the study group. Images that were deemed to be of low quality, or that showed polyps at a high degree of magnification, were removed. Images with a high level of inter-or intra-reviewer disagreement regarding their content were also removed. Lastly, selected images were re-scrutinized to ensure no patient-identifiable data remained within each image. In all, 65 images were selected, 19 of them showing 21 polyps and the remaining 46 with no polyps. Out of the 19 polyp images, 15 were WLE and 4 were NBI, as this modality is preferred for diagnosis rather than for detection of lesions.
All image sizes were either 1920 × 1080 or 854 × 480. In preparation for display to endoscopists, the images were uploaded to a pre-existing digital platform designed for the collection of survey data (Qualtrics, UT, US).
In parallel, the review group of endoscopists agreed on the area of the polyps, for which a binary mask was created using GTCreatorTool [
19]. These manually annotated binary masks were used as ground truth.
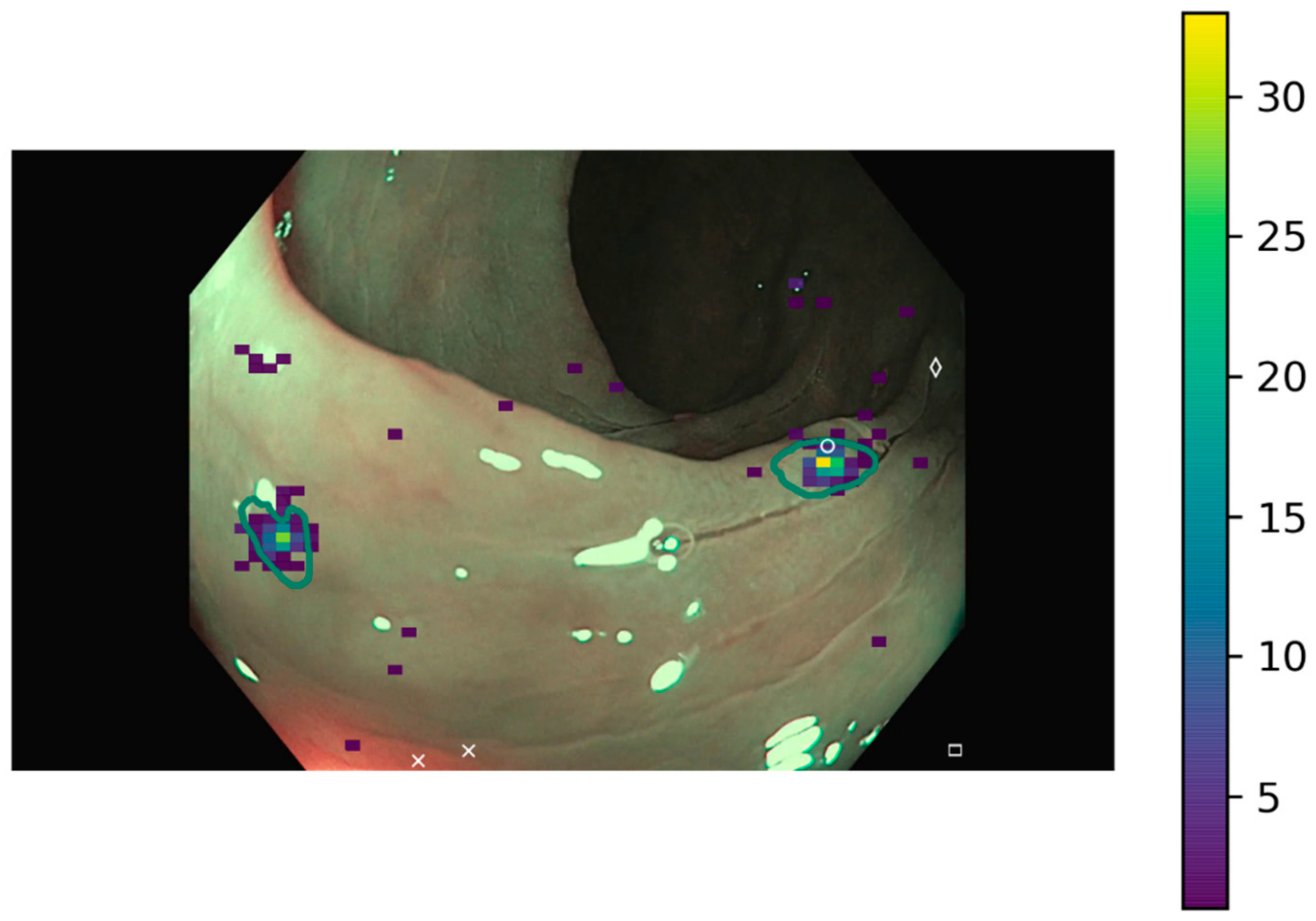
Representative images of the ClinExpPICCOLO dataset are shown in
Figure 1. Both WLE and NBI, showing a polyp or not, are included. In comparison to images from the original PICCOLO dataset, images are not focused on the lesions.
