Abstract
Skin detection involves identifying skin and non-skin areas in a digital image and is commonly used in various applications, such as analyzing hand gestures, tracking body parts, and facial recognition. The process of distinguishing between skin and non-skin regions in a digital image is widely used in a variety of applications, ranging from hand-gesture analysis to body-part tracking to facial recognition. Skin detection is a challenging problem that has received a lot of attention from experts and proposals from the research community in the context of intelligent systems, but the lack of common benchmarks and unified testing protocols has hampered fairness among approaches. Comparisons are very difficult. Recently, the success of deep neural networks has had a major impact on the field of image segmentation detection, resulting in various successful models to date. In this work, we survey the most recent research in this field and propose fair comparisons between approaches, using several different datasets. The main contributions of this work are (i) a comprehensive review of the literature on approaches to skin-color detection and a comparison of approaches that may help researchers and practitioners choose the best method for their application; (ii) a comprehensive list of datasets that report ground truth for skin detection; and (iii) a testing protocol for evaluating and comparing different skin-detection approaches. Moreover, we propose an ensemble of convolutional neural networks and transformers that obtains a state-of-the-art performance.
1. Introduction
People use skin texture and color as crucial clues to understanding the different cultural characteristics of others (age, ethnicity, health, wealth, beauty, etc.). Skin tone in a photograph or video serves as a visual cue that a human is present in that piece of media. As a result, during the past 20 years, much research has been performed on video and image skin detection in the context of intelligent systems. Skin detection, which separates skin and non-skin regions in a digital image, entails performing binary pixel classification and fine segmentation to establish the limits of the skin region. Skin texture and color are important cues that people use to understand different cultural aspects of each other (health, ethnicity, age, beauty, wealth, etc.). The presence of skin color in an image or video indicates the presence of a person in such media. Therefore, over the past two decades, extensive research in the context of professional and intelligent systems has focused on video and image skin detection. Skin detection is the process of distinguishing between skin and non-skin regions in a digital image and consists of performing binary classification of pixels and performing fine segmentation to define skin-region boundaries. It is an advanced process, involving not only model training but many additional methods, including data pre- and postprocessing.
This survey is a revised version of [1]. The aim of this study is to cover the recent literature in deep-learning-based skin segmentation by providing a comprehensive review with specific insights into different aspects of the proposed methods. This includes the training data, the network architectures, loss functions, training strategies, and specific key contributions. Moreover, we propose a new ensemble that is based on convolutional neural networks and transformers and provides a state-of-the-art performance.
Skin detection is used as a preparatory step for medical imaging, such as the detection of skin cancer [2,3], skin diseases in general [4,5], or skin lesions in general [6,7]. It is also adopted for face detection [8] and body tracking [9], hand detection [10], biometric authentication [11], and many others [12,13,14].
This article provides an extensive review of the ways techniques from artificial intelligence, deep learning, and machine learning systems are designed and developed to resolve the problem of skin detection.
The pixel color is one feature that aids in separating skin pixels from non-skin pixels. Still, achieving skin-tone consistency in different lighting, different ethnicity, and a variety of environments and sensing technologies is a highly challenging task.
Additionally, if utilized as an initial step for other applications, skin detection is computationally efficient; invariant to geometric transformations, partial occlusions, or changes in body pose/expression; and can be applied to complex or simulated skin. It is not affected by the background of the capture device.
Pixel intensity depends on scene conditions, such as reflectance and light, that strongly influence color consistency, which is the most influential factor in determining skin color [15]. Some approaches to skin identification include color-constancy-based picture preprocessing techniques (i.e., color-correction techniques based on luminance estimate) and/or dynamic adaption techniques to be effective when lighting conditions vary quickly. A feasible solution is to consider extra data not in the visible spectrum (i.e., infrared images [16] or spectral images [17]), but these sensors require a higher acquisition cost, thus limiting their use for specific applications.
A more specific application for skin detection is hand segmentation, which aims at segmenting the hand profile: this task becomes particularly challenging when the segmentation of a hand is over the face or other portions of skin. Recent approaches to solving these problems are adopting very deep neural network structures and collecting new large-scale datasets on real-life scenes to increase the diversity and complexity [18,19]. New studies try to reduce the size of the network models, refining existing ones, in order to perform with few parameters and increase the inference speed, while achieving high accuracy during the hand-segmentation process [19].
Recent surveys are almost all focused on the adoption of artificial-intelligence techniques for the early detection of skin cancer. They observed the increasing interest of researchers for deep-learning techniques [20,21]. A key point that emerges from this analysis is the number of studies focusing on the automatic detection of lesions [22] or cancer. This is reported in a recent systematic review of the literature [23] which identified 14,224 studies on the early diagnosis of skin cancer published between 1 January 2000, and 9 August 2021, in MEDLINE, Embase, Scopus, and Web of Science. Another systematic review [24] identified 21 open-access datasets containing 106,950 skin-lesion images which can be used for training and testing algorithms for skin cancer diagnosis.
The major contributions of this research work are as follows:
- An exhaustive review of the literature on skin-color-detection approaches, with a detailed description of methods freely available.
- Collection and study of virtually any real skin-detection dataset available in the literature.
- A testing protocol for comparing different approaches for skin detection.
- Four different deep-learning architectures have been trained for skin detection. The proposed ensemble obtains a state-of-the-art performance (the code is made publicly available at https://github.com/LorisNanni (accessed on 26 November 2022)).
2. Methods for Skin Detection
Some skin-detection approaches rely on the assumption that the skin color can be detected in a specific color space from the background color by using clustering rules.
This assumption holds true in constrained environments where both the ethnicity and background color of the people are known, but in complex images taken under unconfined conditions, where the subject has a wide range of human skin tones, it is a very difficult task [25].
The performance of a skin detector is affected by a variety of challenging factors, including the following:
- Age, ethnicity, and other human characteristics. Human racial groupings have skin that ranges in color from white to dark brown; the age-related transition from young to old skin determines a significant variety in tones.
- Shooting conditions connected with acquiring devices’ characteristics and lighting variations have a large effect on the appearance of skin. In general, changes in lighting level or light-source distribution determine the presence of shadows and changes in skin color.
- Skin paint: Tattoos and makeup affect the aspect of the skin.
- Complex background: The presence of skin-colored objects in the background can fool the skin detector.
Existing skin-detection models can be classified according to several aspects of the procedure:
- The presence of preprocessing steps intended to reduce the effects of different acquisition conditions, such as color correction and light removal [26] or dynamic adjustment [27];
- The selection of the most suitable skin-color model [28]. Different color models are evaluated [25,29,30] (e.g., RGB, normalized RGB, the perceptual model, creating new color spaces, and others).
