Deep Learning Approaches for Data Augmentation in Medical Imaging: A Review
Abstract
1. Introduction
2. Background
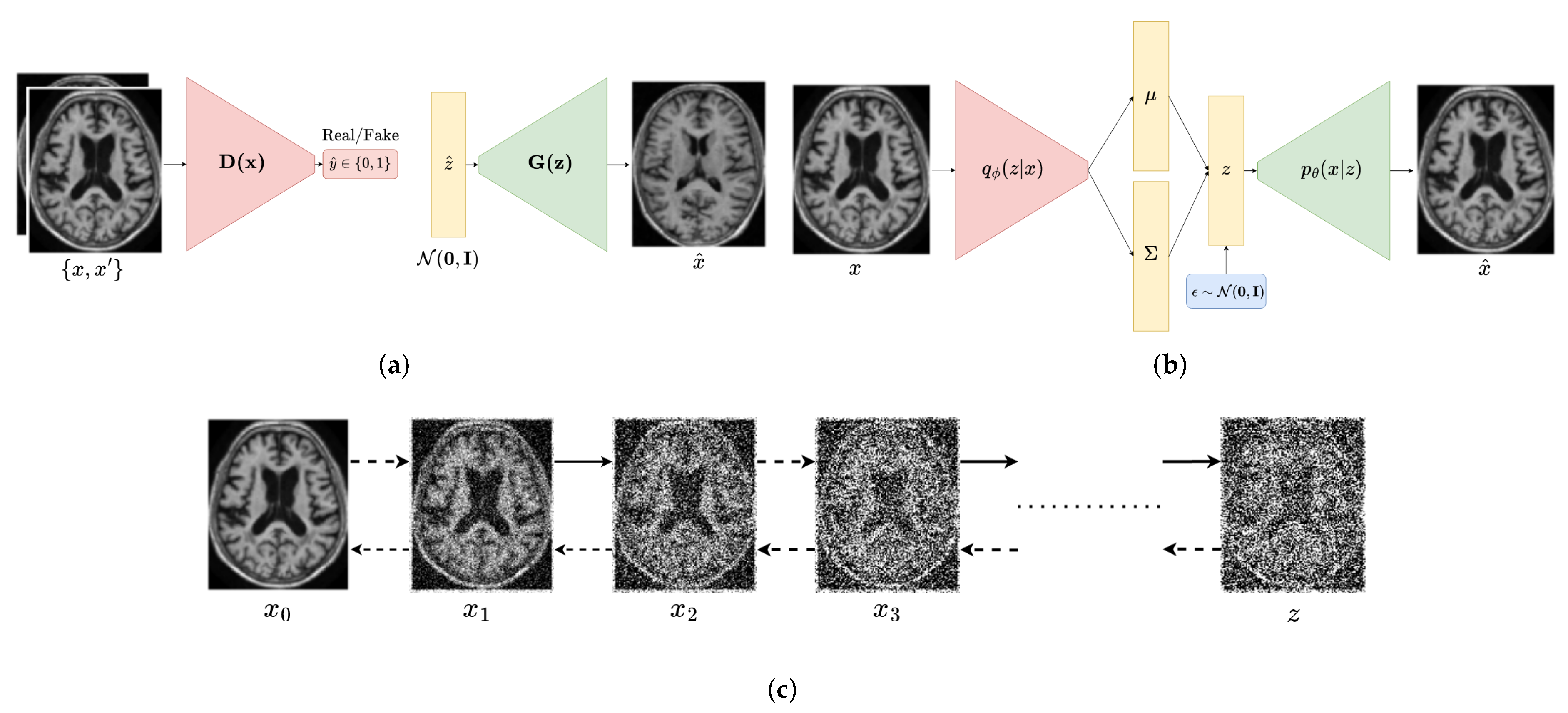
2.1. Generative Adversarial Networks
2.2. Variational Autoencoders
2.3. Diffusion Probabilistic Models
2.4. Exploring the Trade-Offs in Deep Generative Models: The Generative Learning Trilemma
2.4.1. Generative Adversarial Networks
2.4.2. Variational Autoencoders
2.4.3. Diffusion Models
3. Deep Generative Models for Medical Image Augmentation
3.1. Generative Adversarial Networks
3.2. Variational Autoencoders
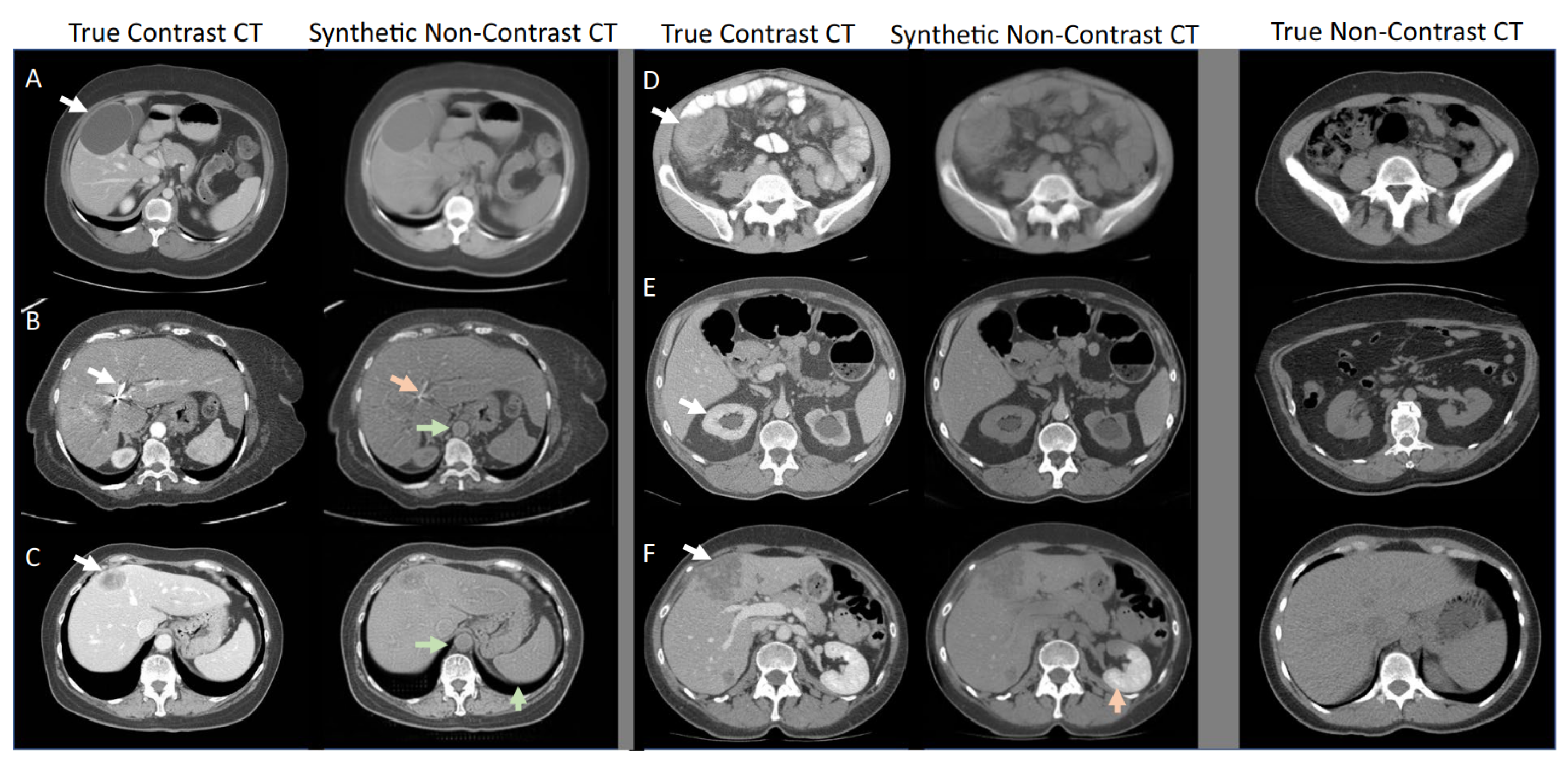
3.3. Diffusion Models
4. Key Findings and Implications
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Amyar, A.; Modzelewski, R.; Vera, P.; Morard, V.; Ruan, S. Weakly Supervised Tumor Detection in PET Using Class Response for Treatment Outcome Prediction. J. Imaging 2022, 8, 130. [Google Scholar] [CrossRef] [PubMed]
- Brochet, T.; Lapuyade-Lahorgue, J.; Huat, A.; Thureau, S.; Pasquier, D.; Gardin, I.; Modzelewski, R.; Gibon, D.; Thariat, J.; Grégoire, V.; et al. A Quantitative Comparison between Shannon and Tsallis–Havrda–Charvat Entropies Applied to Cancer Outcome Prediction. Entropy 2022, 24, 436. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Ruan, S.; Vera, P.; Canu, S. A Tri-Attention fusion guided multi-modal segmentation network. Pattern Recognit. 2022, 124, 108417. [Google Scholar] [CrossRef]
- Chen, X.; Konukoglu, E. Unsupervised detection of lesions in brain MRI using constrained adversarial auto-encoders. arXiv 2018, arXiv:1806.04972. [Google Scholar]
- Lundervold, A.S.; Lundervold, A. An overview of deep learning in medical imaging focusing on MRI. Zeitschrift für Medizinische Physik 2019, 29, 102–127. [Google Scholar] [CrossRef]
- Song, Y.; Zheng, S.; Li, L.; Zhang, X.; Zhang, X.; Huang, Z.; Chen, J.; Wang, R.; Zhao, H.; Chong, Y.; et al. Deep learning enables accurate diagnosis of novel coronavirus (COVID-19) with CT images. IEEE/ACM Trans. Comput. Biol. Bioinform. 2021, 18, 2775–2780. [Google Scholar] [CrossRef]
- Islam, J.; Zhang, Y. GAN-based synthetic brain PET image generation. Brain Inform. 2020, 7, 1–12. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. Commun. ACM 2017, 60, 84–90. [Google Scholar] [CrossRef]
- Zhou, T.; Vera, P.; Canu, S.; Ruan, S. Missing Data Imputation via Conditional Generator and Correlation Learning for Multimodal Brain Tumor Segmentation. Pattern Recognit. Lett. 2022, 158, 125–132. [Google Scholar] [CrossRef]
- Paszke, A.; Gross, S.; Massa, F.; Lerer, A.; Bradbury, J.; Chanan, G.; Killeen, T.; Lin, Z.; Gimelshein, N.; Antiga, L.; et al. Pytorch: An imperative style, high-performance deep learning library. Adv. Neural Inf. Process. Syst. 2019, 32. [Google Scholar]
- Goodfellow, I.J.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative adversarial networks. arXiv 2014, arXiv:1406.2661. [Google Scholar] [CrossRef]
- Sandfort, V.; Yan, K.; Pickhardt, P.J.; Summers, R.M. Data augmentation using generative adversarial networks (CycleGAN) to improve generalizability in CT segmentation tasks. Sci. Rep. 2019, 9, 16884. [Google Scholar] [CrossRef]
- Mahapatra, D.; Bozorgtabar, B.; Garnavi, R. Image super-resolution using progressive generative adversarial networks for medical image analysis. Comput. Med. Imaging Graph. 2019, 71, 30–39. [Google Scholar] [CrossRef]
- Yi, X.; Walia, E.; Babyn, P. Generative adversarial network in medical imaging: A review. Med. Image Anal. 2019, 58, 101552. [Google Scholar] [CrossRef] [PubMed]
- Ali, H.; Biswas, M.R.; Mohsen, F.; Shah, U.; Alamgir, A.; Mousa, O.; Shah, Z. The role of generative adversarial networks in brain MRI: A scoping review. Insights Imaging 2022, 13, 98. [Google Scholar] [CrossRef]
- Chen, Y.; Yang, X.H.; Wei, Z.; Heidari, A.A.; Zheng, N.; Li, Z.; Chen, H.; Hu, H.; Zhou, Q.; Guan, Q. Generative adversarial networks in medical image augmentation: A review. Comput. Biol. Med. 2022, 105382. [Google Scholar] [CrossRef] [PubMed]
- Mescheder, L.; Geiger, A.; Nowozin, S. Which training methods for GANs do actually converge? In Proceedings of the International Conference on Machine Learning, PMLR, Stockholm, Sweden, 10–15 July 2018; pp. 3481–3490. [Google Scholar]
- Kingma, D.P.; Welling, M. Auto-encoding variational bayes. arXiv 2013, arXiv:1312.6114. [Google Scholar]
- Sohl-Dickstein, J.; Weiss, E.; Maheswaranathan, N.; Ganguli, S. Deep unsupervised learning using nonequilibrium thermodynamics. In Proceedings of the International Conference on Machine Learning, PMLR, Lille, France, 6–11 July 2015; pp. 2256–2265. [Google Scholar]
- Ho, J.; Jain, A.; Abbeel, P. Denoising diffusion probabilistic models. Adv. Neural Inf. Process. Syst. 2020, 33, 6840–6851. [Google Scholar]
- Xiao, Z.; Kreis, K.; Vahdat, A. Tackling the generative learning trilemma with denoising diffusion gans. arXiv 2021, arXiv:2112.07804. [Google Scholar]
- Chlap, P.; Min, H.; Vandenberg, N.; Dowling, J.; Holloway, L.; Haworth, A. A review of medical image data augmentation techniques for deep learning applications. J. Med. Imaging Radiat. Oncol. 2021, 65, 545–563. [Google Scholar] [CrossRef]
- Shorten, C.; Khoshgoftaar, T.M. A survey on image data augmentation for deep learning. J. Big Data 2019, 6, 1–48. [Google Scholar] [CrossRef]
- Arjovsky, M.; Chintala, S.; Bottou, L. Wasserstein generative adversarial networks. In Proceedings of the International Conference on Machine Learning, PMLR, Sydney, Australia, 6–11 August 2017; pp. 214–223. [Google Scholar]
- Mirza, M.; Osindero, S. Conditional generative adversarial nets. arXiv 2014, arXiv:1411.1784. [Google Scholar]
- Isola, P.; Zhu, J.Y.; Zhou, T.; Efros, A.A. Image-to-image translation with conditional adversarial networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 1125–1134. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Munich, Germany, 5–9 October 2015; pp. 234–241. [Google Scholar]
- Radford, A.; Metz, L.; Chintala, S. Unsupervised representation learning with deep convolutional generative adversarial networks. arXiv 2015, arXiv:1511.06434. [Google Scholar]
- Karras, T.; Aila, T.; Laine, S.; Lehtinen, J. Progressive growing of gans for improved quality, stability, and variation. arXiv 2017, arXiv:1710.10196. [Google Scholar]
- Zhu, J.Y.; Park, T.; Isola, P.; Efros, A.A. Unpaired image-to-image translation using cycle-consistent adversarial networks. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 2223–2232. [Google Scholar]