2.2. Ethics
Ethical approval was obtained for the creation of the PICCOLO dataset [
18]. The endoscopic images and videos were collected at Hospital Universitario de Basurto (Bilbao, Spain), under ethical approval of the Ethical Committee of the Basque Country (CEIm-E). Patients provided written informed consent using document PI+CES-BIOEF 2017-03, allowing for use of images in research. As this was an observational survey design involving no identifiable participants, additional ethical approval was not sought for the work detailed in this study.
2.3. Deep Learning Models
In this work, four different DL models trained with the PICCOLO dataset have been used (U-Net+VGG16, LinkNet+Densenet121, U-Net+Densenet121, and LinkNet+VGG16) as example. These models are presented in [
18] and show the best generalization capabilities among all tested models for polyp segmentation. These models have an encoder–decoder architecture. While the encoder processes the input image and transform it into a feature vector, the decoder reconstructs it into the binary prediction mask, of equal size to the input image.
2.4. Clinical Performance Baseline
We recruited participants from local and international professional networks, including digital gastroenterology and endoscopy training networks. A unique URL was sent to potential participants, allowing them to take part in the clinical validation stage.
The images were randomly ordered and presented to participants through the online survey on Qualtrics. This allowed images to be displayed to the survey respondent, and for them to indicate the presence of any polyps in the image by either a mouse click if using a computer, or by a tap on the screen if using a mobile device. Due to the maximum allowed image size, 1920 × 1080 images were automatically resized to 900 × 506, while 854 × 480 remained unresized.
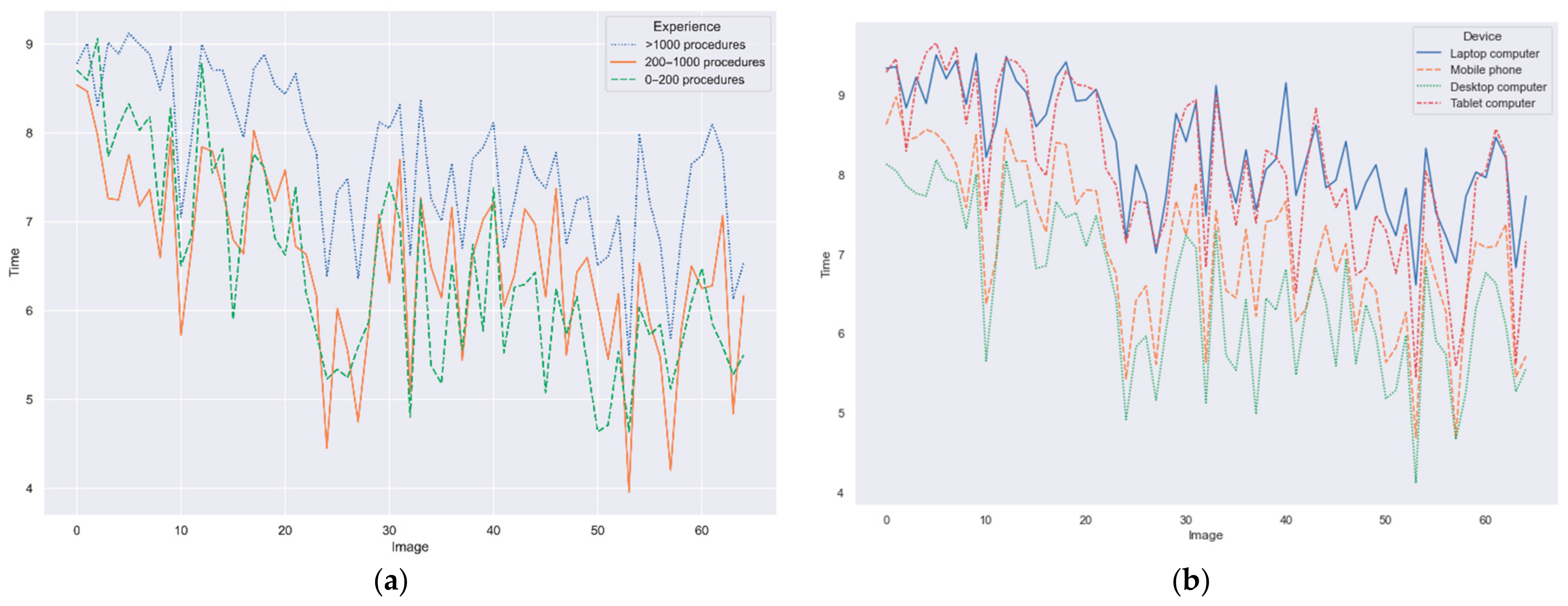
This survey included the instructions to complete it as well as an initial set of demographic questions. Since the survey could be accessed through different devices and, therefore, the displayed image size might vary, the survey also included a question to identify the used device among the following options: desktop computer, laptop computer, tablet computer, and mobile phone.