- The formulation of the problem based on either segmenting the image into human skin regions or treating each pixel as skin or non-skin, regardless of its neighbors. There are few area-based skin-color detection methods [31,32,33,34], including some recent methods (e.g., [35,36]) based on convolutional neural networks.
- The type of approach [37]: Rule-based methods define explicit rules for determining skin color in an appropriate color space; machine learning approaches use nonparametric or parametric learning approaches to estimate the color distribution of the training.
- According to other taxonomies from the field of machine learning [38] that consider the classification step, statistical methods include parametric methods based on Bayes’ rule of mixed models [39] applied at a pixel level. Diffusion-based methods [40,41] extend the analysis to adjacent pixels to improve classification performance. Neural network models [42,43] take into account both color and texture information. Adaptive techniques [44] rely on coordination patterns to adapt to specific conditions (e.g., lighting, skin color, and background). Model calibration often provides performance benefits but increases computation time. Support Vector Machine (SVM)-based systems are parametric models based on SVM classifiers. When the SVM classifier is trained by active learning, this class also repeats the adaptive method [14]. Blending methods are methods based on combining different machine-learning approaches [45]. Finally, hyperspectral models [46] are based on acquisition instruments with hyperspectral capabilities. Despite the benefits of the availability of spectral information, these approaches are not included in this survey, as they only apply to ad hoc datasets.
- Deep-learning methods have shown outstanding potential in dermatology for skin-lesion detection and identification [6]; however, they usually require annotations beforehand and can only classify lesion classes seen in the training set. Moreover, large-scale, open-sourced medical datasets normally have far fewer annotated classes than in real life, further aggravating the problem.
When the detection conditions are controlled, the identification of skin regions is fairly straightforward; for example, in some gesture-recognition applications, hand images are captured by using flatbed scanners and have a dark unsaturated background [47]. For this reason, several simple rule-based methods have been proposed, in addition to approaches based on sophisticated and computationally expensive techniques. These techniques are chosen in particular situations because they are more effective; ready to use; and simple to understand, apply, and reuse. Although they are effective enough, at the same time, simple rule-based methods are typically not even tested against pure skin detection benchmarks, but as a step in more complex tasks (face recognition, hand gesture recognition, etc.). A solution based on a straightforward RGB look-up table is proposed in [47], following a study on different color models, revealing that there is no obvious advantage to using a uniform color space for perception. Older approaches were based on parameterizing color spaces as a preliminary step to detect skin regions [48] or to improve the learning phase, allowing for a reduced number of data in the training phase [49]. More complex approaches perform spatial permutations to deal with the problem of light variations [50]. The creation of new color spaces is reached by introducing linear and nonlinear conversions of RGB color space [30] or applying Principal Component Analysis and a Genetic Algorithm to discover the optimal representation [51]. Recent studies mimic alternate representations of images by developing color-based data augmentations to enrich the dataset with artificial images [29].
When skin detection is performed in uncontrolled situations, the current state-of-the-art is obtained by deep-learning methods [36,52,53]. Often, convolutional neural networks are preferred and implemented in a variety of computer vision tasks, for instance, by applying different structures to identify the most suitable one for skin detection [35,53].
A patch-wise approach is proposed [52], where deep neural networks use image patches as processing units rather than pixels. Another approach [36] integrates fully convolutional neural networks with recurrent neural networks to develop an end-to-end network for human skin detection.
The main problem identified in the analysis of the literature is the heterogeneity of protocols adopted in training and assessing the proposed models. This makes the comparison very difficult, due to the different testing protocols. For instance, recently, a research study compared different deep-learning approaching on different datasets, using different training sets [54]. In this work, we adopted a standard protocol to train the models and validate the results.
Now, we list some of the most interesting approaches proposed in the last twenty years.
- GMM [39] is a simple skin-detection approach based on the Gaussian mixture model that is trained to classify non-skin and skin pixels in the RGB color space.
- Bayes [39] is a fast method based on a Bayesian classifier that is trained to classify skin and non-skin pixels in the RGB color space. The training set is composed of the first 2000 images from the ECU dataset.
- SPL [55] is a pixel-based skin-detection approach that uses a look-up table (LUT) to determine skin probabilities in the RGB domain. For the test image, it is probable that each pixel, x, is occluded, and so apply a threshold, τ, to determine whether it is not occluded/nose.
- Cheddad [56] is a fast pixel-based method that converts the RGB color space into a 1D space by separating the grayscale map from its non-red encoded counterpart. The classification process uses skin probability to define the bottom and upper bounds of the skin cluster, and a classification threshold, τ, determines the outcome.
- Chen [43] is a statistical skin-color method that was designed to be implemented on hardware. The skin region is delineated in a transformed space obtained as the 3D skin cube, whose axes are the difference of two-color channels: sR = R-G, sG = G-B, and sB = R-B.
- SA1 [57], SA2 [44], and SA3 [58] are three skin-detection methods based on spatial analysis. Starting with the skin-probability map obtained with the pixel-color detector, the first step in spatial analysis is to correctly select high-probability pixels, as skin seeds. The second step is to find the shortest path to propagate the “shell” from each seed to each individual pixel. During the enhancement process, all non-adjacent pixels are marked as non-skin. SA2 [44] is an evolution of the previous approach, using both color and textural features to determine the presence of skin: it extracts the textural features from the skin probability maps rather than from the luminance channel. SA3 [58] is a further evolution of the previous spatial analysis approaches that combines probabilistic mapping and local skin-color patterns to describe skin regions.
- DYC [59] is a skin-detection approach which takes into account the lighting conditions. The approach is based on the dynamic definition of the skin cluster range in the YCb and YCr subspaces of YCbCr color space and on the definition of correlation rules between the skin color clusters.
- In [1,60], several deep-learning segmentation approaches are compared: SegNet, U-Net; DeepLabv3+; HarD-NetMSEG (Harmonic Densely Connected Network) (https://github.com/james128333/HarDNet-MSEG, Last access on 5 November 2022); [61] and Polyp-PVT [62], a deep-learning segmentation model based on a transformer encoder, i.e., PVT (Pyramid Vision Transformer) (https://github.com/DengPingFan/Polyp-PVT, Last access on 5 November 2022).
- ALDS [63] is a framework based on probabilistic approach that initially utilizes active contours and watershed merged mask for segmenting out the mole, and, later, the SVM and Neural Classifier are applied for the classification of the segmented mole.
- DNF-OOD [6] applies a non-parametric deep-forest-based approach to the problem of out-of-distribution (OOD) detection
- SANet [64] contains two sub-modules: superpixel average pooling and superpixel attention module. The authors introduce a superpixel average pooling to reformulate the superpixel classification problem as a superpixel segmentation problem, and a superpixel attention module is utilized to focus on discriminative superpixel regions and feature channels.