- Odena, A.; Olah, C.; Shlens, J. Conditional image synthesis with auxiliary classifier gans. arXiv 2016, arXiv:1610.09585. [Google Scholar]
- Larsen, A.B.L.; Sønderby, S.K.; Larochelle, H.; Winther, O. Autoencoding beyond pixels using a learned similarity metric. 2015. arXiv 2015, arXiv:1512.09300. [Google Scholar]
- Higgins, I.; Matthey, L.; Glorot, X.; Pal, A.; Uria, B.; Blundell, C.; Mohamed, S.; Lerchner, A. Early visual concept learning with unsupervised deep learning. arXiv 2016, arXiv:1606.05579. [Google Scholar]
- Kingma, D.P.; Salimans, T.; Jozefowicz, R.; Chen, X.; Sutskever, I.; Welling, M. Improved variational inference with inverse autoregressive flow. Adv. Neural Inf. Process. Syst. 2016, 29. [Google Scholar]
- Zhao, S.; Song, J.; Ermon, S. Infovae: Information maximizing variational autoencoders. arXiv 2017, arXiv:1706.02262. [Google Scholar]
- Razavi, A.; Van den Oord, A.; Vinyals, O. Generating diverse high-fidelity images with vq-vae-2. Adv. Neural Inf. Process. Syst. 2019, 32. [Google Scholar]
- Dhariwal, P.; Nichol, A. Diffusion models beat gans on image synthesis. Adv. Neural Inf. Process. Syst. 2021, 34, 8780–8794. [Google Scholar]
- Salimans, T.; Ho, J. Progressive distillation for fast sampling of diffusion models. arXiv 2022, arXiv:2202.00512. [Google Scholar]
- Kong, Z.; Ping, W. On fast sampling of diffusion probabilistic models. arXiv 2021, arXiv:2106.00132. [Google Scholar]
- Song, J.; Meng, C.; Ermon, S. Denoising diffusion implicit models. arXiv 2020, arXiv:2010.02502. [Google Scholar]
- Han, C.; Hayashi, H.; Rundo, L.; Araki, R.; Shimoda, W.; Muramatsu, S.; Furukawa, Y.; Mauri, G.; Nakayama, H. GAN-based synthetic brain MR image generation. In Proceedings of the IEEE 15th International Symposium on Biomedical Imaging, New York, NY, USA, 16–19 April 2018; pp. 734–738. [Google Scholar]
- Frid-Adar, M.; Diamant, I.; Klang, E.; Amitai, M.; Goldberger, J.; Greenspan, H. GAN-based synthetic medical image augmentation for increased CNN performance in liver lesion classification. Neurocomputing 2018, 321, 321–331. [Google Scholar] [CrossRef]
- Guibas, J.T.; Virdi, T.S.; Li, P.S. Synthetic medical images from dual generative adversarial networks. arXiv 2017, arXiv:1709.01872. [Google Scholar]
- Platscher, M.; Zopes, J.; Federau, C. Image Translation for Medical Image Generation–Ischemic Stroke Lesions. arXiv 2020, arXiv:2010.02745. [Google Scholar] [CrossRef]
- Park, T.; Liu, M.Y.; Wang, T.C.; Zhu, J.Y. Semantic image synthesis with spatially-adaptive normalization. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 16–20 June 2019; pp. 2337–2346. [Google Scholar]
- Yurt, M.; Dar, S.U.; Erdem, A.; Erdem, E.; Oguz, K.K.; Çukur, T. mustGAN: Multi-stream generative adversarial networks for MR image synthesis. Med. Image Anal. 2021, 70, 101944. [Google Scholar] [CrossRef]
- Dar, S.U.; Yurt, M.; Karacan, L.; Erdem, A.; Erdem, E.; Cukur, T. Image synthesis in multi-contrast MRI with conditional generative adversarial networks. IEEE Trans. Med. Imaging 2019, 38, 2375–2388. [Google Scholar] [CrossRef]
- Sun, Y.; Yuan, P.; Sun, Y. MM-GAN: 3D MRI data augmentation for medical image segmentation via generative adversarial networks. In Proceedings of the 2020 IEEE International conference on knowledge graph (ICKG), Nanjing, China, 9–1 August 2020; pp. 227–234. [Google Scholar]
- Han, C.; Rundo, L.; Araki, R.; Nagano, Y.; Furukawa, Y.; Mauri, G.; Nakayama, H.; Hayashi, H. Combining noise-to-image and image-to-image GANs: Brain MR image augmentation for tumor detection. IEEE Access 2019, 7, 156966–156977. [Google Scholar] [CrossRef]
- Kwon, G.; Han, C.; Kim, D.s. Generation of 3D brain MRI using auto-encoding generative adversarial networks. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Shenzhen, China, 13–17 October 2019; pp. 118–126. [Google Scholar]
- Zhuang, P.; Schwing, A.G.; Koyejo, O. Fmri data augmentation via synthesis. In Proceedings of the 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), Venice, Italy, 8–11 April 2019; pp. 1783–1787. [Google Scholar]
- Waheed, A.; Goyal, M.; Gupta, D.; Khanna, A.; Al-Turjman, F.; Pinheiro, P.R. Covidgan: Data augmentation using auxiliary classifier gan for improved covid-19 detection. IEEE Access 2020, 8, 91916–91923. [Google Scholar] [CrossRef] [PubMed]
- Han, C.; Rundo, L.; Araki, R.; Furukawa, Y.; Mauri, G.; Nakayama, H.; Hayashi, H. Infinite brain MR images: PGGAN-based data augmentation for tumor detection. In Neural Approaches to Dynamics of Signal Exchanges; Springer: Singapore, 2019; pp. 291–303. [Google Scholar]
- Sun, L.; Wang, J.; Huang, Y.; Ding, X.; Greenspan, H.; Paisley, J. An adversarial learning approach to medical image synthesis for lesion detection. IEEE J. Biomed. Health Inform. 2020, 24, 2303–2314. [Google Scholar] [CrossRef]
- Wang, Q.; Zhang, X.; Chen, W.; Wang, K.; Zhang, X. Class-aware multi-window adversarial lung nodule synthesis conditioned on semantic features. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Lima, Peru, 4–8 October 2020; pp. 589–598. [Google Scholar]
- Geng, X.; Yao, Q.; Jiang, K.; Zhu, Y. Deep neural generative adversarial model based on VAE+ GAN for disorder diagnosis. In Proceedings of the 2020 International Conference on Internet of Things and Intelligent Applications (ITIA), Zhenjiang, China, 27–29 November 2020; pp. 1–7. [Google Scholar]
- Pang, T.; Wong, J.H.D.; Ng, W.L.; Chan, C.S. Semi-supervised GAN-based radiomics model for data augmentation in breast ultrasound mass classification. Comput. Methods Programs Biomed. 2021, 203, 106018. [Google Scholar] [CrossRef] [PubMed]
- Barile, B.; Marzullo, A.; Stamile, C.; Durand-Dubief, F.; Sappey-Marinier, D. Data augmentation using generative adversarial neural networks on brain structural connectivity in multiple sclerosis. Comput. Methods Programs Biomed. 2021, 206, 106113. [Google Scholar] [CrossRef]
- Shen, T.; Hao, K.; Gou, C.; Wang, F.Y. Mass image synthesis in mammogram with contextual information based on gans. Comput. Methods Programs Biomed. 2021, 202, 106019. [Google Scholar] [CrossRef] [PubMed]
- Ambita, A.A.E.; Boquio, E.N.V.; Naval, P.C. Covit-gan: Vision transformer forcovid-19 detection in ct scan imageswith self-attention gan forDataAugmentation. In Proceedings of the International Conference on Artificial Neural Networks, Bratislava, Slovakia, 14–17 September 2021; pp. 587–598. [Google Scholar]
