Multilevel Multiobjective Particle Swarm Optimization Guided Superpixel Algorithm for Histopathology Image Detection and Segmentation
Abstract
1. Introduction
- The MMPSO algorithm with three different objective functions is used to identify the optimal threshold values for multilevel image thresholding. The MOPSO algorithm is applied for the first time in the field of histopathology image segmentation for multilevel image thresholding. This framework opens a new avenue for researchers to propose segmentation models which include more than one segmentation criterion. It should be noted that in the past, only the PSO algorithm with a single objective function has been used for the segmentation of nuclei regions from histopathology images;
- The proposed MMPSO-S algorithm combines the segmentation output of the MMPSO algorithm and the superpixel clustering algorithm; specifically, the threshold values obtained from the MMPSO algorithm are used to refine the output of the superpixel algorithm. This combined algorithm helps to improve the segmentation results;
- The proposed algorithm is applied to four different H&E-stained histopathology datasets for the detection and segmentation of various ROIs;
- The performance of the proposed method is compared with other single and multiobjective algorithms and also with the existing work performed on the datasets.
2. Related Works
2.1. Image Segmentation Using PSO and Its Variants
2.2. Image Segmentation Using Multiobjective Algorithms
2.3. Image Segmentation Using Superpixel Algorithm
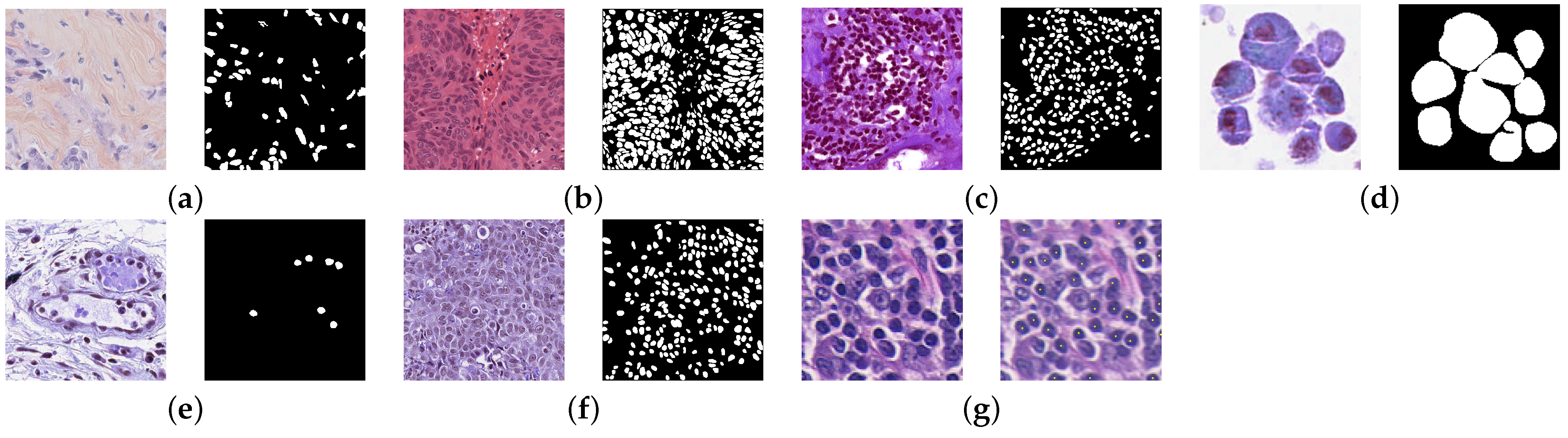
3. Dataset Description
3.1. Triple-Negative Breast Cancer Dataset
3.2. Multi-Organ Nuclei Segmentation Dataset
3.3. Multi-Organ Nuclei Segmentation and Classification Dataset
3.4. Lymphocyte Detection Dataset
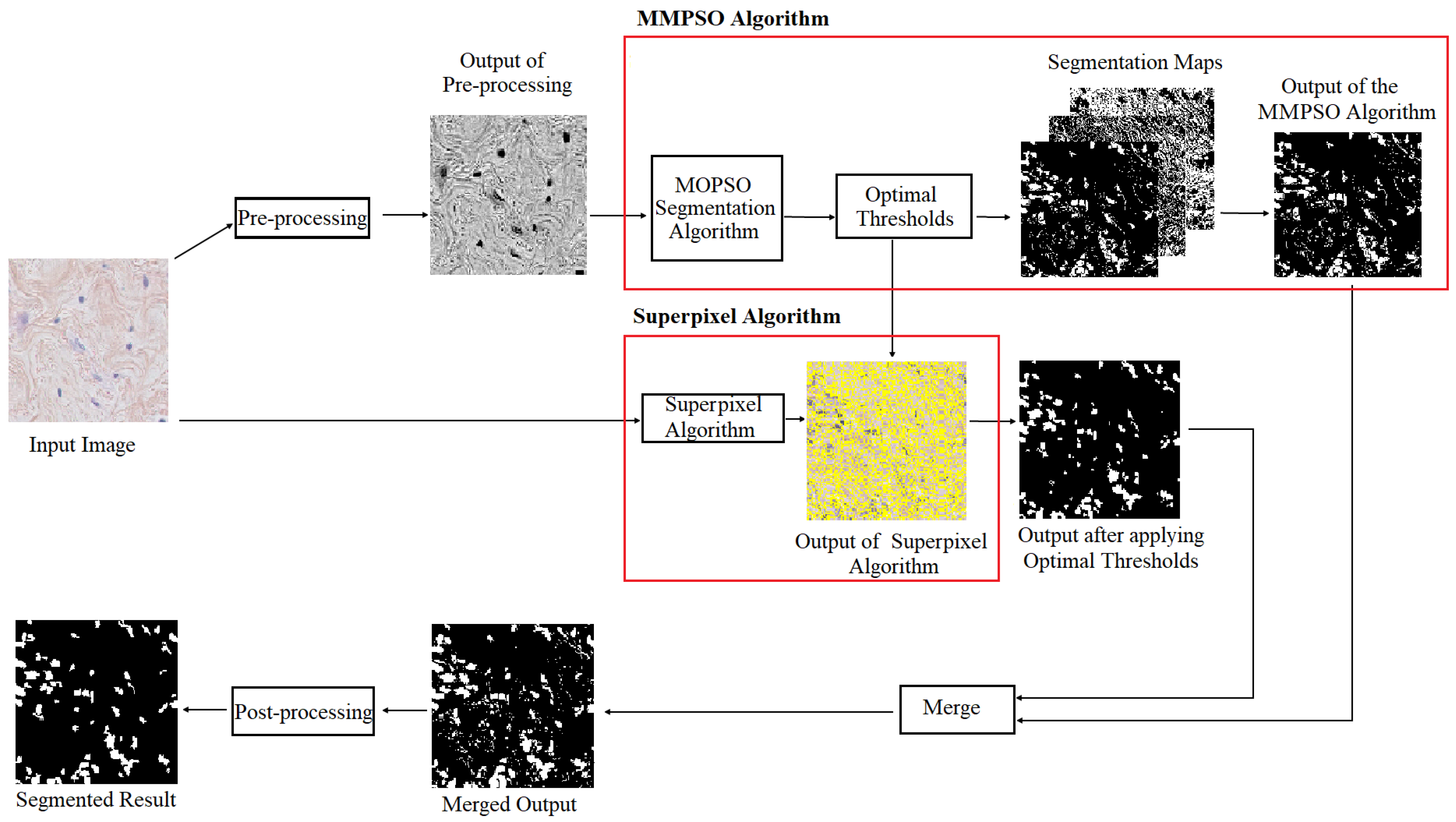
4. The Proposed Method
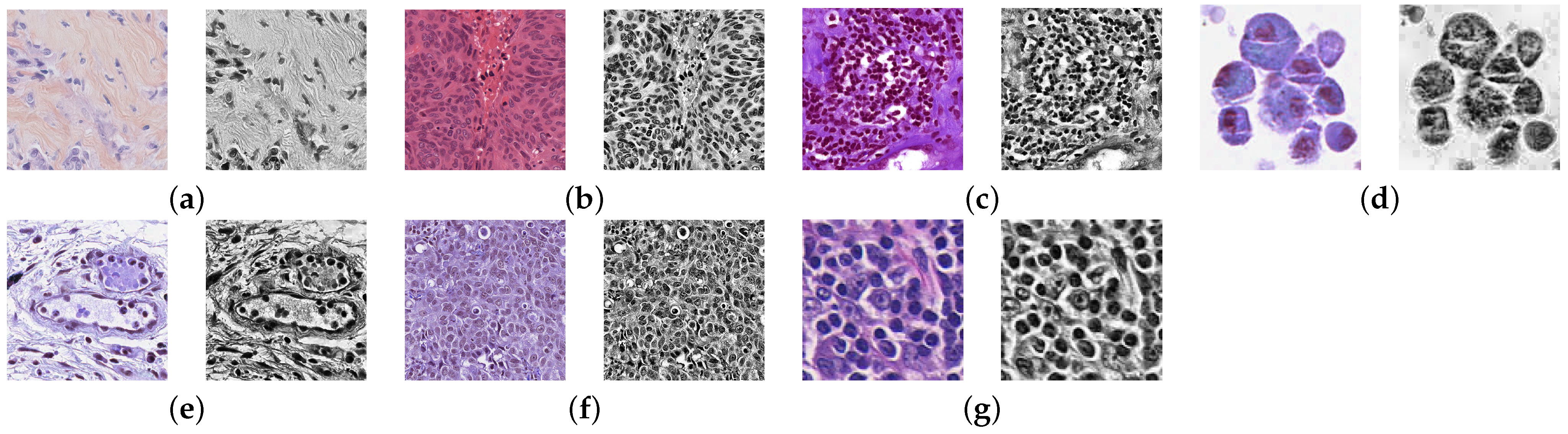
4.1. Pre-Processing
4.2. MMPSO-S Algorithm for Detection and Extraction of ROIs
4.2.1. MMPSO for Multilevel Image Thresholding
- KeyTerms of MOPSO Algorithm
- Decision space: Decision space/search space is the vector space of all decision variables. The search space varies depending on the problem domain.
- Objective space: Objective space is the vector space of all solutions obtained from the evaluation of the decision variables.
- Particles and swarm: Swarm is a collection of particles. Particles are individuals, such as birds or fishes, in the swarm. Let i represent a particle in the swarm and i = 1, 2, …, , where is the population size.
- Position: Each particle i in the search space has two properties, i.e., position and velocity. The position of a particle i is denoted as and is considered as the feasible solution to the optimization problem. It has upper and lower limits, which are the boundary of the search space denoted as .