- OR-Skip-Net [65] is an outer residual skip connection that was designed and implemented to deal with skin segmentation in challenging environments, irrespective of skin color, and to eliminate the cost of the preprocessing. The model is based on a deep convolutional neural network.
- In [29], a new approach for skin detection that performs a color-based data augmentation to enrich the dataset with artificial images to mimic alternate representations of the image is proposed. Data augmentation is performed in the HSV (hue, saturation, and value) space. For each image in a dataset, this approach creates fifteen new images.
- In [30], a different color space is proposed; its goal is to represent the information in images, introducing a linear and nonlinear conversion of the RGB color space through a conversion matrix (W matrix). The W matrix values are optimized to meet two conditions: firstly, maximizing the distance between centers of skin and non-skin classes; and, secondly, minimizing the entropy of each class. The classification step is performed with the adoption of neural networks and an adaptive neuro-fuzzy inference system called Adaptive network-based fuzzy inference system (ANFIS).
- SSS-Net [66] captures the multi-scale contextual information and refines the segmentation results especially along object boundaries. It also reduces the cost of the preprocessing, as well.
- SCMUU [67] stands for skin-color-model updating units, and it performs skin detection by using the similarity of adjacent frames in a video. The method is based on the assumption that the face and other parts of the body have a similar skin color. The color distribution is used to build chrominance components of the YCbCr color space by referring to facial landmarks.
- SKINNY [68] is a U-net based model. The model has more depth levels; it uses wider convolutional kernels for the expansive path and employs inception modules alongside dense blocks to strengthen feature propagation. In such a way, the model is able to increase the multi-scale analysis range.
A rough classification of the most used methods is reported in Table 1.

Table 1.
Rough classification of the tested approaches.
Hand Segmentation
As is the case in skin detection, deep-learning methods are used for hand segmentation to achieve a cutting-edge performance. Current state-of-the-art approaches for human hand detection [69] have achieved great success by making good use of multiscale and contextual information, but still remain unsatisfactory for hand segmentation, especially in complex scenarios. In this context, deep approaches have faced some difficulties, such as the clutter in the background that hinders the reliable detection of hand gestures in real-world environments. Moreover, frequently the task described in literature is not clear: for instance, some studies report a hand segmentation task but in the empirical analysis the authors used a mask to recognize the whole arm [70]; this affects the final results, as makes the goal being a skin-segmentation task rather than a hand-detection one.
Among the several recent studies focused on hand segmentation, we cite the following:
- Refined U-net [19]: The authors proposed a refinement of U-net that performs with a few parameters and increases the inference speed, while achieving high accuracy during the hand-segmentation process.
- CA-FPN [69] stands for Context Attention Feature Pyramid Network and is a model designed for human hand detection. In this method, a novel Context Attention Module (CAM) is inserted into the feature pyramid networks. The CAM is designed to capture relative contextual information for hands and build long-range dependencies around hands.
In this work, we did not make a complete survey of hand segmentation, but we treated the task as a subtask for skin segmentation and used some datasets collected for this task to show the robustness of the proposed ensemble of skin detectors. We show that the proposed method gives a good performance in this domain without ad hoc training.
3. Materials and Methods
This section presents some of the most interesting models and methods for training used in the field of skin detection. We also report a brief overview of all the main available loss functions developed for skin segmentation. Some of the following approaches have been included for the creation of the proposed ensemble.
3.1. Deep Learning for Semantic Image Segmentation
In order to solve the problem of semantic segmentation, several deep-learning models have been proposed in the specialized literature.
Semantic segmentation aims to identify objects in an image and their relative boundaries. Therefore, the main purpose is to assign classes at the pixel level, which is a task achieved thanks to FCNs (Fully Convolutional Networks). An FCN has very high performance, and unlike convolutional neural network (CNN), it uses a fully convolutional last layer instead of a fully connected layer. [71]. An FCN and autoencoder are combined to obtain a deconvolutional network such as the U-Net. The U-Net represents the first attempt to use autoencoders in image-segmentation operations. Autoencoders can shrink the input while increasing the number of features used to describe the input space. Another symbolic example can be found in SegNet [72].
DeepLab [73] is of a set of autoencoder models provided by Google and has shown excellent results in semantic segmentation applications [73,74,75,76]. The key features included to ensure better performance comprehend an advanced convolution to reduce merging and transition effects and significantly increase resolution; information is obtained by the Atrous Spatial Pyramid Pooling of different scales, and a combination of CNNs and probabilistic graphical models can determine object boundaries. In this work, we adopted an extension of the suite developed by Google DeepLabV3+ [75]. We found two major innovations in DeepLabV3+: first, a 1x1 Convolution and Packet Normalization in Atrous Spatial Pyramid Pooling; and, second, a set of parallel and cascaded convolution scaling modules. One of the main features of this extension is a depth-roll and spot-roll decoder. Different depths at the same location but different channels use the same channel at different locations in a point. We can consider other features of the model structure to achieve a different design for your framework. In fact, the architecture model itself is only a used choice. Here, we consider ResNet101 [77] as the backbone for DeepLabV3+; ResNet101 is a very popular CNN that obtains residual functions by referencing block inputs (for a complete list of CNN structures please refer to [78]). It is pretrained on the VOC segmentation dataset and then tuned by using the parameters specified on the github page (https://github.com/matlab-deep-learning/pretrained-deeplabv3plus (accessed on 1 January 2020) We adopted the same parameters to prevent overfitting (i.e., the same parameters in all the training datasets):
- Initial learning rate = 0.01;
- Number of epoch = 10 (using the simple data augmentation approach, DA1; see Section 3.3) or 15 (the latter more complex data augmentation approach, DA2 (see Section 3.3), since the slower convergence using this larger augmented training set);
- Momentum = 0.9;
- L2 Regularization = 0.005;
- Learning Rate Drop Period = 5;
- Learning Rate Drop Factor = 0.2;
- Shuffle training images every epoch;
- Optimizer = SGD (stochastic gradient descent).