- Hirte, A.U.; Platscher, M.; Joyce, T.; Heit, J.J.; Tranvinh, E.; Federau, C. Realistic generation of diffusion-weighted magnetic resonance brain images with deep generative models. Magn. Reson. Imaging 2021, 81, 60–66. [Google Scholar] [CrossRef]
- Kaur, S.; Aggarwal, H.; Rani, R. MR image synthesis using generative adversarial networks for Parkinson’s disease classification. In Proceedings of the International Conference on Artificial Intelligence and Applications, Jiangsu, China, 15–17 October 2021; pp. 317–327. [Google Scholar]
- Guan, Q.; Chen, Y.; Wei, Z.; Heidari, A.A.; Hu, H.; Yang, X.H.; Zheng, J.; Zhou, Q.; Chen, H.; Chen, F. Medical image augmentation for lesion detection using a texture-constrained multichannel progressive GAN. Comput. Biol. Med. 2022, 145, 105444. [Google Scholar] [CrossRef]
- Ahmad, B.; Sun, J.; You, Q.; Palade, V.; Mao, Z. Brain Tumor Classification Using a Combination of Variational Autoencoders and Generative Adversarial Networks. Biomedicines 2022, 10, 223. [Google Scholar] [CrossRef]
- Pombo, G.; Gray, R.; Cardoso, M.J.; Ourselin, S.; Rees, G.; Ashburner, J.; Nachev, P. Equitable modelling of brain imaging by counterfactual augmentation with morphologically constrained 3d deep generative models. Med. Image Anal. 2022, 102723. [Google Scholar] [CrossRef]
- Neff, T.; Payer, C.; Stern, D.; Urschler, M. Generative adversarial network based synthesis for supervised medical image segmentation. In Proceedings of the OAGM and ARW Joint Workshop, Vienna, Austria, 10–12 May 2017; p. 4. [Google Scholar]
- Mok, T.C.; Chung, A. Learning data augmentation for brain tumor segmentation with coarse-to-fine generative adversarial networks. In Proceedings of the International MICCAI Brainlesion Workshop, Granada, Spain, 16 September 2018; pp. 70–80. [Google Scholar]
- Shin, H.C.; Tenenholtz, N.A.; Rogers, J.K.; Schwarz, C.G.; Senjem, M.L.; Gunter, J.L.; Andriole, K.P.; Michalski, M. Medical image synthesis for data augmentation and anonymization using generative adversarial networks. In Proceedings of the International Workshop on Simulation and Synthesis in Medical Imaging, Granada, Spain, 16 September 2018; pp. 1–11. [Google Scholar]
- Jiang, J.; Hu, Y.C.; Tyagi, N.; Zhang, P.; Rimner, A.; Deasy, J.O.; Veeraraghavan, H. Cross-modality (CT-MRI) prior augmented deep learning for robust lung tumor segmentation from small MR datasets. Med. Phys. 2019, 46, 4392–4404. [Google Scholar] [CrossRef]
- Jiang, Y.; Chen, H.; Loew, M.; Ko, H. COVID-19 CT image synthesis with a conditional generative adversarial network. IEEE J. Biomed. Health Inform. 2020, 25, 441–452. [Google Scholar] [CrossRef] [PubMed]
- Qasim, A.B.; Ezhov, I.; Shit, S.; Schoppe, O.; Paetzold, J.C.; Sekuboyina, A.; Kofler, F.; Lipkova, J.; Li, H.; Menze, B. Red-GAN: Attacking class imbalance via conditioned generation. Yet another medical imaging perspective. In Proceedings of the Medical Imaging with Deep Learning, Montreal, QC, Canada, 6–9 July 2020; pp. 655–668. [Google Scholar]
- Shi, H.; Lu, J.; Zhou, Q. A novel data augmentation method using style-based GAN for robust pulmonary nodule segmentation. In Proceedings of the 2020 Chinese Control and Decision Conference (CCDC), Hefei, China, 22–24 August 2020; pp. 2486–2491. [Google Scholar]
- Shen, Z.; Ouyang, X.; Xiao, B.; Cheng, J.Z.; Shen, D.; Wang, Q. Image synthesis with disentangled attributes for chest X-ray nodule augmentation and detection. Med. Image Anal. 2022, 102708. [Google Scholar] [CrossRef] [PubMed]
- Chartsias, A.; Joyce, T.; Dharmakumar, R.; Tsaftaris, S.A. Adversarial image synthesis for unpaired multi-modal cardiac data. In Proceedings of the International Workshop on Simulation and Synthesis in Medical Imaging, Québec City, QC, Canada, 10 September 2017; pp. 3–13. [Google Scholar]
- Wolterink, J.M.; Dinkla, A.M.; Savenije, M.H.; Seevinck, P.R.; van den Berg, C.A.; Išgum, I. Deep MR to CT synthesis using unpaired data. In Proceedings of the International Workshop on Simulation and Synthesis in Medical Imaging, Québec City, QC, Canada, 10 September 2017; pp. 14–23. [Google Scholar]
- Nie, D.; Trullo, R.; Lian, J.; Wang, L.; Petitjean, C.; Ruan, S.; Wang, Q.; Shen, D. Medical image synthesis with deep convolutional adversarial networks. IEEE Trans. Biomed. Eng. 2018, 65, 2720–2730. [Google Scholar] [CrossRef]
- Armanious, K.; Jiang, C.; Fischer, M.; Küstner, T.; Hepp, T.; Nikolaou, K.; Gatidis, S.; Yang, B. MedGAN: Medical image translation using GANs. Comput. Med. Imaging Graph. 2020, 79, 101684. [Google Scholar] [CrossRef]
- Yang, H.; Lu, X.; Wang, S.H.; Lu, Z.; Yao, J.; Jiang, Y.; Qian, P. Synthesizing multi-contrast MR images via novel 3D conditional Variational auto-encoding GAN. Mob. Netw. Appl. 2021, 26, 415–424. [Google Scholar] [CrossRef]
- Sikka, A.; Skand; Virk, J.S.; Bathula, D.R. MRI to PET Cross-Modality Translation using Globally and Locally Aware GAN (GLA-GAN) for Multi-Modal Diagnosis of Alzheimer’s Disease. arXiv 2021, arXiv:2108.02160. [Google Scholar]
- Amirrajab, S.; Lorenz, C.; Weese, J.; Pluim, J.; Breeuwer, M. Pathology Synthesis of 3D Consistent Cardiac MR Images Using 2D VAEs and GANs. In Proceedings of the International Workshop on Simulation and Synthesis in Medical Imaging, Singapore, 18 September 2022; pp. 34–42. [Google Scholar]
- Pesteie, M.; Abolmaesumi, P.; Rohling, R.N. Adaptive augmentation of medical data using independently conditional variational auto-encoders. IEEE Trans. Med. Imaging 2019, 38, 2807–2820. [Google Scholar] [CrossRef]
- Chadebec, C.; Thibeau-Sutre, E.; Burgos, N.; Allassonnière, S. Data augmentation in high dimensional low sample size setting using a geometry-based variational autoencoder. IEEE Trans. Pattern Anal. Mach. Intell. 2022, 45, 2879–2896. [Google Scholar] [CrossRef]
- Huo, J.; Vakharia, V.; Wu, C.; Sharan, A.; Ko, A.; Ourselin, S.; Sparks, R. Brain Lesion Synthesis via Progressive Adversarial Variational Auto-Encoder. In Proceedings of the International Workshop on Simulation and Synthesis in Medical Imaging, Singapore, 18 September 2022; pp. 101–111. [Google Scholar]
- Gulrajani, I.; Ahmed, F.; Arjovsky, M.; Dumoulin, V.; Courville, A.C. Improved training of wasserstein gans. Adv. Neural Inf. Process. Syst. 2017, 30. [Google Scholar]