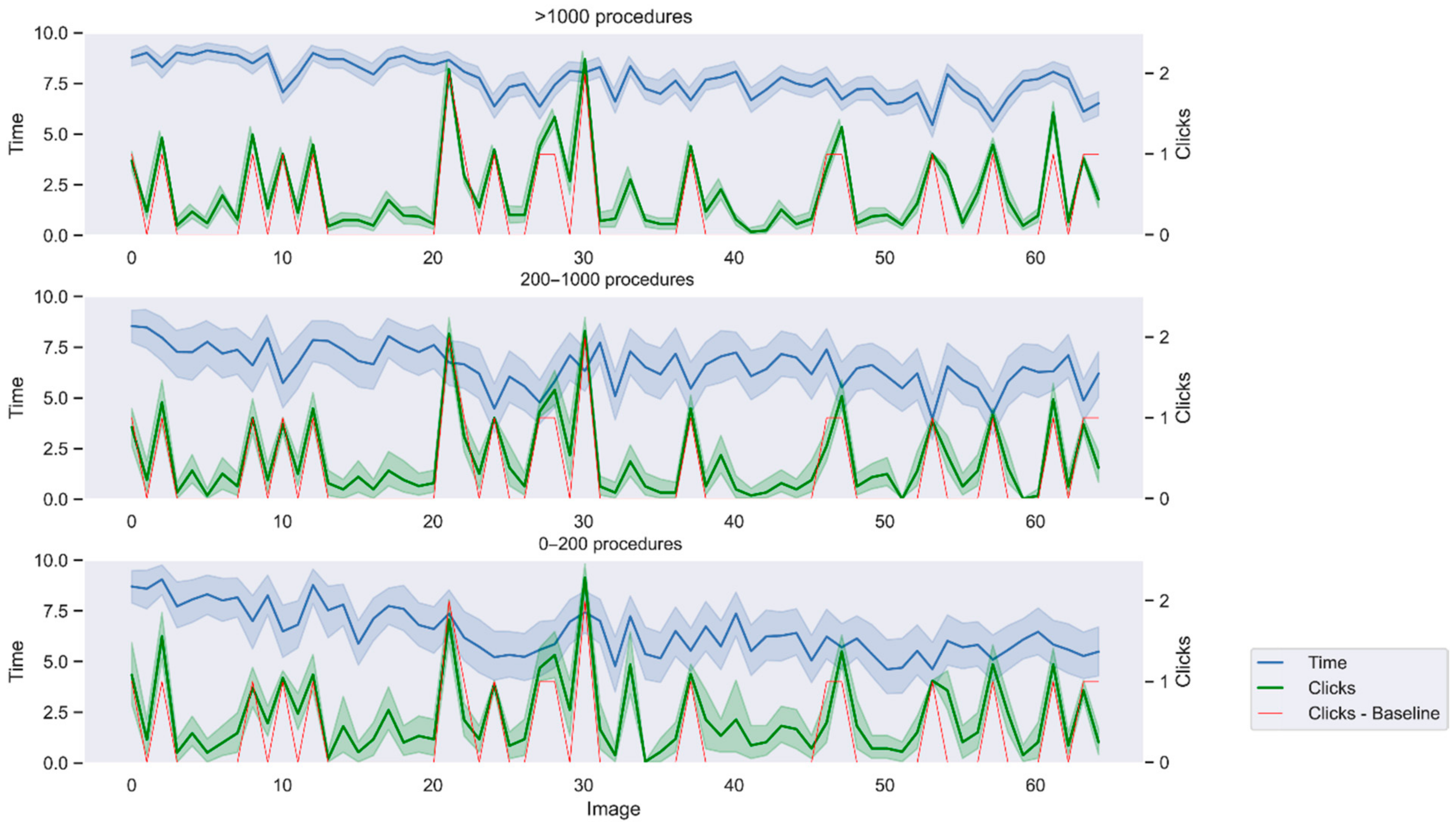
A limit of three clicks per image was set to avoid unlimited attempts. If a user performed more than three clicks on the same image, the fourth click would replace the first one and so on. In addition, a timer per question was also included. A time limit of 10 s was established. During this time, the user could click polyps on the image and move to the next image, otherwise the survey automatically skipped to the next image when the time ran out. Time and clicks are automatically recoded by Qualtrics. Solutions were not displayed to not bias user behavior depending on whether the click correctly located a polyp or not. Lastly, we also analyzed factors that might influence the results, such as prevalence bias or time pressure.