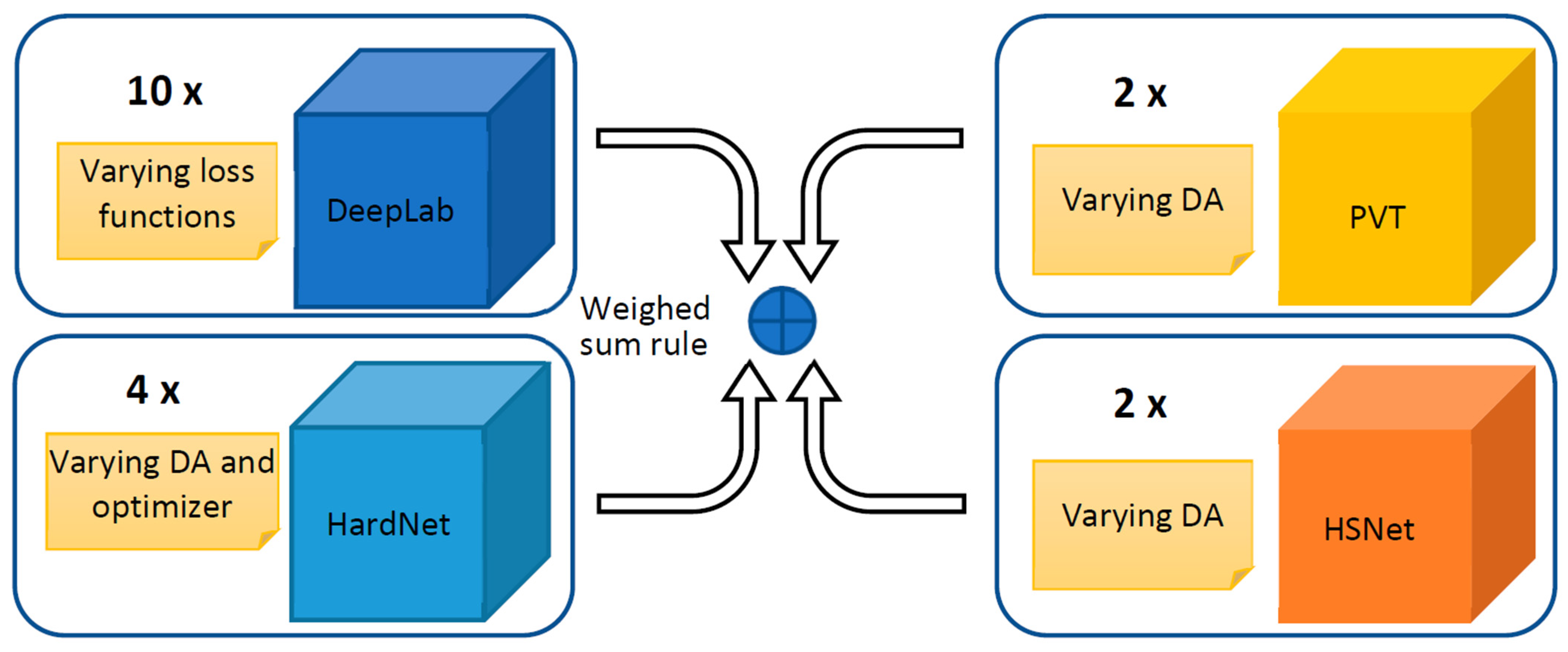
An ensemble is a group of models that work together to improve performance by combining their predictions. A strong ensemble is made up of models that are individually accurate and diverse in their mistakes. In order to boost diversity, we present an ensemble based on different architectures: DeepLabV3+, HarDNet-MSEG [61], Polyp-PVT [62], and Hybrid Semantic Network (HSN) [79]. Moreover, models with the same architecture are differentiated in the training phase by varying the data augmentation, the loss function, or the optimizer. In Figure 1, a schema of the proposed ensemble is reported.
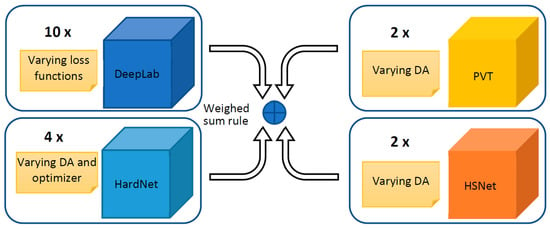
Figure 1.
A general schema of our ensemble approach: DeepLabV3+ and HarDNet-MSEG are CNN-based networks, Polyp-PVT is transformer based, and HSNet is a hybrid.
The HarD-Net-MSEG (Harmonic Densely Connected Network) [61] is a model influenced by densely connected networks that can reduce memory consumption by diminishing aggregation with the reduction of most connection layers to the DenseNet layer. Moreover, the input/output channel ratio is balanced (due to increased connections) as the layer channel width increases.
Polyp-PVT [62] is based on a pure convolutional network of transformers that aims to achieve high-resolution displays from microscopic inputs. The computational cost of the model decreases with the depth of the model through progressive pyramidal reduction. The Spatial Reduction Focusing (SRA) layer was introduced to further reduce the computational complexity of the system. The decoder part is based on a cascaded fusion module (CFM) used to collect the semantic and location information of foreground pixels from high-level features; a camouflage identification module (CIM) is applied to capture skin information disguised in low-level features; and a similarity aggregation module (SAM) is used to extend the pixel features of the skin area with high-level semantic position information to the entire image, thereby effectively fusing cross-level features.
The Hybrid Semantic Network [79] leverages transformers and convolutional neural networks. HSNs include the Cross-Semantic Attention Module (CSA), Hybrid Semantic Complement Module (HSC), and Multi-Scale Prediction Module (MSP). The authors introduced a new CSA module, which fills the gap between low-level and high-level functions by an interactive mechanism that replaces the two semantics of different NNs. Moreover, HSN adopts a new HSC module that captures both long-range dependencies and local scene details, using the two-way architecture of a transformer and CNN. In addition, the MSP module can learn weights for combining prediction masks at the decoder stage.
HardNet-MSEG, PVT-Polyp, and HSNet network topologies are trained by using the structure loss function, which is the sum of weighted IoU loss and weighted binary cross-entropy (BCE) loss, where weights are related to pixel importance (which is calculated according to the difference between the center pixel and its surroundings). We employed the Adam or SGD optimization algorithms for HardNet-MSEG and AdamW for PVT-Polyp and HSNet. The learning rate is 1 × 10−4 for HardNet-MSEG and PVT-Polyp and 5e-5 for HSNet (decaying to 5 × 10−6 after 30 epochs). The whole network is trained in an end-to-end manner for 100 epochs with a batch size of 20 for HardNet-MSEG and 8 for PVT-Polyp and HSNet. The output prediction map is generated after a sigmoid operation.
Notice that, in the original code of PVT, HardNet-MSEG, and HSN, each output map is normalized between [0, 1], so we avoid that normalization in the test phase (otherwise, it always finds a foreground region).
3.2. Loss Functions
Loss functions play an important role in any statistical model; they define what is and what is not a good prediction, so the choice of the right loss function determines the quality of the estimator.
In general, loss functions affect the training duration and model performance. In semantic segmentation operations, pixel cross-entropy is one of the most common loss functions. It works at the pixel level and checks whether the predicted signature of a given pixel matches the correct answer.
An unbalanced dataset with respect to labels is one of the main problems for this approach, and it can be solved by adopting a counterweight. A recent study offered a comprehensive review of image segmentation and loss functions [80].