- Imran, A.-A.-Z.; Terzopoulos, D. Multi-adversarial variational autoencoder networks. In Proceedings of the 2019 18th IEEE International Conference On Machine Learning And Applications (ICMLA), Boca Raton, FL, USA, 16–19 December 2019; pp. 777–782. [Google Scholar]
- Qiang, N.; Dong, Q.; Liang, H.; Ge, B.; Zhang, S.; Sun, Y.; Zhang, C.; Zhang, W.; Gao, J.; Liu, T. Modeling and augmenting of fMRI data using deep recurrent variational auto-encoder. J. Neural Eng. 2021, 18, 0460b6. [Google Scholar] [CrossRef]
- Madan, Y.; Veetil, I.K.; V, S.; EA, G.; KP, S. Synthetic Data Augmentation of MRI using Generative Variational Autoencoder for Parkinson’s Disease Detection. In Evolution in Computational Intelligence; Springer: Berlin/Heidelberg, Germany, 2022; pp. 171–178. [Google Scholar]
- Chadebec, C.; Allassonnière, S. Data Augmentation with Variational Autoencoders and Manifold Sampling. In Deep Generative Models, and Data Augmentation, Labelling, and Imperfections; Springer: Berlin/Heidelberg, Germany, 2021; pp. 184–192. [Google Scholar]
- Liang, J.; Chen, J. Data augmentation of thyroid ultrasound images using generative adversarial network. In Proceedings of the 2021 IEEE International Ultrasonics Symposium (IUS), Xi’an, China, 12–15 September 2021; pp. 1–4. [Google Scholar]
- Gan, M.; Wang, C. Esophageal optical coherence tomography image synthesis using an adversarially learned variational autoencoder. Biomed. Opt. Express 2022, 13, 1188–1201. [Google Scholar] [CrossRef]
- Hu, Q.; Li, H.; Zhang, J. Domain-Adaptive 3D Medical Image Synthesis: An Efficient Unsupervised Approach. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Singapore, 18–22 September 2022; pp. 495–504. [Google Scholar]
- Biffi, C.; Oktay, O.; Tarroni, G.; Bai, W.; Marvao, A.D.; Doumou, G.; Rajchl, M.; Bedair, R.; Prasad, S.; Cook, S.; et al. DLearning interpretable anatomical features through deep generative models: Application to cardiac remodeling. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Grenada, Spain, 16–20 September 2018; pp. 464–471. [Google Scholar]
- Volokitin, A.; Erdil, E.; Karani, N.; Tezcan, K.C.; Chen, X.; Gool, L.V.; Konukoglu, E. Modelling the distribution of 3D brain MRI using a 2D slice VAE. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Lima, Peru, 4–8 October 2020; pp. 657–666. [Google Scholar]
- Huang, Q.; Qiao, C.; Jing, K.; Zhu, X.; Ren, K. Biomarkers identification for Schizophrenia via VAE and GSDAE-based data augmentation. Comput. Biol. Med. 2022, 105603. [Google Scholar] [CrossRef] [PubMed]
- Beetz, M.; Banerjee, A.; Sang, Y.; Grau, V. Combined Generation of Electrocardiogram and Cardiac Anatomy Models Using Multi-Modal Variational Autoencoders. In Proceedings of the 2022 IEEE 19th International Symposium on Biomedical Imaging (ISBI), Kolkata, India, 28–31 March 2020; pp. 1–4. [Google Scholar]
- Sundgaard, J.V.; Hannemose, M.R.; Laugesen, S.; Bray, P.; Harte, J.; Kamide, Y.; Tanaka, C.; Paulsen, R.R.; Christensen, A.N. Multi-modal data generation with a deep metric variational autoencoder. arXiv 2022, arXiv:2202.03434. [Google Scholar] [CrossRef] [PubMed]
- Pinaya, W.H.; Tudosiu, P.D.; Dafflon, J.; Da Costa, P.F.; Fernandez, V.; Nachev, P.; Ourselin, S.; Cardoso, M.J. Brain imaging generation with latent diffusion models. In Proceedings of the MICCAI Workshop on Deep Generative Models, Singapore, 22 September 2022; pp. 117–126. [Google Scholar]
- Rombach, R.; Blattmann, A.; Lorenz, D.; Esser, P.; Ommer, B. High-resolution image synthesis with latent diffusion models. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, New Orleans, LA, USA, 18–24 June 2022; pp. 10684–10695. [Google Scholar]
- Mao, X.; Li, Q.; Xie, H.; Lau, R.; Wang, Z.; Smolley, S. Least squares generative adversarial networks. arXiv 2016, arXiv:1611.04076. [Google Scholar]
- Heusel, M.; Ramsauer, H.; Unterthiner, T.; Nessler, B.; Hochreiter, S. Gans trained by a two time-scale update rule converge to a local nash equilibrium. Adv. Neural Inf. Process. Syst. 2017, 30. [Google Scholar]
- Fernandez, V.; Pinaya, W.H.L.; Borges, P.; Tudosiu, P.D.; Graham, M.S.; Vercauteren, T.; Cardoso, M.J. Can segmentation models be trained with fully synthetically generated data? In Proceedings of the International Workshop on Simulation and Synthesis in Medical Imaging, Singapore, 18–22 September 2022; pp. 79–90. [Google Scholar]
- Isensee, F.; Petersen, J.; Klein, A.; Zimmerer, D.; Jaeger, P.F.; Kohl, S.; Wasserthal, J.; Koehler, G.; Norajitra, T.; Wirkert, S.; et al. nnu-net: Self-adapting framework for u-net-based medical image segmentation. arXiv 2018, arXiv:1809.10486. [Google Scholar]
- Lyu, Q.; Wang, G. Conversion Between CT and MRI Images Using Diffusion and Score-Matching Models. arXiv 2022, arXiv:2209.12104. [Google Scholar]
- Song, Y.; Sohl-Dickstein, J.; Kingma, D.P.; Kumar, A.; Ermon, S.; Poole, B. Score-based generative modeling through stochastic differential equations. arXiv 2020, arXiv:2011.13456. [Google Scholar]
- Nyholm, T.; Svensson, S.; Andersson, S.; Jonsson, J.; Sohlin, M.; Gustafsson, C.; Kjellén, E.; Söderström, K.; Albertsson, P.; Blomqvist, L.; et al. MR and CT data with multiobserver delineations of organs in the pelvic area—Part of the Gold Atlas project. Med. Phys. 2018, 45, 1295–1300. [Google Scholar] [CrossRef]
- Dorjsembe, Z.; Odonchimed, S.; Xiao, F. Three-Dimensional Medical Image Synthesis with Denoising Diffusion Probabilistic Models. In Proceedings of the Medical Imaging with Deep Learning, Zurich, Switzerland, 18–22 September 2022. [Google Scholar]
- Packhäuser, K.; Folle, L.; Thamm, F.; Maier, A. Generation of anonymous chest radiographs using latent diffusion models for training thoracic abnormality classification systems. arXiv 2022, arXiv:2211.01323. [Google Scholar]
- Moghadam, P.A.; Van Dalen, S.; Martin, K.C.; Lennerz, J.; Yip, S.; Farahani, H.; Bashashati, A. A Morphology Focused Diffusion Probabilistic Model for Synthesis of Histopathology Images. arXiv 2022, arXiv:2209.13167. [Google Scholar]
- Chambon, P.; Bluethgen, C.; Delbrouck, J.B.; Van der Sluijs, R.; Połacin, M.; Chaves, J.M.Z.; Abraham, T.M.; Purohit, S.; Langlotz, C.P.; Chaudhari, A. RoentGen: Vision-Language Foundation Model for Chest X-ray Generation. arXiv 2022, arXiv:2211.12737. [Google Scholar]
- Wolleb, J.; Sandkühler, R.; Bieder, F.; Cattin, P.C. The Swiss Army Knife for Image-to-Image Translation: Multi-Task Diffusion Models. arXiv 2022, arXiv:2204.02641. [Google Scholar]