- Velocity: Velocity of a particle defines its ability to move in the search space, which allows the particle to update its position. The upper and the lower limits of the velocity are denoted as .
- Objective function: It is also known as the fitness function/cost function. The objective function maps an element from the decision space to the objective space. The objective function is evaluated using the position and the outcome is a real number known as the cost value or the fitness value. In the case of MOPSO, the outcome of all objective functions form a vector.
- Local best: The local best value for a particle is the position value which gives the best fitness value in the whole history of its movement. It is denoted by .
- Feasible solution set: A solution that satisfies all the constraints of an MOP is called a feasible solution. A set of all feasible solutions is called the feasible solution set.
- Non-dominated solution: A feasible solution is non-dominated if there does not exist another feasible solution better than the current one in some objective function without worsening another objective function.
- External repository: It is a storage space to store all the best particles (non-dominated solutions) [10]. This repository is often known as an external archive and is denoted by A. External repository has a maximum size (). To avoid the high computational cost of searching and updating the external repository, its size is limited.
- Leader: From the external repository, one solution (L) is selected as the leader for the entire swarm and its position is taken as the value.
- Initialization Phase of MOPSO Algorithm
- Objective Functions
- (a)
- Otsu’s multilevel thresholding: Otsu’s method is an unsupervised and non-parametric threshold selection method [40]. In Otsu’s method, the threshold is selected by the discriminant criterion, that is to maximize the between-class variance among segmented regions/classes [43]. Otsu’s objective function () for the multilevel grayscale image segmentation is given by Equation (4).
- (b)
- Kapur’s multilevel thresholding: Kapur’s entropy is a generalization of Shannon’s entropy. In Kapur’s method, the threshold is selected by the discriminant criterion, that is to maximize the between-class entropy [41,44]. Kapur’s objective function () for the multilevel segmentation of grayscale images is given by Equation (5).where is the Kapur’s entropy of the region i and is given by Equation (6).
- (c)
- Renyi’s multilevel thresholding: Renyi’s entropy is a generalized form of Shannon’s entropy with a parameter used to evaluate the randomness of a system. When = 1, Renyi’s entropy is equal to Shannon’s entropy [42]. Renyi’s objective function () for the multilevel segmentation of grayscale images is given by Equation (7).where is the Renyi’s entropy of the region i and is given by Equation (8).For better segmented output, the above objective functions must be maximized.
- Iterative Phase of MOPSO Algorithm
- Leader selection from the external archive:The repository with the non-dominated solutions is mapped to an adaptive grid with a grid size comprising hypercubes [10]. Each non-dominated solution from the archive is placed in the hypercube by considering its fitness values as the coordinates. A hypercube can hold number of non-dominated solutions where . The following steps are used to select a leader from the non-dominated solutions:
- (a)
- The fitness value of a hypercube is calculated by dividing any number x ( 1) by the number of particles in that hypercube.
- (b)
- A roulette wheel algorithm is used to select a hypercube using the fitness values.
- (c)
- If the selected hypercube has one particle, then the particle is set as the leader of the swarm. Otherwise, if the number of particles in the selected hypercube is greater than 1 (i.e., ), then one particle is chosen randomly and is set as the leader of the swarm.
- Update position and velocity of each particle:Once the leader is selected, the velocity and position of all the particles in the swarm are updated using Equations (9) and (10).where is known as the inertia parameter, is the position of the particle i at time t and is the velocity of the particle i at time t, is the position of the particle i at time , and is the velocity of the particle i at time . are the positive constants known as acceleration coefficients. are the random numbers in the range (0, 1). In this work, are set to 1.3, 0.5, and 0.5, respectively.
- Compute fitness values for each particle:Once the position value of each particle i in the swarm is updated, the fitness values of each particle are calculated for all the fitness functions.
- Update the local best value of each particle:If the current value of a particle i is dominated by the new position value of the particle, then the current value of the particle is replaced with the value. Otherwise, the current value of the particle i is kept as it is. If neither the current value nor the new position value of a particle are dominating each other, then one of the values is randomly selected as the .
- Update the external repository:The non-dominated particles are identified based on the pareto dominance condition [10]. The non-dominated particles are compared with the particles already existing in the external archive in order to decide their inclusion to the external archive. The MOPSO algorithm follows four rules to add a non-dominated particle to the archive:
- (a)
- If the archive is empty, then the new particle is added to the archive.
- (b)
- If the particle is dominated by any of the particles in the archive, then the new particle is discarded.
- (c)
- If none of the particles in the archive dominate the new particle, and if the archive has enough space, then the new particle is added to the archive. During the entry, any particle in the archive dominated by the new particle is deleted from the archive.
- (d)
- If none of the particles in the archive dominate the new particle and the archive does not have enough space, then the particle from the most crowded hypercube is removed and the new particle is inserted in the archive. During the time of entry, any particle in the archive that is dominated by the new particle is removed from the archive.
- Apply mutation operator to the particles:The relevance of the mutation operator in the MOPSO algorithm is to allow the algorithm to explore the search space with a high exploratory capability. During the initial iterations of the algorithm, the mutation operator affects all the particles in the search space; however, the number of particles affected by the operator decreases as the number of iterations increases. In this work, the mutation rate () is set to 0.1.