2.5. Calibration Survey
A calibration survey was also created to measure the error of the different input devices. Four black images showing three white crosses each at predefined location were uploaded to Qualtrics. Two images were 1920 × 1080 and the other two were 854 × 480 to account for the image size of endoscopic images. Participants were asked to click in the cross center. Error was measured as the Euclidean distance between each of the three input points and the center of the closest white cross.
2.6. Evaluating the Results
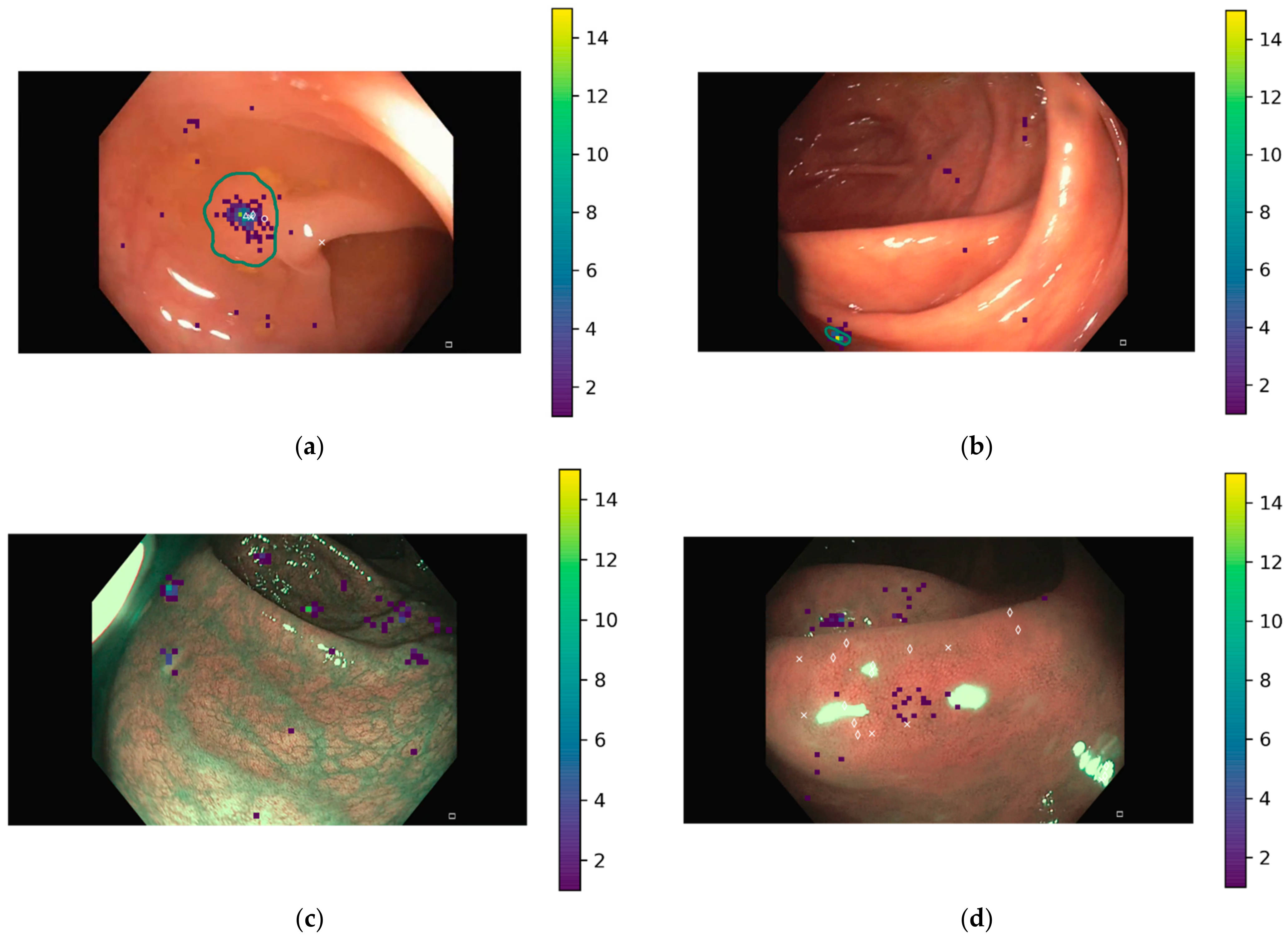
Both the clicks from the questionnaire and the predicted binary masks by the DL models were processed to identify whether the polyps were correctly localized or not. To liken the binary masks of the DL models with the clicks provided by gastroenterologists, centroids of each region in the binary masks were calculated and considered as the location points by the DL methods.