- Sagers, L.W.; Diao, J.A.; Groh, M.; Rajpurkar, P.; Adamson, A.S.; Manrai, A.K. Improving dermatology classifiers across populations using images generated by large diffusion models. arXiv 2022, arXiv:2211.13352. [Google Scholar]
- Peng, W.; Adeli, E.; Zhao, Q.; Pohl, K.M. Generating Realistic 3D Brain MRIs Using a Conditional Diffusion Probabilistic Model. arXiv 2022, arXiv:2212.08034. [Google Scholar]
- Ali, H.; Murad, S.; Shah, Z. Spot the fake lungs: Generating synthetic medical images using neural diffusion models. In Proceedings of the Artificial Intelligence and Cognitive Science: 30th Irish Conference, AICS 2022, Munster, Ireland, 8–9 December 2022; pp. 32–39. [Google Scholar]
- Saeed, S.U.; Syer, T.; Yan, W.; Yang, Q.; Emberton, M.; Punwani, S.; Clarkson, M.J.; Barratt, D.C.; Hu, Y. Bi-parametric prostate MR image synthesis using pathology and sequence-conditioned stable diffusion. arXiv 2023, arXiv:2303.02094. [Google Scholar]
- Weber, T.; Ingrisch, M.; Bischl, B.; Rügamer, D. Cascaded Latent Diffusion Models for High-Resolution Chest X-ray Synthesis. arXiv 2023, arXiv:2303.11224. [Google Scholar]
- Khader, F.; Mueller-Franzes, G.; Arasteh, S.T.; Han, T.; Haarburger, C.; Schulze-Hagen, M.; Schad, P.; Engelhardt, S.; Baessler, B.; Foersch, S.; et al. Medical Diffusion–Denoising Diffusion Probabilistic Models for 3D Medical Image Generation. arXiv 2022, arXiv:2211.03364. [Google Scholar]
- Özbey, M.; Dar, S.U.; Bedel, H.A.; Dalmaz, O.; Özturk, Ş.; Güngör, A.; Çukur, T. Unsupervised medical image translation with adversarial diffusion models. arXiv 2022, arXiv:2207.08208. [Google Scholar]
- Meng, X.; Gu, Y.; Pan, Y.; Wang, N.; Xue, P.; Lu, M.; He, X.; Zhan, Y.; Shen, D. A Novel Unified Conditional Score-based Generative Framework for Multi-modal Medical Image Completion. arXiv 2022, arXiv:2207.03430. [Google Scholar]
- Kim, B.; Ye, J.C. Diffusion deformable model for 4D temporal medical image generation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Singapore, 18–22 September 2022; pp. 539–548. [Google Scholar]
- Kazerouni, A.; Aghdam, E.K.; Heidari, M.; Azad, R.; Fayyaz, M.; Hacihaliloglu, I.; Merhof, D. Diffusion models for medical image analysis: A comprehensive survey. arXiv 2022, arXiv:2211.07804. [Google Scholar]
- Abdollahi, B.; Tomita, N.; Hassanpour, S. Data Augmentation in Training Deep Learning Models for Medical Image Analysis; Springer: Berlin/Heidelberg, Germany, 2020; pp. 167–180. [Google Scholar]
- Huang, H.; Li, Z.; He, R.; Sun, Z.; Tan, T. Introvae: Introspective variational autoencoders for photographic image synthesis. Adv. Neural Inf. Process. Syst. 2018, 31. [Google Scholar]
- Amyar, A.; Ruan, S.; Vera, P.; Decazes, P.; Modzelewski, R. RADIOGAN: Deep convolutional conditional generative adversarial network to generate PET images. In Proceedings of the 2020 7th International Conference on Bioinformatics Research and Applications, Berlin, Germany, 13–15 September 2020; pp. 28–33. [Google Scholar]
- Bullitt, E.; Zeng, D.; Gerig, G.; Aylward, S.; Joshi, S.; Smith, J.K.; Lin, W.; Ewend, M.G. Vessel tortuosity and brain tumor malignancy: A blinded study1. Acad. Radiol. 2005, 12, 1232–1240. [Google Scholar] [CrossRef]
- Staal, J.; Abràmoff, M.D.; Niemeijer, M.; Viergever, M.A.; Van Ginneken, B. Ridge-based vessel segmentation in color images of the retina. IEEE Trans. Med. Imaging 2004, 23, 501–509. [Google Scholar] [CrossRef] [PubMed]
- Bernard, O.; Lalande, A.; Zotti, C.; Cervenansky, F.; Yang, X.; Heng, P.A.; Cetin, I.; Lekadir, K.; Camara, O.; Ballester, M.A.G.; et al. Deep learning techniques for automatic MRI cardiac multi-structures segmentation and diagnosis: Is the problem solved? IEEE Trans. Med. Imaging 2018, 37, 2514–2525. [Google Scholar] [CrossRef]
- Wang, X.; Peng, Y.; Lu, L.; Lu, Z.; Bagheri, M.; Summers, R.M. Chestx-ray8: Hospital-scale chest x-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 2097–2106. [Google Scholar]
- Bai, W.; Shi, W.; de Marvao, A.; Dawes, T.J.; O’Regan, D.P.; Cook, S.A.; Rueckert, D. A bi-ventricular cardiac atlas built from 1000+ high resolution MR images of healthy subjects and an analysis of shape and motion. Med. Image Anal. 2015, 26, 133–145. [Google Scholar] [CrossRef] [PubMed]
- Van Ginneken, B.; Stegmann, M.B.; Loog, M. Segmentation of anatomical structures in chest radiographs using supervised methods: A comparative study on a public database. Med. Image Anal. 2006, 10, 19–40. [Google Scholar] [CrossRef] [PubMed]
- Van Essen, D.C.; Smith, S.M.; Barch, D.M.; Behrens, T.E.; Yacoub, E.; Ugurbil, K.; WU-Minn HCP Consortium. The WU-Minn human connectome project: An overview. Neuroimage 2013, 80, 62–79. [Google Scholar] [CrossRef]
- Kermany, D.S.; Goldbaum, M.; Cai, W.; Valentim, C.C.; Liang, H.; Baxter, S.L.; McKeown, A.; Yang, G.; Wu, X.; Yan, F.; et al. Identifying medical diagnoses and treatable diseases by image-based deep learning. Cell 2018, 172, 1122–1131. [Google Scholar] [CrossRef]
- Groh, M.; Harris, C.; Soenksen, L.; Lau, F.; Han, R.; Kim, A.; Koochek, A.; Badri, O. Evaluating deep neural networks trained on clinical images in dermatology with the fitzpatrick 17k dataset. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Nashville, TN, USA, 20–25 June 2021; pp. 1820–1828. [Google Scholar]
- Yang, X.; He, X.; Zhao, J.; Zhang, Y.; Zhang, S.; Xie, P. COVID-CT-dataset: A CT scan dataset about COVID-19. arXiv 2020, arXiv:2003.13865. [Google Scholar]
- Soares, E.; Angelov, P.; Biaso, S.; Froes, M.H.; Abe, D.K. SARS-CoV-2 CT-scan dataset: A large dataset of real patients CT scans for SARS-CoV-2 identification. medRxiv 2020. [Google Scholar] [CrossRef]
- Johnson, A.E.; Pollard, T.J.; Greenbaum, N.R.; Lungren, M.P.; Deng, C.y.; Peng, Y.; Lu, Z.; Mark, R.G.; Berkowitz, S.J.; Horng, S. MIMIC-CXR-JPG, a large publicly available database of labeled chest radiographs. arXiv 2019, arXiv:1901.07042. [Google Scholar]
- Jones, S.; Tillin, T.; Park, C.; Williams, S.; Rapala, A.; Al Saikhan, L.; Eastwood, S.V.; Richards, M.; Hughes, A.D.; Chaturvedi, N. Cohort Profile Update: Southall and Brent Revisited (SABRE) study: A UK population-based comparison of cardiovascular disease and diabetes in people of European, South Asian and African Caribbean heritage. Int. J. Epidemiol. 2020, 49, 1441–1442e. [Google Scholar] [CrossRef]
- Saha, A.; Twilt, J.; Bosma, J.; van Ginneken, B.; Yakar, D.; Elschot, M.; Veltman, J.; Fütterer, J.; de Rooij, M.; Huisman, H. Artificial Intelligence and Radiologists at Prostate Cancer Detection in MRI: The PI CAI Challenge. In Proceedings of the RSNA, Chicago, IL, USA, 27 November–1 December 2022. [Google Scholar]