- Obtaining the Optimal Threshold Values
- Generating segmentation maps:
4.2.2. Segmentation by Superpixel Algorithm
- Refining the clusters:
4.3. Combining Segmentation Maps and Post-Processing
| Algorithm 1: MMPSO algorithm. |
Input: Pre-processed image Output: Segmentation map Parameters: MMPSO parameters, i.e., , , k, , , , and
|
| Algorithm 2: Superpixel algorithm, cluster refinement, output merging, and post-processing. |
Input: Histopathology colour image, and output image from MOPSO algorithm Output: Image after post-processing Parameters: Expected number of superpixels to be generated , Cluster centres
|
5. Results and Discussion
5.1. Parameter Tuning
5.2. Segmentation Performance
- Segmentation Performance on the MoNuSeg Dataset:
- Segmentation Performance on the TNBC Dataset:
- Segmentation performance on the MoNuSAC dataset:
- Detection performance on the LD dataset:
- Discussion:
5.3. Normalised Execution Time
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A. Evaluation Metrics
References
- Sikandar, A. Histopathology: An Old Yet Important Technique in Modern Science. In Histopathology; Srivastava, S., Ed.; IntechOpen: Rijeka, Croatia, 2018; Chapter 1. [Google Scholar] [CrossRef]
- Hayakawa, T.; Prasath, S.; Kawanaka, H.; Aronow, B.; Tsuruoka, S. Computational Nuclei Segmentation Methods in Digital Pathology: A Survey. Arch. Comput. Methods Eng. 2021, 28, 1–13. [Google Scholar] [CrossRef]
- Sapna, S.; Renuka, A. Computer-aided system for Leukocyte nucleus segmentation and Leukocyte classification based on nucleus characteristics. Int. J. Comput. Appl. 2020, 42, 622–633. [Google Scholar] [CrossRef]
- Angel Arul Jothi, J.; Mary Anita Rajam, V. A survey on automated cancer diagnosis from histopathology images. Artif. Intell. Rev. 2017, 48, 31–81. [Google Scholar] [CrossRef]
- Pei, G. Digital Orthopedics; Springer: Ansterdam, The Netherlands, 2018. [Google Scholar]
- Jiao, L.; Shang, R.; Liu, F.; Zhang, W. Chapter 3—Theoretical basis of natural computation. In Brain and Nature-Inspired Learning Computation and Recognition; Jiao, L., Shang, R., Liu, F., Zhang, W., Eds.; Elsevier: Amsterdam, The Netherlands, 2020; pp. 81–95. [Google Scholar] [CrossRef]
- Coello, C.; Toscano Pulido, G.; Lechuga, M. Handling Multiple Objectives With Particle Swarm Optimization. IEEE Trans. Evol. Comput. 2004, 8, 256–279. [Google Scholar] [CrossRef]
- Emmerich, M.T.; Deutz, A.H. A Tutorial on Multiobjective Optimization: Fundamentals and Evolutionary Methods. Nat. Comput. Int. J. 2018, 17, 585–609. [Google Scholar] [CrossRef]
- Kennedy, J.; Eberhart, R. Particle swarm optimization. In Proceedings of the ICNN’95—International Conference on Neural Networks, Perth, WA, Australia, 27 November–1 December1995; Volume 4, pp. 1942–1948. [Google Scholar] [CrossRef]
- Coello Coello, C.; Lechuga, M. MOPSO: A proposal for multiple objective particle swarm optimization. In Proceedings of the 2002 Congress on Evolutionary Computation. CEC’02 (Cat. No.02TH8600), Honolulu, HI, USA, 12–17 May2002; Volume 2, pp. 1051–1056. [Google Scholar] [CrossRef]
- Parsopoulos, K.; Vrahatis, M. Multi-Objective Particle Swarm Optimization Approaches; IGI Global: Hershey, PA, USA, 2008; pp. 20–42. [Google Scholar] [CrossRef]
- Ibrahim, A.; El-kenawy, E.S. Image Segmentation Methods Based on Superpixel Techniques: A Survey. J. Comput. Sci. Inf. Syst. 2020, 1, 1–10. [Google Scholar]
- Jothi, J.A.A.; Rajam, V.M.A. Segmentation of Nuclei from Breast Histopathology Images Using PSO-based Otsu’s Multilevel Thresholding. In Artificial Intelligence and Evolutionary Algorithms in Engineering Systems; Suresh, L.P., Dash, S.S., Panigrahi, B.K., Eds.; Springer: New Delhi, India, 2015; pp. 835–843. [Google Scholar]
- Liu, Y.; Mu, C.; Kou, W.; Liu, J. Modified Particle Swarm Optimization-Based Multilevel Thresholding for Image Segmentation. Soft Comput. 2015, 19, 1311–1327. [Google Scholar] [CrossRef]