In this section, we detail some of the most used loss functions in the segmentation field. Table 2 reports all the mathematical formulation of the following loss functions:

Table 2.
Mathematical formalization of the adopted loss functions.
- Dice Loss is a commonly accepted measure for models used for semantic segmentation. It is derived from the Sorensen–Dice ratio coefficients that test how similar two images are. The value range is [0, 1].
- Tversky Loss [81] deals with a common problem in machine learning and image segmentation that manifests as unbalanced classes in dataset, meaning that one class dominates the other.
- Focal Tversky Loss: The cross-entropy (CE) function is designed to limit the inequality between two probability distributions. Several variants of CE have been proposed in the literature, including, for example, focal loss [82] and binary cross-entropy. The first uses a modulation coefficient y > 0 to allow the model to focus on rough patterns rather than correctly classified patterns. The second is an adaptation of CE applied to a binary classification problem (i.e., a problem with only two classes).
- Focal Generalized Dice Loss allows users to focus on a limited ROI to reduce the weight of ordinary samples. This is achieved by regulating the modulating factor.
- Log-Cosh-Type Loss is a combination of Dice Loss and Log-Cos. Log-Cosh function is commonly applied with the purpose of smoothing the curve in regression applications.
- SSIM Loss [83] is obtained from the structural similarity (SSIM) index [84], usually adopted to evaluate the quality of an image.
- Cross-entropy: The cross-entropy loss (CE) function provides a measure of the difference between two probability distributions. The aim is to minimize these differences and avoid deviations between small and large areas. This can be problematic when working with unbalanced datasets. Thus, a weighted cross-entropy loss and a better-balanced classification for unbalanced scenarios were introduced [85]. The weighted binary cross-entropy formula is given in (14).
- Intersection-over-Union (IoU) loss is another well-known loss function, which was introduced for the first time in [86].
- Structure Loss is based on the combination of weighted Intersect-over-Union and weighted binary-crossed entropy. In Table 2, Formula (19) refers to structure loss, while Formula (20) is a simple variation that wants to give more importance to the binary-crossed entropy loss.
- Boundary Enhancement Loss is a loss proposed in [87] which explicitly focus on the boundary areas during training. This loss has very good performances, as it does not require any pre- or postprocessing of the image nor a particular net in order to work. In [60], the authors propose to combine it with Dice Loss and weighted cross-entropy loss.
- Contour-aware loss was proposed for the first time in [88]. It consists of a weighted binary cross-entropy loss where the weights are obtained with the aim of giving more importance to the borders of the image. In the loss, a morphological gradient edge detector was employed. Basically, the difference between the dilated and the eroded label map is evaluated. Then, for smoothing purposes, the Gaussian blur was applied.
In Table 2, T represents the image of the correct answer; Y is the prediction for the output image; K is the number of classes; M is the number of pixels; and Tkm and Ykm are, respectively, the ground truth value and the prediction value for the pixel m belonging to the class k.
Some works [89,90,91] show that varying the loss function is a good technique for generating diversity among outcomes and creating robust ensembles.
3.3. Data Augmentation
Different methods can be applied to the original dataset to increase the amount of data available for training the system. We applied these techniques to the training set on both input samples and masks. We adopted the two data augmentation techniques defined in [60]:
- DA1, base data augmentation consisting of horizontal and vertical flip, 90° rotation.
- DA2, this technique performs a set of operations to the original images in order to derive new ones. These operations comprehend shadowing, color mapping, vertical, or horizontal flipping, and others.
4. Performance Evaluation
4.1. Performance Indicators
Since skin segmentation and hand segmentation are binary classification problems, we can evaluate their performance by using standard measures for general classification problems [92], such as, precision, accuracy, recall, F1 measure, kappa, receiver operating characteristic (ROC) curve, area under the curve, etc. However, due to the specific nature of this problem, which relies on pixel-level classification and disproportionate distribution, the following metrics are usually considered for performance evaluation: confusion matrix, F1 measure (Dice), Intersection over Union (IoU), true-positive rate (TPR), and false-positive rate (FPR).
The confusion matrix is obtained by comparing the actual predictions to the expected ones and determining, at the pixel level, the number of true negatives (tn), false negatives (fn), true positives (tp), and false positives (fp). Precision is the percentage of correctly classified pixels out of all pixels classified as skins, and recall measures the model’s ability to detect positive samples.
In Table 3, we report the mathematical formalization of the metrics.

Table 3.
Performance indicators.
We used F1/Dice in this paper for skin segmentation and IoU for hand segmentation, because they are widely used in the related literature.
4.2. Skin Detection Evaluation: Datasets
There are several well-known color image datasets that are offered with ground truth to aid research in the field of skin detection. For a fair empirical evaluation of skin-detection systems, it is imperative to employ a uniform and representative benchmark. Some of the most popular datasets are listed in Table 4, and each of them is briefly described in this section.
- Compaq [39] is one of the first and most widely used large-scale skin datasets, consisting of images collected from web browsing. The original dataset was composed of 9731 images containing skin pixels and 8965 images with no skin pixels. Moreover, only 4675 skin images come with a ground truth.
- TDSD [93] contains 555 images with highly imprecise annotations produced with automatic labeling.
- Chile [94] contains 103 images with different lighting conditions and complex backgrounds. The ground truth is manually interpreted with moderate accuracy. The ECU Skin dataset [95] is a collection of 4000 color images with a relatively high ground-truth annotation. It is particularly challenging because they contain a wide variety of lighting conditions, background scenes, and skin types.
- Schmugge [96] is a collection of 845 images with accurate annotations on the three classes (skinned/non-skinned/unrelated). The dataset includes images come from different face datasets (i.e., the University of Chile database, the UOPB dataset, and the AR face dataset).
- Feeval [15] is a low-quality dataset composed of 8991 frames extracted from 25 online videos. The image quality is very low, as well as the precision of the annotations.
- The MCG skin database [97] contains 1000 images selected from the Internet, including blurred backgrounds, various ambient lights, and various human beings. Ground truths have been obtained by hand marking, but it is not accurate, as sometimes eyes, eyebrows, and even wrists are marked with skin.
- The VMD [98] contains 285 images; it is usually implemented to recognize human activity. The images cover a wide range of lighting levels and conditions.
- The SFA dataset [99] contains 1118 manually labeled images (with moderate accuracy).
- Pratheepan [100] contains 78 images randomly downloaded from Google.
- The HGR [58] contains 1558 images representing Polish and American Sign Language gestures with controlled and uncontrolled backgrounds.
- The SDD [101] contains 21,000 images, some images taken from a video and some others taken from a popular face dataset with different lighting conditions and with different skin colors of people around the world.