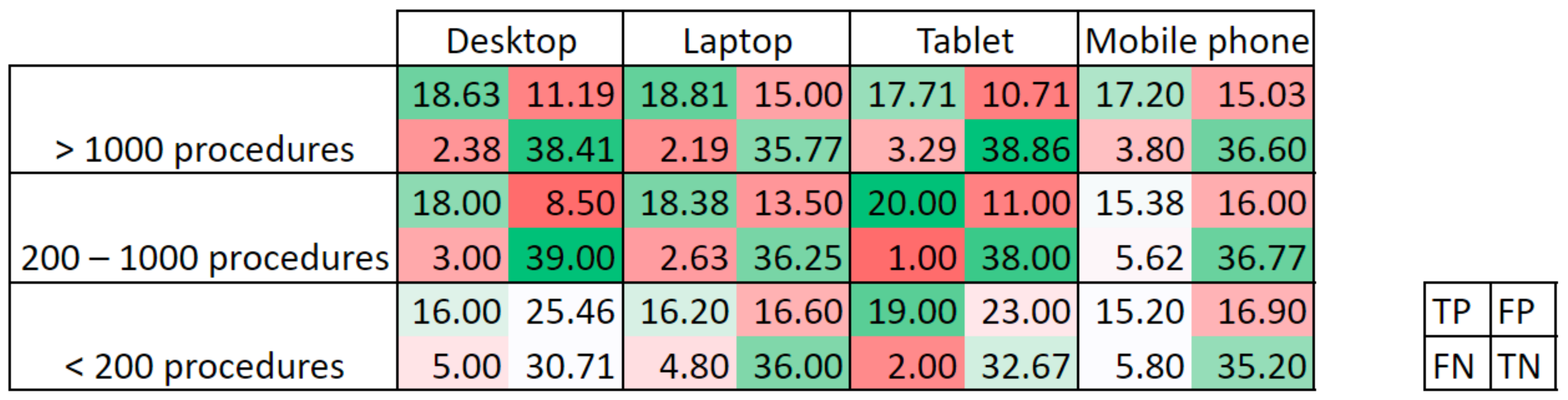
Next, the (x,y) coordinates of up to three clicks provided by Qualtrics and those of all centroids were analyzed to obtain the confusion matrix for each user/model and image. Points were labelled as:
True positive (TP), if the point lay within the area delimitated as polyp in the ground truth. If more than one point was in the polyp area, only one TP was counted. Calibration errors have been considered to correct the location points depending on the input device and image size. Therefore, if the point was within the corresponding error distance from the polyp area indicated in the ground truth, it was also considered as TP;
False positive (FP), if the point lay outside the area delimitated as polyp in the ground truth.
On the other hand, true negatives (TN) were considered when the ground truth was a black image, and no point was given, while false negatives (FN) were considered when no point lay within the polyp area in the ground truth.