- Chakraborty, R.; Sushil, R.; Garg, M.L. An Improved PSO-Based Multilevel Image Segmentation Technique Using Minimum Cross-Entropy Thresholding. Arab. J. Sci. Eng. 2019, 44, 3005–3020. [Google Scholar] [CrossRef]
- Li, Y.; Wang, S.; Xiao, J. Image Segmentation Based on Dynamic Particle Swarm Optimization for Crystal Growth. Sensors 2018, 18, 3878. [Google Scholar] [CrossRef]
- Saini, B.; Gupta, S.; Kaur, T. A comparative study on Kapur’s and Tsallis entropy for multilevel thresholding of MR images via particle swarm optimisation technique. Int. J. Comput. Syst. Eng. 2018, 4, 156. [Google Scholar] [CrossRef]
- Xia, P.; Lin, Y.; Li-Hua, Z. An Improved PSO-FCM Algorithm for Image Segmentation. IOP Conf. Ser. Earth Environ. Sci. 2019, 267, 042081. [Google Scholar] [CrossRef]
- Suresh, S.; Lal, S. Multilevel thresholding based on Chaotic Darwinian Particle Swarm Optimization for segmentation of satellite images. Appl. Soft Comput. 2017, 55, 503–522. [Google Scholar] [CrossRef]
- Tang, Q.; Gao, S.; Liu, Y.; Yu, F. Infrared image segmentation algorithm for defect detection based on FODPSO. Infrared Phys. Technol. 2019, 102, 103051. [Google Scholar] [CrossRef]
- Guo, F.; Peng, H.; Zou, B.; Rongchang, Z.; Liu, X. Localization and segmentation of optic disk with the fractional-order Darwinian particle swarm optimization algorithm. IET Image Process. 2018, 12, 1303–1312. [Google Scholar] [CrossRef]
- Zhe, L.; Bao, X.; Yuqing, S.; Hu, L.; Qingfeng, L. An Improved Unsupervised Image Segmentation Method Based on Multi-Objective Particle Swarm Optimization Clustering Algorithm. Comput. Mater. Contin. 2019, 58, 451–461. [Google Scholar] [CrossRef]
- Maryam, H.; Mustapha, A.; Younes, J. A multilevel thresholding method for image segmentation based on multiobjective particle swarm optimization. In Proceedings of the 2017 International Conference on Wireless Technologies, Embedded and Intelligent Systems (WITS), Fez, Morocco, 19–20 April 2017; pp. 1–6. [Google Scholar] [CrossRef]
- Hinojosa, S.; Oliva, D.; Cuevas, E.; Pajares, G.; Zaldivar, D.; Cisneros, M. Reducing overlapped pixels: A multi-objective color thresholding approach. Soft Comput. 2020, 24, 6787–6807. [Google Scholar] [CrossRef]
- Pham, T.X.; Siarry, P.; Oulhadj, H. A multi-objective optimization approach for brain MRI segmentation using fuzzy entropy clustering and region-based active contour methods. Magn. Reson. Imaging 2019, 61, 41–65. [Google Scholar] [CrossRef]
- Elaziz, M.A.; Oliva, D.; Ewees, A.A.; Xiong, S. Multi-level thresholding-based grey scale image segmentation using multi-objective multi-verse optimizer. Expert Syst. Appl. 2019, 125, 112–129. [Google Scholar] [CrossRef]
- Oliva, D.; Abd Elaziz, M.; Hinojosa, S. Image Segmentation as a Multiobjective Optimization Problem. In Metaheuristic Algorithms for Image Segmentation: Theory and Applications; Springer International Publishing: Cham, Switzerland, 2019; pp. 157–179. [Google Scholar] [CrossRef]
- Sağ, T.; Çunkaş, M. Color image segmentation based on multiobjective artificial bee colony optimization. Appl. Soft Comput. 2015, 34, 389–401. [Google Scholar] [CrossRef]
- Albayrak Abdulkadir, B.G. A Hybrid Method of Superpixel Segmentation Algorithm and Deep Learning Method in Histopathological Image Segmentation. In Proceedings of the Innovations in Intelligent Systems and Applications INISTA, Thessaloniki, Greece, 3–5 July 2018; pp. 1–5. [Google Scholar] [CrossRef]
- Albayrak, A.; Bilgin, G. Automatic cell segmentation in histopathological images via two-staged superpixel-based algorithms. Med. Biol. Eng. Comput. 2018, 57, 653–665. [Google Scholar] [CrossRef]
- Ding, S.; Cong, L.; Wang, L.; Zhang, A.; Jia, W. Image Segmentation Algorithm Based on Superpixel Clustering. IET Image Process. 2018, 12, 2030–2035. [Google Scholar] [CrossRef]
- Zhang, S.; You, Z.; Wu, X. Plant disease leaf image segmentation based on superpixel clustering and EM algorithm. Neural Comput. Appl. 2019, 31, 1225–1232. [Google Scholar] [CrossRef]