- VT-AAST [102] is a color-image database for benchmarking face detection and includes 66 images with precise ground truth.
- The Abdominal Skin Dataset [18] consists of 1400 abdominal images collected by using Google image search and then manually segmented. The dataset preserves the diversity of different ethnic groups and avoids the racial bias implicit in segmentation algorithms: 700 images represent dark-skinned people, and 700 images represent light-skinned people. Additionally, 400 images represent individuals with high body mass index (BMI), evenly distributed between light and dark skins. The dataset also took into account other inter-individual variation, such as hair and tattoo coverage, and external variation, such as shadows, when preparing the dataset.

Table 4.
Some of the most used datasets per skin detection.
Table 4.
Some of the most used datasets per skin detection.
| Name (Abbr.) | Ref. | Images | Ground Truth | Download | Year |
|---|---|---|---|---|---|
| Compaq (CMQ) | [39] | 4675 | Semi-supervised | currently not available | 2002 |
| TDSD | [93] | 555 | Imprecise | http://lbmedia.ece.ucsb.edu/research/skin/skin.htm (accessed on 26 November 2022) | 2004 |
| UChile (UC) | [94] | 103 | Medium Precision | http://agami.die.uchile.cl/skindiff/ (accessed on 26 November 2022) | 2004 |
| ECU | [95] | 4000 | Precise | http://www.uow.edu.au/~phung/download.html (currently not available) (accessed on 26 November 2022) | 2005 |
| VT-AAST (VT) | [102] | 66 | Precise | ask to the authors | 2007 |
| Schmugge (SCH) | [96] | 845 | Precise (3 classes) | https://www.researchgate.net/publication/257620282_skin_image_Data_set_with_ground_truth (accessed on 26 November 2022) | 2007 |
| Feeval | [15] | 8991 | Low quality, imprecise | http://www.feeval.org/Data-sets/Skin_Colors.html (accessed on 26 November 2022) | 2009 |
| MCG | [97] | 1000 | Imprecise | http://mcg.ict.ac.cn/result_data_02mcg_skin.html (ask the authors) (accessed on 26 November 2022) | 2011 |
| Pratheepan (PRAT) | [100] | 78 | Precise | http://web.fsktm.um.edu.my/~cschan/downloads_skin_dataset.html (accessed on 26 November 2022) | 2012 |
| VDM | [98] | 285 | Precise | http://www-vpu.eps.uam.es/publications/SkinDetDM/ (accessed on 26 November 2022) | 2013 |
| SFA | [99] | 1118 | Medium Precision | http://www1.sel.eesc.usp.br/sfa/ (accessed on 26 November 2022) | 2013 |
| HGR | [44,58] | 1558 | Precise | http://sun.aei.polsl.pl/~mkawulok/gestures/ (accessed on 26 November 2022) | 2014 |
| SDD | [101] | 21,000 | Precise | Not available | 2015 |
| Abdominal Skin Dataset | [18] | 1400 | Precise | https://github.com/MRE-Lab-UMD/abd-skin-segmentation (accessed on 26 November 2022) | 2019 |
4.3. Hand-Detection Evaluation: Datasets
Similar to the skin-detection task, we adopted some well-known color-image datasets equipped with ground truth for hand detection. Notice that we do not want to review the datasets of hand segmentation; instead we chose two known ones to show the strength of the proposed ensemble. In Table 5, two datasets are summarized, and, in this section, a brief description of each of them is given.

Table 5.
Some of the most used datasets per hand detection.
- EgoYouTubeHands (EYTH) [70] dataset: It comprehends images extracted from YouTube videos. Specifically, authors downloaded three videos with an egocentric point of view and annotated one frame every five frames. The user in the video interacts with other people and performs several activities. The dataset has 1290 frames with hand annotation at the pixel level, where the environment, number of participants, hand sizes, and other factors vary among different images.
- GeorgiaTech Egocentric Activity dataset (GTEA) [103]: The dataset contains images from videos about four different subjects performing seven daily activities. Originally, the dataset was built for activity recognition in the same environment. The original dataset has 663 images with pixel-level hand annotations, considering hand till arm. Arms have been removed for a fair training, as already achieved in previous works (e.g., [70]).
It is important to notice that the use of the GTEA dataset is far from homogeneous in the literature, and this creates several issues in the comparison of the results among different studies. For instance, some research studies do not remove arms in the training phase. This makes the task a skin-segmentation task in which the performance is higher, but that should not be compared with results about hand segmentation. We emphasize the importance of a single standard protocol for these cases that should be adopted by all those proposing a solution for this problem.
5. Experimental Results
We performed an empirical evaluation to assess the performance of our proposal compared with the state-of-the-art models. We adopted the same methods for both skin and hand segmentation.
The performance of classifiers is affected by the amount of data used for the training phase, and ensembles are no exception. In this work, we employed DA1 and DA2 (see Section 3.3) on the training set and maintained the test sets as they are. Notice that, for skin segmentation only, the first 2000 images of ECU are used as the training set, and the other images of ECU make up one of the test sets used for assessing the performance.
HardNet-MSEG is trained with two different optimizers, stochastic gradient descent (SGD), denoted as H_S; and Adam, denoted as H_A. The ensemble FH is the fusion of HarDNet-MSEG trained with both the optimizers. PVT and HSN are trained by using the AdamW optimizer (as suggested in their original papers). The loss function for HarDNet-MSEG, HSN, and PVT is the same as the one in the original papers (structure Loss).
- PVT(2), sum rule between PVT combined with DA1 and PVT combined with DA2;
- HSN(2) is similar to PVT(2), i.e., sum rule between one HSN combined with DA1 and one HSN combined with DA2;
- FH(2), sum rule among two H_S (one combined with DA1, the latter with DA2) and two H_A (one combined with DA1, the latter with DA2);
- FH(4) computes FH(2) twice, and the output is aggregated by using the sum rule.
- FH(2) + 2 × PVT(2), weighted sum rule between PVT(2) and FH(2); the weight of PVT(2) is assigned so that its importance in the ensemble is the same of FH(2) (notice that FH(2) consists of four networks, while PVT(2) is built by only two networks).
- FH(4) + 4 × PVT(2), weighted sum rule between PVT(2) and FH(4); the weight of PVT(2) is assigned so that its importance in the ensemble is the same of FH(4).
- AllM = ELossMix2(10) + (10/4) × FH(2) + (10/2) × PVT(2), weighted sum rule among ElossMix2(10), FH(2), and PVT(2); as in the previous ensemble, the weights are assigned so that each ensemble member has the same importance. ELossMix2(10) is an ensemble, combined by sum rule, of ten stand-alone DeepLabV3+ segmentators with Resnet101 backbone (pretrained as detailed before using VOC); the ten networks are obtained by coupling five loss, vix.: LDiceBES, Comb1, Comb2, and Comb3 (see Table 2 for loss definitions) one time, using DA1, and another time, using DA2.