- Kynkäänniemi, T.; Karras, T.; Laine, S.; Lehtinen, J.; Aila, T. Improved precision and recall metric for assessing generative models. Adv. Neural Inf. Process. Syst. 2019, 32. [Google Scholar]
- Wang, Z.; Bovik, A.C.; Sheikh, H.R.; Simoncelli, E.P. Image quality assessment: From error visibility to structural similarity. IEEE Trans. Image Process. 2004, 13, 600–612. [Google Scholar] [CrossRef] [PubMed]
- Ledig, C.; Theis, L.; Huszár, F.; Caballero, J.; Cunningham, A.; Acosta, A.; Aitken, A.; Tejani, A.; Totz, J.; Wang, Z.; et al. Photo-realistic single image super-resolution using a generative adversarial network. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 4681–4690. [Google Scholar]
- Zhang, R.; Isola, P.; Efros, A.A.; Shechtman, E.; Wang, O. The unreasonable effectiveness of deep features as a perceptual metric. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 18–22 June 2018; pp. 586–595. [Google Scholar]
- Salimans, T.; Goodfellow, I.; Zaremba, W.; Cheung, V.; Radford, A.; Chen, X. Improved techniques for training gans. Adv. Neural Inf. Process. Syst. 2016, 29. [Google Scholar]
- Sudre, C.H.; Li, W.; Vercauteren, T.; Ourselin, S.; Jorge Cardoso, M. Generalised dice overlap as a deep learning loss function for highly unbalanced segmentations. In Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support, Proceedings of the Third International Workshop, DLMIA 2017, and 7th International Workshop, ML-CDS 2017, Held in Conjunction with MICCAI 2017, Québec City, QC, Canada, 14 September 2017; Springer: Berlin/Heidelberg, Germany, 2017; pp. 240–248. [Google Scholar]
- Rockafellar, R.T.; Wets, R.J.B. Variational Analysis; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2009; Volume 317. [Google Scholar]
- Gretton, A.; Borgwardt, K.M.; Rasch, M.J.; Schölkopf, B.; Smola, A. A kernel two-sample test. J. Mach. Learn. Res. 2012, 13, 723–773. [Google Scholar]
- Cover, T.; Hart, P. Nearest neighbor pattern classification. IEEE Trans. Inf. Theory 1967, 13, 21–27. [Google Scholar] [CrossRef]
- Bounliphone, W.; Belilovsky, E.; Blaschko, M.B.; Antonoglou, I.; Gretton, A. A test of relative similarity for model selection in generative models. arXiv 2015, arXiv:1511.04581. [Google Scholar]
- Vaserstein, L.N. Markov processes over denumerable products of spaces, describing large systems of automata. Probl. Peredachi Informatsii 1969, 5, 64–72. [Google Scholar]
- Fawcett, T. An introduction to ROC analysis. Pattern Recognit. Lett. 2006, 27, 861–874. [Google Scholar] [CrossRef]
- Nguyen, X.; Wainwright, M.J.; Jordan, M.I. Estimating divergence functionals and the likelihood ratio by convex risk minimization. IEEE Trans. Inf. Theory 2010, 56, 5847–5861. [Google Scholar] [CrossRef]
- Sheikh, H.R.; Bovik, A.C. A visual information fidelity approach to video quality assessment. First Int. Workshop Video Process. Qual. Metrics Consum. Electron. 2005, 7, 2117–2128. [Google Scholar]
- Wang, Z.; Bovik, A.C. A universal image quality index. IEEE Signal Process. Lett. 2002, 9, 81–84. [Google Scholar] [CrossRef]
- Tavse, S.; Varadarajan, V.; Bachute, M.; Gite, S.; Kotecha, K. A Systematic Literature Review on Applications of GAN-Synthesized Images for Brain MRI. Future Internet 2022, 14, 351. [Google Scholar] [CrossRef]
- Ramesh, A.; Pavlov, M.; Goh, G.; Gray, S.; Voss, C.; Radford, A.; Chen, M.; Sutskever, I. Zero-shot text-to-image generation. In Proceedings of the International Conference on Machine Learning, PMLR, Online, 18–24 July 2021; pp. 8821–8831. [Google Scholar]
- Saharia, C.; Chan, W.; Saxena, S.; Li, L.; Whang, J.; Denton, E.L.; Ghasemipour, K.; Gontijo Lopes, R.; Karagol Ayan, B.; Salimans, T.; et al. Photorealistic text-to-image diffusion models with deep language understanding. Adv. Neural Inf. Process. Syst. 2022, 35, 36479–36494. [Google Scholar]
- Kang, M.; Zhu, J.Y.; Zhang, R.; Park, J.; Shechtman, E.; Paris, S.; Park, T. Scaling up GANs for Text-to-Image Synthesis. arXiv 2023, arXiv:2303.05511. [Google Scholar]
- Sauer, A.; Karras, T.; Laine, S.; Geiger, A.; Aila, T. Stylegan-t: Unlocking the power of gans for fast large-scale text-to-image synthesis. arXiv 2023, arXiv:2301.09515. [Google Scholar]
- Delgado, J.M.D.; Oyedele, L. Deep learning with small datasets: Using autoencoders to address limited datasets in construction management. Appl. Soft Comput. 2021, 112, 107836. [Google Scholar] [CrossRef]
- Caterini, A.L.; Doucet, A.; Sejdinovic, D. Hamiltonian variational auto-encoder. Adv. Neural Inf. Process. Syst. 2018, 31. [Google Scholar]
- He, Y.; Wang, L.; Yang, F.; Clarysse, P.; Robini, M.; Zhu, Y. Effect of different configurations of diffusion gradient directions on accuracy of diffusion tensor estimation in cardiac DTI. In Proceedings of the 16th IEEE International Conference on Signal Processing (ICSP), Beijing, China, 21–24 October 2022; Volume 1, pp. 437–441. [Google Scholar]
- Talo, M.; Baloglu, U.B.; Yıldırım, Ö.; Acharya, U.R. Application of deep transfer learning for automated brain abnormality classification using MR images. Cogn. Syst. Res. 2019, 54, 176–188. [Google Scholar] [CrossRef]
- Ren, P.; Xiao, Y.; Chang, X.; Huang, P.Y.; Li, Z.; Gupta, B.B.; Chen, X.; Wang, X. A survey of deep active learning. ACM Comput. Surv. (CSUR) 2021, 54, 1–40. [Google Scholar] [CrossRef]
- Rahimi, S.; Oktay, O.; Alvarez-Valle, J.; Bharadwaj, S. Addressing the exorbitant cost of labeling medical images with active learning. In Proceedings of the International Conference on Machine Learning in Medical Imaging and Analysis, Barcelona, Spain, 24–25 May 2021; p. 1. [Google Scholar]
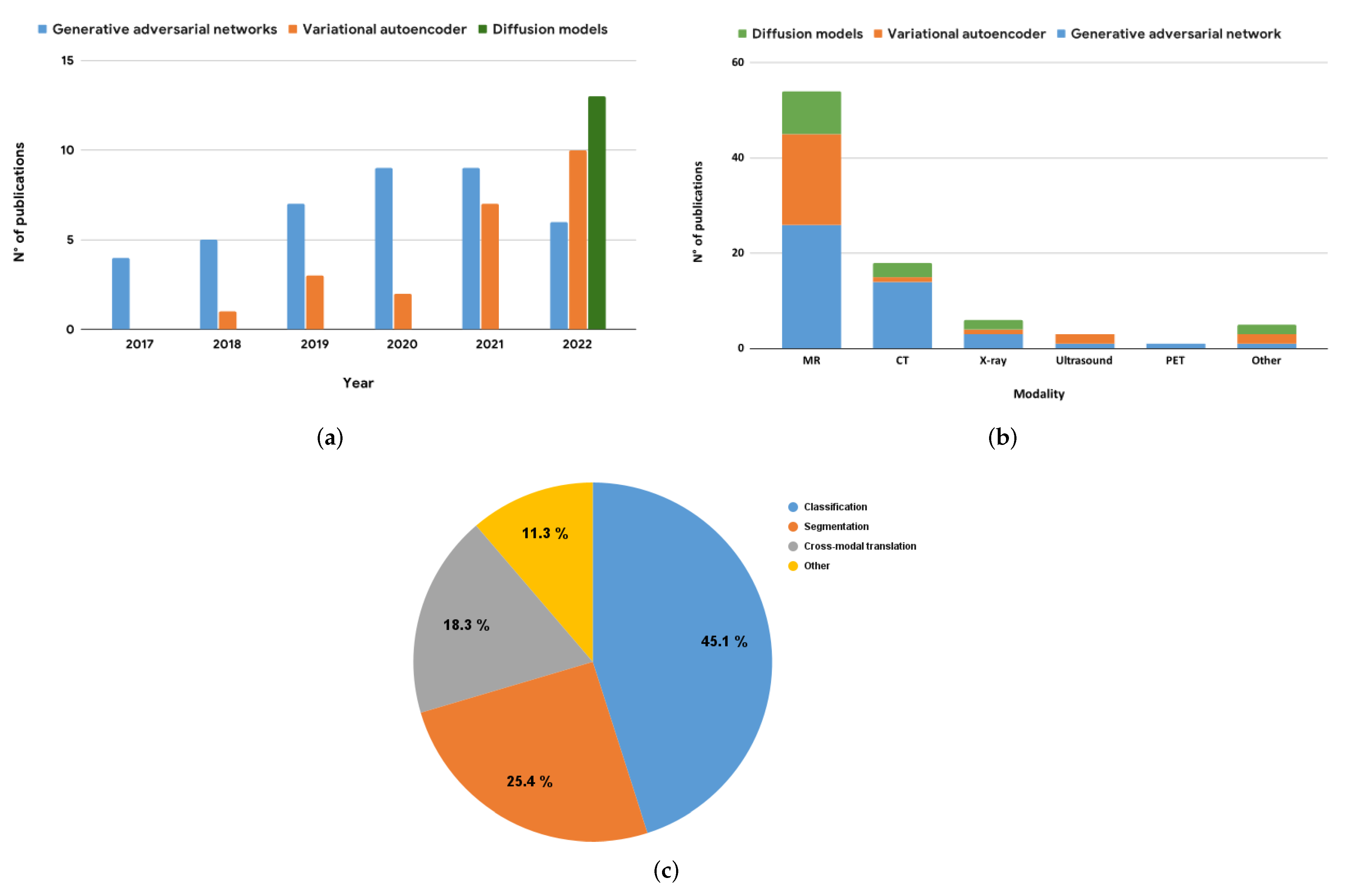

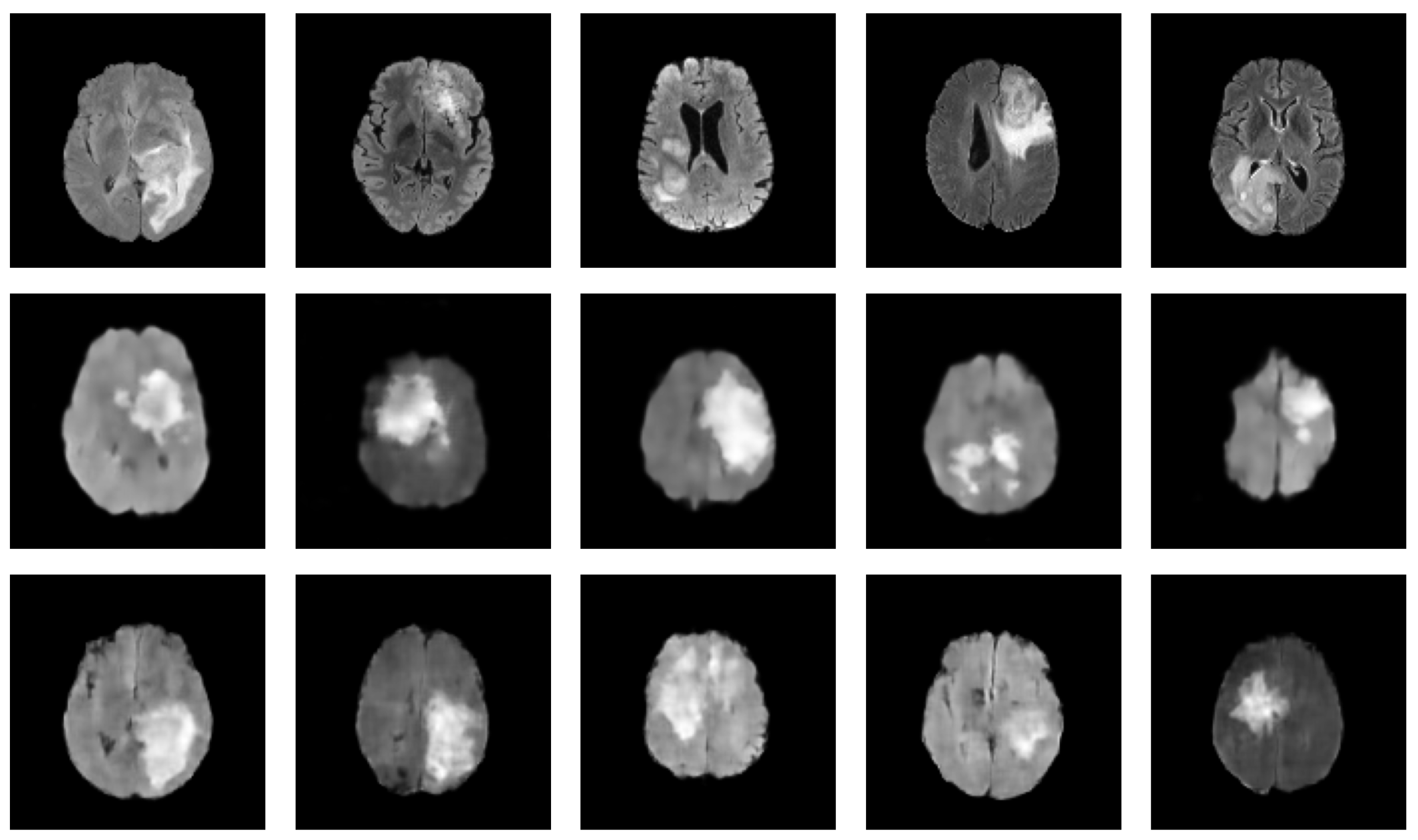
| Reference | Architecture | Hybrid Status | Dataset | Modality | 3D | Eval. Metrics |
|---|---|---|---|---|---|---|
| Classification | ||||||
| [42] | DCGAN, ACGAN | Private | CT | Sens., Spec. | ||
| [41] | DCGAN, WGAN | BraTS2016 | MR | Acc. | ||
| [49] | PGGAN, MUNIT | BraTS2016 | MR | ✓ | Acc., Sens., Spec., | |
| [50] | AE-GAN | Hybrid (V + G) | BraTS2018, ADNI | MR | ✓ | MMD, MS-SSIM |
| [51] | ICW-GAN | OpenfMRI, HCP | MR | ✓ | Acc., Prec., F1 | |
| NeuroSpin, IBC | Recall | |||||
| [52] | ACGAN | IEEE CCX | X-ray | Acc., Sens., Spec. | ||
| Prec., Recall, F1 | ||||||