- Naylor, P.; Laé, M.; Reyal, F.; Walter, T. Segmentation of Nuclei in Histopathology Images by Deep Regression of the Distance Map. IEEE Trans. Med. Imaging 2018, 38, 448–459. [Google Scholar] [CrossRef] [PubMed]
- Kumar, N.; Verma, R.; Anand, D.; Zhou, Y.; Onder, O.F.; Tsougenis, E.; Chen, H.; Heng, P.A.; Li, J.; Hu, Z.; et al. A Multi-Organ Nucleus Segmentation Challenge. IEEE Trans. Med Imaging 2020, 39, 1380–1391. [Google Scholar] [CrossRef] [PubMed]
- Kumar, N.; Verma, R.; Sharma, S.; Bhargava, S.; Vahadane, A.; Sethi, A. A Dataset and a Technique for Generalized Nuclear Segmentation for Computational Pathology. IEEE Trans. Med. Imaging 2017, 36, 1550–1560. [Google Scholar] [CrossRef]
- Verma, R.; Kumar, N.; Patil, A.; Kurian, N.C.; Rane, S.; Graham, S.; Vu, Q.D.; Zwager, M.; Raza, S.E.A.; Rajpoot, N.; et al. MoNuSAC2020: A Multi-organ Nuclei Segmentation and Classification Challenge. IEEE Trans. Med Imaging 2021, 40, 3413–3423. [Google Scholar] [CrossRef]
- Andrew, J.; Anant, M. Deep learning for digital pathology image analysis: A comprehensive tutorial with selected use cases. J. Pathol. Inform. 2016, 7, 29. [Google Scholar] [CrossRef]
- Ogiela, M.; Tadeusiewicz, R. Preprocessing medical images and their overall enhancement. Stud. Comput. Intell. 2008, 84, 65–97. [Google Scholar] [CrossRef]
- Zuiderveld, K. Contrast Limited Adaptive Histogram Equalization. In Graphics Gems IV; Academic Press Professional, Inc.: New York, NY, USA, 1994; pp. 474–485. [Google Scholar]
- Otsu, N. A Threshold Selection Method from Gray-Level Histograms. IEEE Trans. Syst. Man Cybern. 1979, 9, 62–66. [Google Scholar] [CrossRef]
- Kapur, J.; Sahoo, P.; Wong, A. A new method for gray-level picture thresholding using the entropy of the histogram. Comput. Vis. Graph. Image Process. 1985, 29, 273–285. [Google Scholar] [CrossRef]
- Sahoo, P.; Wilkins, C.; Yeager, J. Threshold selection using Renyi’s entropy. Pattern Recognit. 1997, 30, 71–84. [Google Scholar] [CrossRef]
- Angel Arul Jothi, J.; Mary Anita Rajam, V. Effective segmentation and classification of thyroid histopathology images. Appl. Soft Comput. 2016, 46, 652–664. [Google Scholar] [CrossRef]
- Lang, C.; Jia, H. Kapur’s Entropy for Color Image Segmentation Based on a Hybrid Whale Optimization Algorithm. Entropy 2019, 21, 318. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, B. Chapter 2—Frame Fields. In Elementary Differential Geometry, 2nd ed.; O’Neill, B., Ed.; Academic Press: Boston, MA, USA, 2006; pp. 43–99. [Google Scholar] [CrossRef]
- Achanta, R.; Shaji, A.; Smith, K.; Lucchi, A.; Fua, P.; Süsstrunk, S. SLIC Superpixels Compared to State-of-the-Art Superpixel Methods. IEEE Trans. Pattern Anal. Mach. Intell. 2012, 34, 2274–2282. [Google Scholar] [CrossRef] [PubMed]
- Bao, J.; Yin, J.; Yang, J. Superpixel-based segmentation for multi-temporal PolSAR images. In Proceedings of the 2017 Progress in Electromagnetics Research Symposium—Fall (PIERS-FALL), Singapore, 19–22 November 2017; pp. 654–658. [Google Scholar] [CrossRef]
- Heris, M.K. MOEA/D in MATLAB. 2015. Available online: https://yarpiz.com/95/ypea124-moead (accessed on 20 March 2023).
- Heris, M.K. NSGA2 in MATLAB. 2015. Available online: https://yarpiz.com/56/ypea120-nsga2 (accessed on 20 March 2023).
- Tillett, J.; Rao, T.; Sahin, F.; Rao, R. Darwinian Particle Swarm Optimization. In Proceedings of the Indian International Conference on Artificial Intelligence IICAI, Pune, India, 20–22 December 2005; pp. 1474–1487. [Google Scholar]
- Couceiro, M.; Ghamisi, P. Fractional Order Darwinian Particle Swarm Optimization: Applications and Evaluation of an Evolutionary Algorithm; Springer Publishing Company, Incorporated: Cham, Switzerland, 2015. [Google Scholar]
- Lagree, A.; Mohebpour, M.; Meti, N.; Saednia, K.; Lu, F.I.; Slodkowska, E.; Gandhi, S.; Rakovitch, E.; Shenfield, A.; Sadeghi-Naini, A.; et al. A review and comparison of breast tumor cell nuclei segmentation performances using deep convolutional neural networks. Sci. Rep. 2021, 11, 8025. [Google Scholar] [CrossRef] [PubMed]