- AllM_H = ELossMix2(10) + (10/4) × FH(2) + (10/2) × PVT(2) + (10/2) × HSN(2), similar to the previous one but with the add-on of HSN(2).
5.1. Skin Segmentation
Due to the lack of a common evaluation standard, it is very difficult to compare different approaches fairly. Most published works are tested on self-collected datasets, which are frequently unavailable for further comparison. In many cases, the testing protocol is not clearly explained; many datasets are of low quality; and the accuracy of the ground truth is in doubt because lips, mouths, rings, and bracelets have occasionally been mistakenly classified as skin. Table 6 reports the performance of the different models on 10 different datasets collected for benchmarking purposes; in the last column, the average Dice is reported.

Table 6.
Performance (Dice) of different approaches in 10 datasets for skin detection. The bold represents the best performance.
From Table 6, it is clear that combining different topologies boosts the performance: the best average result is obtained by AllM_H, which combines transformers (i.e., PVT and HSN) with CNN-based models (i.e., HardNet/DeepLabV3+).
It is interesting to observe the behavior of ensembles with PVT: the PVT with DA1 ensemble obtained a higher performance on the UC dataset than its counterpart, PVT with DA2; the opposite happened on the CMQ dataset, where the PVT with DA2 ensemble obtained a higher performance than its counterpart, PVT with DA1. Meanwhile, the fusion of these two PVTs performs as the best of the two approaches on both situations.
We present a comparison of our methods with some previously proposed methods in the literature in Table 7: this is helpful for illustrating how performance changes over time. Be aware that, here, we report results only form a subset of the datasets previously considered in Table 6, because some datasets were not tested in previous works based on handcrafted methods. Table 7 shows that the adoption of deep learning in this domain is primarily responsible for the significant improvement in performance; approaches from 2002 and 2014 give results that are comparable.

Table 7.
Comparison with the literature. The bold represents the best performance.
5.2. Hand Segmentation
In this section, we report the results from the empirical analysis performed for the hand-segmentation task. We also provide an ablation study that shows the importance of adopting an ensemble based on DeepLabV3+; this ablation study, for the skin segmentation, was already reported in [60].
Each ensemble is made up of N models (N = 1 denotes a stand-alone model) which differ only for the randomization in the training process. We employed the standard Dice Loss for all the methods. As a standard metric adopted in the literature to evaluate the different models, in Table 8, we report the resulting IoU. In particular, we tested the following approaches:

Table 8.
Performance (IoU) of the proposed ensembles in the five benchmark datasets; the last column, AVG, reports the average performance. We report the resulting IoU because this is the standard metric adopted to evaluate the different models. The bold represents the best performance.
- RN18 a stand-alone DeepLabV3+ segmentators with backbone Resnet18 (pretrained in ImageNet);
- ERN18(N) is an ensemble of N RN18 networks (pretrained in ImageNet);
- RN50 a stand-alone DeepLabV3+ segmentators with backbone Resnet50 (pretrained in ImageNet);
- ERN50(N) is an ensemble of N RN50 networks;
- RN101 a stand-alone DeepLabV3+ segmentators with backbone Resnet101 (pretrained as detailed in before using VOC);
- ERN101(N) is an ensemble of N RN101 networks.
It is possible to notice from the results that the ensembles are performing well but not surprisingly. In this set of experiments, ERN101 is the best model.
In Table 9, the performances of RN101, with different loss functions, are reported and compared with the Dice Loss as the baseline and DA1 as the data-augmentation method. The following methods are reported (see Table 2 for loss definitions):

Table 9.
Performance of RN101, with different loss functions. The bold represents the best performance.
- ELoss101(10) is an ensemble, combined by sum rule, of 10 RN101, each coupled with data-augmentation DA1 and a given loss function; the final fusion is given by 2 × + 2 × + 2 × Comb1 + 2 × Comb2 + 2 × Comb3, where, with 2 × , we mean two different RN101 trained by using the loss function.
- ELossMix(10) is an ensemble that is similar to the previous one, but here data augmentation is used to increase diversity: the networks coupled with the loss used in ELoss101(10) ( , Comb1, Comb2, and Comb3) are trained one time, using DA1, and another time, using DA2 (i.e., 5 networks each trained two times, so we have an ensemble of 10 networks);
- ELossMix2(10) is similar to the previous ensemble, but it used LDiceBES instead of .
In Table 10, the previous ensembles are compared with the different models considered in Table 6 for the skin-detection problem. It can be noticed from the results that ELossMix2(10) obtained better results than HardNet, HSN, and PVT. The ensemble is the best trade-off, considering both skin and hand segmentation.

Table 10.
Performance of different models on the two datasets. The bold represents the best performance.
We also compared our models with some baselines (see Table 11). In particular, we noticed the following:

Table 11.
Performance comparison with state-of-the-art.
- Some approaches adopt ad hoc pretraining for hand segmentation, so the performance improves, but it becomes difficult to tell whether the improvement is related to model choice or better pretraining;
- Others use additional training images, making performance comparison unfair.
The proposed ensemble approximates the state-of-the-art, without optimizing the model or performing any domain-specific tuning for hand segmentation. Comparisons among different methods in this case is not easy. As already mentioned before, many methods have higher performance because during the pretraining phase they do not omit other parts of the body (e.g., arms or head) or they add different images during the training phase, making the comparison among performance unfair. For example, [74] reports an IoU of 0.848 without external training data and 0.880 adding examples to the original training data; moreover, in [74] for GTEA dataset also the skin of forearms is considered as foreground. In [76], their method is pretrained using PASCAL person parts (more suited for this specific task); even in [104], for GTEA dataset also the skin of forearms is considered as foreground.
6. Conclusions and Future Research Directions
In this paper, we proposed a new ensemble for combining different skin-detector approaches, a testing protocol for fair evaluation of handcrafted and deep-learned methods, and a comprehensive comparison of different approaches performed on several different datasets. We reviewed the latest available approaches, trained and tested four popular deep-learning models for data segmentation on this classification problem, and proposed a new ensemble that obtains state-of-the-art performance for skin segmentation.