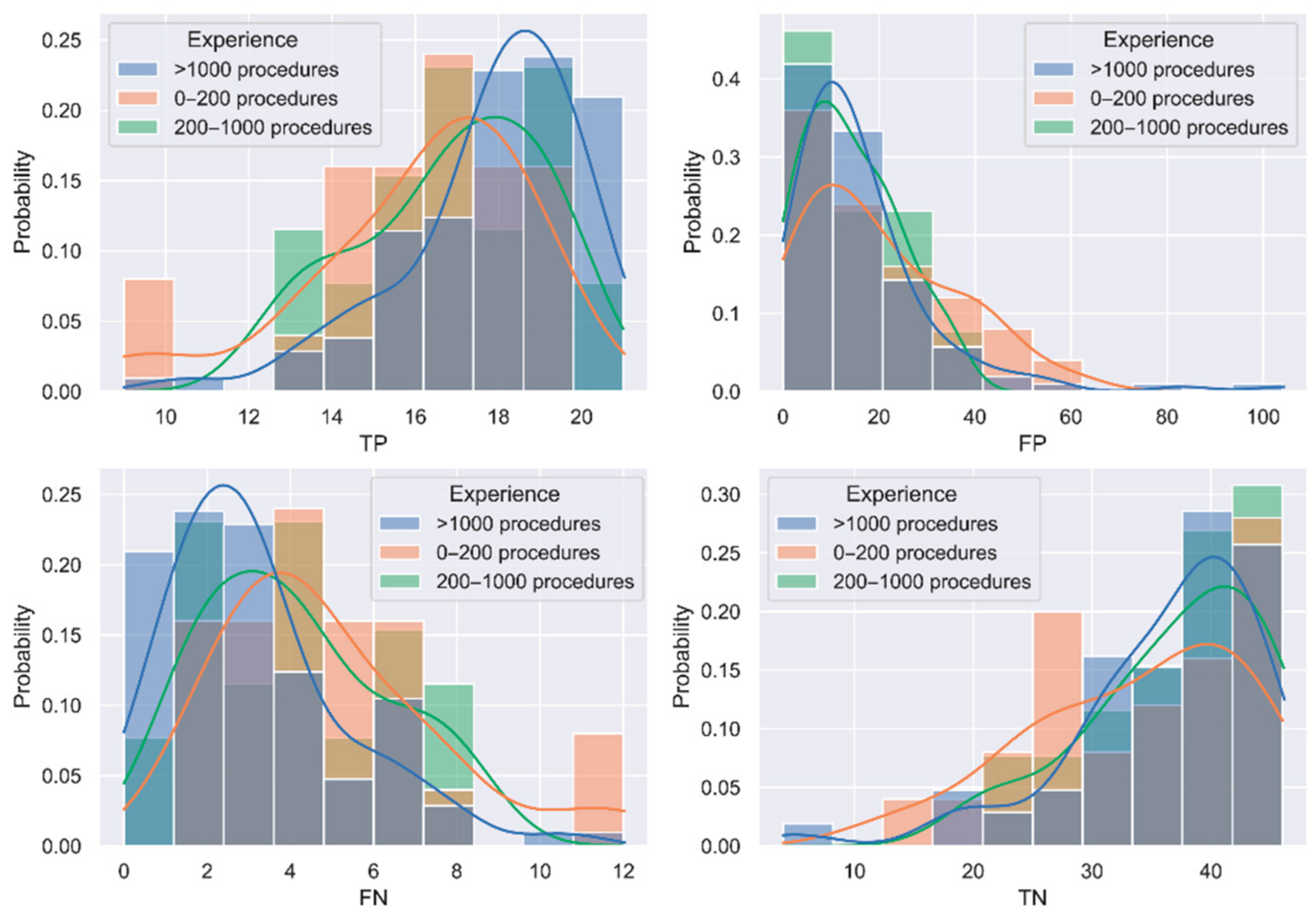
With this convention, a maximum value of 21 TP and 46 TN could be obtained. Based on the elements of the confusion matrix, a set of eight metrics have been calculated:
Furthermore, a simulated ADR was calculated per participant/model. In this case, the original video from which each polyp frame was extracted was considered, as they originated from different patients. In all, there were images from seven unique videos and detection was considered when at least one polyp in the video was identified (i.e., a polyp in any of the images of the video has at least one true positive).
2.7. Statistical Analysis
Results of the questionnaires and DL models have been statistically analyzed to identify differences between groups.
Interquartile ranges (IQR) have been calculated for the confusion matrix elements per group of experience, to identify outliers. In this case, outliers are considered those observations 1.5 times the IQR below the first quartile or above the third quartile.
A permutation test implemented in Python 3.7 was used to identify whether differences between groups were statistically significant without assuming any data distribution. One million iterations in the permutation test are calculated and significance is evaluated at p-value < 0.05.