- Alemi Koohbanani, N.; Jahanifar, M.; Zamani Tajadin, N.; Rajpoot, N. NuClick: A deep learning framework for interactive segmentation of microscopic images. Med. Image Anal. 2020, 65, 101771. [Google Scholar] [CrossRef]




| Algorithms | Method | Images |
|---|---|---|
| PSO and its variants | PSO-based Otsu’s multilevel thresholding [13] | Histopathology images |
| PSO-based clustering method [14] | Histopathology images | |
| PSO-based multilevel thresholding [15] | Grayscale images and medical images | |
| Dynamic PSO [16] | Real crystal growth images | |
| PSO using Kapur’s and Tsallis entropy [17] | Normal brain MRI | |
| PSO-FCM algorithm [18] | Ultrasonic teeth images | |
| DPSO [19] | Satellite images | |
| FODPSO [20] | Infrared images | |
| FODPSO [21] | Retinal images | |
| Multiobjective algorithms | UISMOPC [22] | Standard images |
| MOPSO [23] | Standard images | |
| Multiobjective colour thresholding [24] | Standard images | |
| Multiobjective optimization [25] | Simulated MRI and MRI | |
| Multiobjective multiverse optimization [26] | Natural grayscale images | |
| Multiobjective grey wolf optimization [27] | Natural grayscale images | |
| Multiobjective artificial bee colony [28] | Standard images | |
| Superpixel algorithm | SLIC and CNN [29] | Histopathology images |
| SLIC and clustering algorithm [30] | Histopathology images | |
| Superpixel algorithm and clustering algorithm [31] | Satellite images | |
| superpixel and EM [32] | Plant disease leaves images |
| Dataset | Task | Total Images | Image Format |
|---|---|---|---|
| TNBC [33] | Segmentation of nuclei cells | 50 | .png |
| MoNuSeg [34,35] | Nuclei segmentation from multiple organs | 44 | .tif |
| MoNuSAC [36] | Segmentation of lymphocytes | 146 | .tif |
| Segmentation of macrophages | 58 | .tif | |
| Segmentation of neutrophils | 94 | .tif | |
| Segmentation of epithelial cells | 96 | .tif | |
| LD [37] | Detection of lymphocyte cells | 100 | .tif |
| Parameters | Variable | Values |
|---|---|---|
| Population size | 150 | |
| Maximum no. of iterations | 150 | |
| No. of thresholds | k | 2 |
| Position range | [0, 255] | |
| Velocity range | [−5, 5] | |
| Repository size | 30 |
| Algorithm | F-Measure | Dice Value | Jaccard Value |
|---|---|---|---|
| PSO | 0.59 | 0.59 | 0.44 |
| DPSO | 0.62 | 0.62 | 0.45 |
| FODPSO | 0.62 | 0.62 | 0.46 |
| MOPSO(Kapur + Otsu) | 0.69 | 0.69 | 0.54 |
| MOPSO(Renyi + Otsu) | 0.61 | 0.61 | 0.45 |
| MOPSO(Kapur + Renyi) | 0.69 | 0.69 | 0.54 |
| MOPSO(Kapur + Otsu + Renyi) | 0.71 | 0.71 | 0.55 |
| Superpixel algorithm | 0.59 | 0.59 | 0.43 |
| MOEA/D | 0.38 | 0.38 | 0.24 |
| NSGA2 | 0.58 | 0.58 | 0.43 |
| Otsu threshold [52] | 0.03 | - | 0.05 |
| Watershed transform [52] | 0.09 | - | 0.08 |
| The ImageJ2-Fiji package [52] | 0.18 | - | 0.34 |
| Region growing [53] | - | 0.37 | 0.16 |
| Active contour [53] | - | 0.58 | 0.28 |
| MMPSO-S | 0.72 | 0.72 | 0.56 |
| Algorithm | F-Measure | Dice Value | Jaccard Value |
|---|---|---|---|
| PSO | 0.61 | 0.61 | 0.46 |
| DPSO | 0.61 | 0.61 | 0.46 |
| FODPSO | 0.61 | 0.61 | 0.46 |
| MOPSO(Kapur + Otsu) | 0.57 | 0.57 | 0.42 |
| MOPSO(Renyi + Otsu) | 0.63 | 0.63 | 0.46 |
| MOPSO(Kapur + Renyi) | 0.60 | 0.60 | 0.44 |
| MOPSO(Kapur + Otsu + Renyi) | 0.64 | 0.64 | 0.47 |
| Superpixel algorithm | 0.54 | 0.54 | 0.38 |
| MOEA/D | 0.24 | 0.24 | 0.14 |
| NSGA2 | 0.58 | 0.58 | 0.42 |
| MMPSO-S | 0.65 | 0.65 | 0.49 |
| Dataset | Algorithm | F-Measure | Dice Value | Jaccard Value |
|---|---|---|---|---|
| MoNuSAC-L | PSO | 0.68 | 0.68 | 0.53 |
| DPSO | 0.66 | 0.66 | 0.50 | |
| FODPSO | 0.67 | 0.67 | 0.52 | |
| MOPSO(Kapur + Otsu) | 0.68 | 0.68 | 0.52 | |
| MOPSO(Renyi + Otsu) | 0.67 | 0.67 | 0.51 | |
| MOPSO(Renyi + Kapur) | 0.67 | 0.67 | 0.51 | |
| MOPSO(Kapur + Renyi + Otsu) | 0.70 | 0.70 | 0.54 | |