| [53] | PGGAN | BraTS2016 | MR | Acc., Sens., Spec. | ||
| [54] | ANT-GAN | BraTS2018 | MR | Acc. | ||
| [55] | MG-CGAN | LIDC-IDRI | CT | Acc., F1 | ||
| [56] | FC-GAN | Hybrid (V + G) | ADHD, ABIDE | MR | Acc., Sens., Spec., AUC | |
| [57] | TGAN | Private | Ultrasound | Acc., Sens., Spec. | ||
| [58] | AAE | Private | MR | Prec., Recall, F1 | ||
| [59] | DCGAN, InfillingGAN | DDSM | CT | LPIPS, Recall | ||
| [60] | SAGAN | COVID-CT, SARS-COV2 | CT | Acc. | ||
| [61] | StyleGAN | Private | MR | - | ||
| [62] | DCGAN | PPMI | MR | Acc., Spec., Sens. | ||
| [63] | TMP-GAN | CBIS-DDMS, Private | CT | Prec., Recall, F1, AUC | ||
| [64] | VAE-GAN | Hybrid (V + G) | Private | MR | Acc., Sens., Spec. | |
| [65] | CounterSynth | UK Biobank, OASIS | MR | ✓ | Acc., MSE, SSIM, MAE | |
| Segmentation | ||||||
| [43] | CGAN | DRIVE | Fundus photography | KLD, F1 | ||
| [66] | DCGAN | SCR | X-ray | Dice, Hausdorff | ||
| [67] | CB-GAN | BraTS2015 | MR | Dice, Prec., Sens. | ||
| [68] | Pix2Pix | BraTS2015, ADNI | MR | ✓ | Dice | |
| [12] | CycleGAN | NIHPCT | CT | Dice | ||
| [69] | CM-GAN | Private | MR | KLD, Dice | ||
| hausdorff | ||||||
| [70] | CGAN | COVID-CT | CT | FID, PSNR, SSIM, RMSE | ||
| [71] | Red-GAN | BraTS2015, ISIC | MR | Dice | ||
| [44] | Pix2Pix, SPADE, CycleGAN | Private | MR | Dice | ||
| [72] | StyleGAN | LIDC-IDRI | CT | Dice, Pres., Sens. | ||
| [73] | DCGAN, GatedConv | Private | X-ray | MAE, PSNR, SSIM, FID, AUC | ||
| Cross-modal translation | ||||||
| [74] | CycleGAN | Private | MR ↔ CT | ✓ | Dice | |
| [75] | CycleGAN | Private | MR → CT | MAE, PSNR | ||
| [76] | Pix2Pix | ADNI, Private | MR → CT | ✓ | MAE, PSNR, Dice | |
| [77] | MedGAN | Private | PET → CT | SSIM, PSNR, MSE | ||
| VIF, UQI, LPIPS | ||||||
| [47] | pGAN, CGAN | BraTS2015, MIDAS, IXI | T1 ⟷ T2 | SSIM, PSNR | ||
| [69] | CM-GAN | Private | MR | KLD, Dice | ||
| hausdorff | ||||||
| [46] | mustGAN | IXI, ISLES | T1 ↔ T2 ↔ PD | SSIM, PSNR | ||
| [78] | CAE-ACGAN | Hybrid (V + G) | Private | CT → MR | ✓ | PSNR, SSIM, MAE |
| [79] | GLA-GAN | ADNI | MR → PET | SSIM, PSNR, MAE | ||
| Acc., F1 | ||||||
| Other | ||||||
| [80] | VAE-CGAN | Hybrid (V + G) | ACDC | MR | ✓ | - |
| Reference | Architecture | Hybrid Status | Dataset | Modality | 3D | Eval. Metrics |
|---|---|---|---|---|---|---|
| Classification | ||||||
| [81] | ICVAE | Private | MR | Acc., Sens., Spec. | ||
| Ultrasound | Dice, Hausdroff, … | |||||
| [51] | CVAE | OpenfMRI, HCP | MR | ✓ | Acc., Prec., F1 | |
| NeuroSpin, IBC | Recall | |||||
| [82] | GA-VAE | ADNI, AIBL | MR | ✓ | Acc., Spec., Sens. | |
| [85] | MAVENs | Hybrid (V + G) | APCXR | X-ray | FID, F1 | |
| [61] | IntroVAE | Hybrid (V + G) | Private | MR | - | |
| [86] | DR-VAE | HCP | MR | - | ||
| [64] | VAE-GAN | Hybrid (V + G) | Private | MR | Acc., Sens., Spec. | |
| [87] | VAE | Private | MR | Acc. | ||
| [88] | RH-VAE | OASIS | MR | ✓ | Acc. | |
| Segmentation | ||||||
| [89] | VAE-GAN | Hybrid (V + G) | Private | Ultrasound | MMD, 1-NN, MS-SSIM | |
| [90] | AL-VAE | Hybrid (V + G) | Private | OCT 1 | MMD, MS, WD | |
| [83] | PA-VAE | Hybrid (V + G) | Private | MR | ✓ | PSNR, SSIM, Dice |
| NMSE, Jacc., … | ||||||
| Cross-modal translation | ||||||
| [78] | CAE-ACGAN | Hybrid (V + G) | Private | CT → MR | ✓ | PSNR, SSIM, MAE |
| [91] | 3D-UDA | Private | FLAIR ↔ T1 ↔ T2 | ✓ | SSIM, PSNR, Dice | |
| Other | ||||||
| [92] | CVAE | ACDC, Private | MR | ✓ | - | |
| [92] | CVAE | Private | MR | ✓ | Dice, Hausdorff | |
| [93] | Slice-to-3D-VAE | HCP | MR | ✓ | MMD, MS-SSIM | |
| [94] | GS-VDAE | MLSP | MR | Acc. | ||
| [80] | VAE-CGAN | Hybrid (V + G) | ACDC | MR | ✓ | - |
| [95] | MM-VAE | UK Biobank | MR | ✓ | MMD | |
| [96] | DM-VAE | Private | Otoscopy | - |
| Reference | Architecture | Hybrid Status | Dataset | Modality | 3D | Eval. Metrics |
|---|---|---|---|---|---|---|
| Classification | ||||||
| [97] | CLDM | UK Biobank | MR | ✓ | FID, MS-SSIM | |
| [106] | DDPM | ICTS | MR | ✓ | MS-SSIM | |
| [107] | LDM | CXR8 | X-ray | AUC | ||
| [108] | MF-DPM | TCGA | Dermoscopy | Recall | ||
| [109] | RoentGen | Hybrid (D + V) | MIMIC-CXR | X-ray | Accuracy | |
| [110] | IITM-Diffusion | BraTS2020 | MR | - | ||
| [111] | DALL-E2 | Fitzpatrick | Dermoscopy | Accuracy | ||
| [112] | CDDPM | ADNI | MR | ✓ | MMD, MS-SSIM, FID | |
| [113] | DALL-E2 | Private | X-ray | - | ||
| [114] | DDPM | OPMR | MR | ✓ | Acc., Dice | |
| [115] | LDM | MaCheX | X-ray | MSE, PSNR, SSIM | ||
| Segmentation | ||||||
| [116] | DDPM | ADNI, MRNet, | MR, CT | Dice | ||
| LIDC-IDRI | ||||||
| [101] | brainSPADE | Hybrid (V + G + D) | SABRE, BraTS2015 | MR | Dice, Accuracy | |
| OASIS, ABIDE | Precision, Recall | |||||
| [110] | IITM-Diffusion | BraTS2020 | MR | - | ||
| Cross-modal translation | ||||||
| [117] | SynDiff | Hybrid (D + G) | IXI, BraTS2015 | CT → MR | PSNR, SSIM | |
| MRI-CT-PTGA | ||||||
| [118] | UMM-CSGM | BraTS2019 | FLAIR ↔ T1 ↔ T1c ↔ T2 | PSNR, SSIM, MAE | ||
| [103] | CDDPM | MRI-CT-PTGA | CT ↔ MR | PSNR, SSIM | ||
| Other | ||||||
| [119] | DDM | ACDC | MR | ✓ | PSNR, NMSE, DICE |
| Abbreviation | Reference | Availability | Dataset | Modality | Anatomy |
|---|---|---|---|---|---|
| ADNI | UC | Alzheimers disease neuroimaging Initiative | MR, PET | Brain | |
| BraTS2015 | Public | Brain tumor segmentation challenge | MR | Brain | |
| BraTS2016 | Public | Brain tumor segmentation challenge | MR | Brain | |
| BraTS2017 | Public | Brain tumor segmentation challenge | MR | Brain | |
| BraTS2019 | Public | Brain tumor segmentation challenge | MR | Brain | |
| BraTS2020 | Public | Brain tumor segmentation challenge | MR | Brain | |
| IEEE CCX | Public | IEEE Covid Chest X-ray dataset | X-ray | Lung | |
| UK Biobank | UC | UK Biobank | MR | Brain, Heart | |
| NIHPCT | Public | National Institutes of Health Pancreas-CT dataset | CT | Kidney | |
| DataDecathlon | Public | Medical Segmentation Decathlon dataset | CT | Liver, Spleen | |
| MIDAS | [124] | Public | Michigan institute for data science | MR | Brain |
| IXI | Public | Information eXtraction from Images Dataset | MR | Brain | |
| DRIVE | [125] | Public | Digital Retinal Images for Vessel Extraction | Fundus photography | Retinal fundus |
| ACDC | [126] | Public | Automated Cardiac Diagnosis Challenge | MR | Heart |
| MRI-CT PTGA | [105] | Public | MRI-CT Part of the Gold Atlas project | CT, MR | Pelvis |
| ICTS | [50] | Public | National Taiwan University Hospital’s Intracranial Tumor Segmentation dataset | MR | Brain |