| Superpixel algorithm | 0.47 | 0.47 | 0.32 | |
| MOEA/D | 0.42 | 0.42 | 0.27 | |
| NSGA2 | 0.40 | 0.40 | 0.31 | |
| MMPSO-S | 0.70 | 0.70 | 0.55 | |
| MoNuSAC-M | PSO | 0.55 | 0.55 | 0.38 |
| DPSO | 0.56 | 0.56 | 0.40 | |
| FODPSO | 0.58 | 0.58 | 0.41 | |
| MOPSO(Kapur + Otsu) | 0.59 | 0.59 | 0.43 | |
| MOPSO(Renyi + Otsu) | 0.57 | 0.57 | 0.40 | |
| MOPSO(Renyi + Kapur) | 0.57 | 0.57 | 0.41 | |
| MOPSO(Kapur + Renyi + Otsu) | 0.63 | 0.63 | 0.47 | |
| Superpixel algorithm | 0.34 | 0.34 | 0.23 | |
| MOEA/D | 0.31 | 0.31 | 0.20 | |
| NSGA2 | 0.62 | 0.62 | 0.48 | |
| MMPSO-S | 0.65 | 0.65 | 0.48 | |
| MoNuSAC-N | PSO | 0.44 | 0.44 | 0.29 |
| DPSO | 0.45 | 0.45 | 0.30 | |
| FODPSO | 0.47 | 0.47 | 0.31 | |
| MOPSO(Kapur + Otsu) | 0.50 | 0.50 | 0.35 | |
| MOPSO(Renyi + Otsu) | 0.51 | 0.51 | 0.36 | |
| MOPSO(Renyi + Kapur) | 0.46 | 0.46 | 0.31 | |
| MOPSO(Kapur + Renyi + Otsu) | 0.50 | 0.50 | 0.34 | |
| Superpixel algorithm | 0.44 | 0.44 | 0.29 | |
| MOEA/D | 0.12 | 0.12 | 0.07 | |
| NSGA2 | 0.35 | 0.35 | 0.23 | |
| MMPSO-S | 0.53 | 0.53 | 0.38 | |
| MoNuSAC-E | PSO | 0.52 | 0.52 | 0.35 |
| DPSO | 0.57 | 0.57 | 0.40 | |
| FODPSO | 0.59 | 0.59 | 0.42 | |
| MOPSO(Kapur + Otsu) | 0.61 | 0.61 | 0.44 | |
| MOPSO(Renyi + Otsu) | 0.58 | 0.58 | 0.41 | |
| MOPSO(Renyi + Kapur) | 0.59 | 0.59 | 0.42 | |
| MOPSO(Kapur + Renyi + Otsu) | 0.62 | 0.62 | 0.46 | |
| Superpixel algorithm | 0.37 | 0.37 | 0.25 | |
| MOEA/D | 0.18 | 0.18 | 0.11 | |
| NSGA2 | 0.50 | 0.50 | 0.34 | |
| MMPSO-S | 0.63 | 0.63 | 0.47 |
| Algorithm | Precision | Recall | F-Measure |
|---|---|---|---|
| PSO | 0.84 | 0.96 | 0.90 |
| DPSO | 0.85 | 0.99 | 0.92 |
| FODPSO | 0.86 | 0.99 | 0.92 |
| MOPSO(Kapur + Otsu) | 0.87 | 0.96 | 0.91 |
| MOPSO(Kapur + Renyi) | 0.88 | 0.94 | 0.91 |
| MOPSO(Renyi + Otsu) | 0.87 | 0.96 | 0.91 |
| MOPSO(Kapur + Otsu + Renyi) | 0.93 | 0.99 | 0.96 |
| Superpixel algorithm | 0.78 | 0.83 | 0.80 |
| NSGA2 | 0.86 | 0.94 | 0.90 |
| MOEA/D | 0.78 | 0.71 | 0.74 |
| MMPSO-S | 0.96 | 0.99 | 0.98 |
| Algorithm | TNBC | MoNuSeg | MoNuSAC- L | MoNuSAC- M | MoNuSAC- N | MoNuSAC- E | LD |
|---|---|---|---|---|---|---|---|
| PSO | 0.021 | 0.036 | 0.022 | 0.021 | 0.021 | 0.038 | 0.011 |
| DPSO | 0.186 | 0.203 | 0.203 | 0.196 | 0.192 | 0.200 | 0.207 |
| FODPSO | 0.241 | 0.238 | 0.276 | 0.246 | 0.265 | 0.255 | 0.280 |
| MOPSO(Kapur + Otsu) | 0.102 | 0.098 | 0.104 | 0.102 | 0.092 | 0.091 | 0.098 |
| MOPSO(Renyi + Otsu) | 0.100 | 0.094 | 0.098 | 0.096 | 0.098 | 0.085 | 0.092 |
| MOPSO(Kapur + Renyi) | 0.096 | 0.072 | 0.093 | 0.089 | 0.091 | 0.087 | 0.086 |
| MOPSO(Kapur + Otsu + Renyi) | 0.112 | 0.103 | 0.084 | 0.110 | 0.095 | 0.097 | 0.101 |
| Superpixel algorithm | 0.015 | 0.031 | 0.020 | 0.014 | 0.018 | 0.031 | 0.016 |
| MMPSO-S | 0.126 | 0.125 | 0.100 | 0.126 | 0.128 | 0.116 | 0.109 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kanadath, A.; Angel Arul Jothi, J.; Urolagin, S. Multilevel Multiobjective Particle Swarm Optimization Guided Superpixel Algorithm for Histopathology Image Detection and Segmentation. J. Imaging 2023, 9, 78. https://doi.org/10.3390/jimaging9040078
Kanadath A, Angel Arul Jothi J, Urolagin S. Multilevel Multiobjective Particle Swarm Optimization Guided Superpixel Algorithm for Histopathology Image Detection and Segmentation. Journal of Imaging. 2023; 9(4):78. https://doi.org/10.3390/jimaging9040078
Chicago/Turabian StyleKanadath, Anusree, J. Angel Arul Jothi, and Siddhaling Urolagin. 2023. "Multilevel Multiobjective Particle Swarm Optimization Guided Superpixel Algorithm for Histopathology Image Detection and Segmentation" Journal of Imaging 9, no. 4: 78. https://doi.org/10.3390/jimaging9040078
APA StyleKanadath, A., Angel Arul Jothi, J., & Urolagin, S. (2023). Multilevel Multiobjective Particle Swarm Optimization Guided Superpixel Algorithm for Histopathology Image Detection and Segmentation. Journal of Imaging, 9(4), 78. https://doi.org/10.3390/jimaging9040078