| CXR8 | [127] | Public | ChestX-ray8 | X-ray | Lung |
| C19CT | Public | COVID-19 CT segmentation dataset | CT | Lung | |
| TCGA | Private | The Cancer Genome Atlas Program | Microscopy | - | |
| UKDHP | [128] | UC | UK Digital Heart Project | MR | Heart |
| SCR | [129] | Public | SCR database: Segmentation in Chest Radiographs | X-ray | Lung |
| HCP | [130] | Public | Human connectom project dataset | MR | Brain |
| AIBL | UC | Australian Imaging Biomarkers and Lifestyle Study of Ageing | MR, PET | Brain | |
| OpenfMRI | Public | OpenfMRI | MR | Brain | |
| IBC | Public | Individual Brain Charting | MR | Brain | |
| NeuroSpin | Private | Institut des sciences du vivant Frédéric Joliot | MR | Brain | |
| OASIS | Public | The Open Access Series of Imaging Studies | MR | Brain | |
| APCXR | [131] | Public | The anterior-posterior Chest X-Ray dataset | X-ray | Lung |
| Fitzpatrick | [132] | Public | Fitzpatrick17k dataset | Dermoscopy | Skin |
| ISIC | Public | The International Skin Imaging Collaboration dataset | Dermoscopy | Skin | |
| DDSM | Public | The Digital Database for Screening Mammography | CT | Breast | |
| CBIS-DDMS | Public | Curated Breast Imaging Subset of DDSM | CT | Breast | |
| LIDC-IDRI | Public | The Lung Image Database Consortium (LIDC) and Image Database Resource Initiative (IDRI) | CT | Lung | |
| COVID-CT | [133] | Public | - | CT | Lung |
| SARS-COV2 | [134] | Public | CT | Lung | |
| MIMIC-CXR | [135] | Public | Massachusetts Institute of Technology | CT | Lung |
| PPMI | Public | Parkinson’s Progression Markers Initiative | MR | Brain | |
| ADHD | Public | Attention Deficit Hyperactivity Disorder | MR | Brain | |
| MRNet | Public | MRNet dataset | MR | Knee | |
| MLSP | Public | MLSP 2014 Schizophrenia Classification Challenge | MR | Brain | |
| SABRE | [136] | Public | The Southall and Brent Revisited cohort | MR | Brain, Heart |
| ABIDE | Public | The Autism Brain Imaging Data Exchange | MR | Brain | |
| OPMR | [137] | Public | Open-source prostate MR data | MR | Pelvis |
| MaCheX | [115] | Public | Massive Chest X-ray Dataset | X-ray | Lung |
| Abbrv. | Reference | Metric Name | Description |
|---|---|---|---|
| Dice | [143] | Sørensen–Dice coefficient | A measure of the similarity between two sets of data, calculated as twice the size of the intersection of the two sets divided by the sum of the sizes of the two sets |
| Hausdorff | [144] | Hausdorff distance | A measure of the similarity between two sets of points in a metric space |
| FID | [100] | Fréchet inception distance | A measure of the distance between the distributions of features extracted from real and generated images, based on the activation patterns of a pretrained inception model |
| IS | [142] | Inception score | A measure of the quality and diversity of generated images, based on the activation patterns of a pretrained Inception model |
| MMD | [145] | Maximum mean discrepancy | A measure of the difference between two probability distributions, defined as the maximum value of the difference between the two means |
| 1-NN | [146] | 1-nearest neighbor score | A method for classification or regression that involves finding the data point in a dataset that is most similar to a given query point |
| (MS-)SSIM | [139] | (Multi-scale) structural similarity | A measure of the similarity between two images based on their structural information, taking into account luminance, contrast, and structure. |
| MS | [147] | Mode score | A measure of the quality of samples generated with two probabilistic generative models based on the difference in maximum mean discrepancies between a reference distribution and simulated distribution |
| WD | [148] | Wasserstein distance | A measure of the distance between two probability distributions, defined as the minimum amount of work required to transform one distribution into the other |
| PSNR | [138] | Peak signal-to-noise ratio | A measure of the quality of an image or video, based on the ratio between the maximum possible power of a signal and the power of the noise that distorts the signal |
| (N)MSE | - | (Normalized) mean squared error | A measure of the average squared difference between the predicted and actual values |
| Jacc. | [143] | Jaccard index | A measure of the overlap between two sets of data, calculated as the ratio of the area of intersection to the area of union |
| MAE | - | Mean absolute error | A measure of the average magnitude of the errors between the predicted and actual values |
| AUC | [149] | Area under the curve | A measure of the performance of a binary classifier, calculated as the area under the receiver operating characteristic curve |
| LPIPS | [141] | Learned perceptual image patch similarity | An evaluation metric that measures the distance between two images in a perceptual space based on the activation of a deep CNN |
| KLD | [150] | Kullback–Leibler divergence | A measure of the difference between two probability distributions, often used to compare the similarity of the distributions, with a smaller KL divergence indicating a greater similarity |
| VIF | [151] | Visual information fidelity | A measure that quantifies the Shannon information that is shared between the reference and the distorted image |
| UQI | [152] | Universal quality index | A measure of the quality of restored images. It is based on the principle that the quality of an image can be quantified using the correlation between the original and restored images |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kebaili, A.; Lapuyade-Lahorgue, J.; Ruan, S. Deep Learning Approaches for Data Augmentation in Medical Imaging: A Review. J. Imaging 2023, 9, 81. https://doi.org/10.3390/jimaging9040081
Kebaili A, Lapuyade-Lahorgue J, Ruan S. Deep Learning Approaches for Data Augmentation in Medical Imaging: A Review. Journal of Imaging. 2023; 9(4):81. https://doi.org/10.3390/jimaging9040081
Chicago/Turabian StyleKebaili, Aghiles, Jérôme Lapuyade-Lahorgue, and Su Ruan. 2023. "Deep Learning Approaches for Data Augmentation in Medical Imaging: A Review" Journal of Imaging 9, no. 4: 81. https://doi.org/10.3390/jimaging9040081
APA StyleKebaili, A., Lapuyade-Lahorgue, J., & Ruan, S. (2023). Deep Learning Approaches for Data Augmentation in Medical Imaging: A Review. Journal of Imaging, 9(4), 81. https://doi.org/10.3390/jimaging9040081