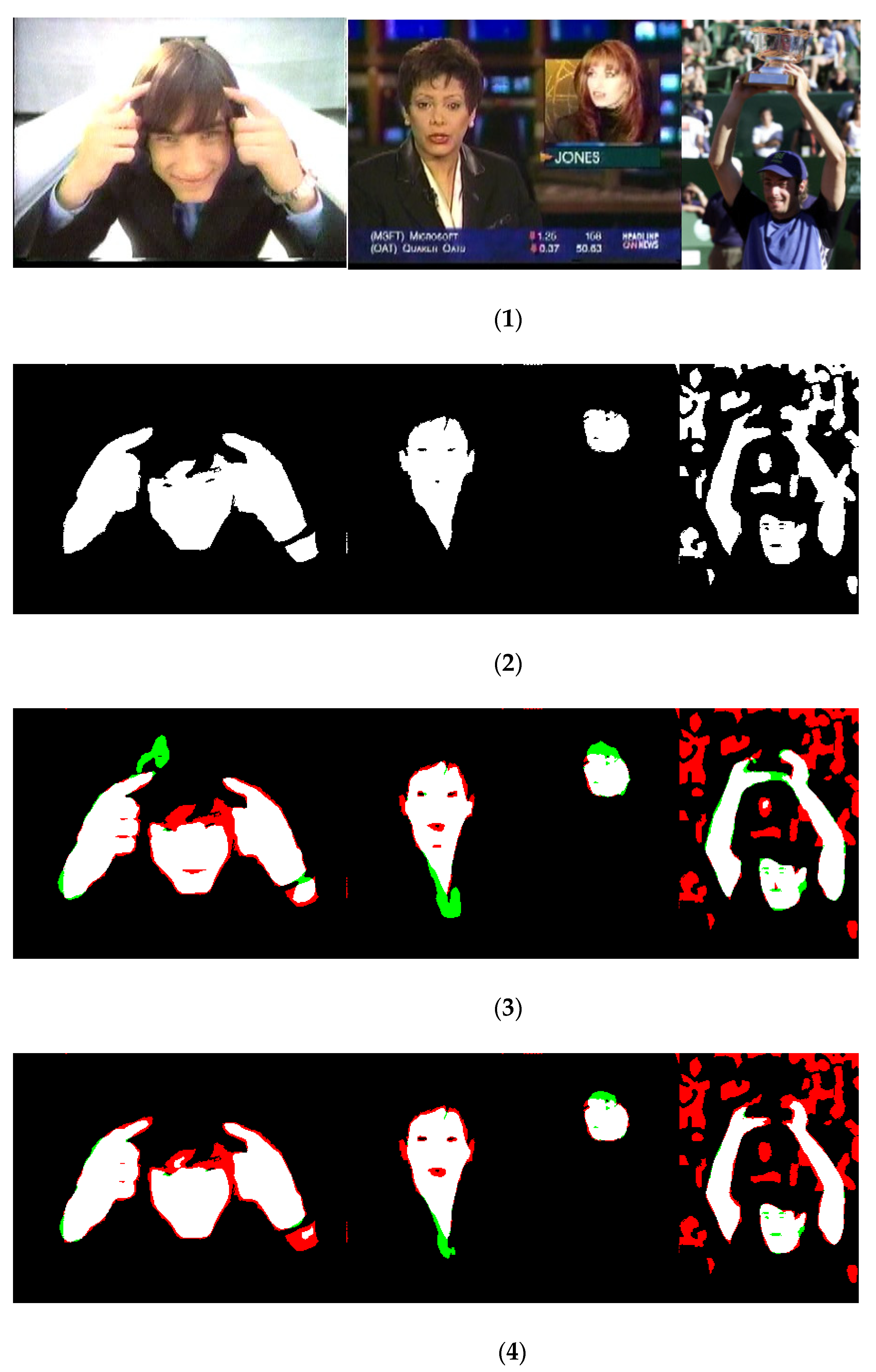
Empirical evidence indicates that CNNs/transformers work very well for skin segmentation and outperform all previous methods based on hand-crafted approaches: our extensive experiments carried out in several different datasets clearly demonstrate the supremacy of these deep-learned approaches. Furthermore, the proposed ensemble performs very well compared to other previous approaches. Some inference masks are shown in Figure 2: they demonstrate that our ensemble model produces better boundary results and makes more accurate predictions with respect to the best stand-alone model.

Figure 2.
Inference results on the UV dataset; each line contains (1) original images, (2) ground truth, (3) result from PVT_DA2 (i.e., the best stand-alone approach), and (4) AllM_H (the best ensemble). False-positive pixels are in green, while the false negatives are in red.
In conclusion, we showed that skin detection is a very difficult problem that cannot be solved by individual methods. The performance of many skin-detection methods depends on the color space used, the parameters used, the nature of the data, the characteristics of the image, the shape of the distribution, the size of the training sample, the presence of data noise, etc. New methods based on deep learning are less affected by these problems.
The advent of deep learning has led to the rapid development of image segmentation, with new models introduced in recent years [76]. These new models require a lot of data with respect to traditional computer vision techniques. Therefore, it is recommended to collect and label large datasets with people from different regions of the world for future research.
Moreover, further research is needed to develop lightweight architectures that can run on resource-constrained hardware without compromising performance.
Author Contributions
Conceptualization, L.N. and A.L. (Andrea Loreggia); software, A.L. (Alessandra Lumini) and A.D.; writing—review and editing, all the authors. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Links are provided in the paper.
Acknowledgments
We would like to acknowledge the support that NVIDIA provided us through the GPU Grant Program. We used a donated TitanX GPU to train CNNs used in this work.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Lumini, A.; Nanni, L. Fair comparison of skin detection approaches on publicly available datasets. Expert Syst. Appl. 2020, 160, 113677. [Google Scholar] [CrossRef]
- Han, S.S.; Park, I.; Eun Chang, S.; Lim, W.; Kim, M.S.; Park, G.H.; Chae, J.B.; Huh, C.H.; Na, J.I. Augmented Intelligence Dermatology: Deep Neural Networks Empower Medical Professionals in Diagnosing Skin Cancer and Predicting Treatment Options for 134 Skin Disorders. J. Investig. Dermatol. 2020, 140, 1753–1761. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.R.H.; Pavlova, M.; Famouri, M.; Wong, A. Cancer-Net SCa: Tailored deep neural network designs for detection of skin cancer from dermoscopy images. BMC Med. Imaging 2022, 22, 143. [Google Scholar] [CrossRef] [PubMed]
- Maniraju, M.; Adithya, R.; Srilekha, G. Recognition of Type of Skin Disease Using CNN. In Proceedings of the 2022 First International Conference on Artificial Intelligence Trends and Pattern Recognition (ICAITPR), Hyderabad, India, 10–12 March 2022; pp. 1–4. [Google Scholar]
- Zhao, M.; Kawahara, J.; Abhishek, K.; Shamanian, S.; Hamarneh, G. Skin3D: Detection and longitudinal tracking of pigmented skin lesions in 3D total-body textured meshes. Med. Image Anal. 2022, 77, 102329. [Google Scholar] [CrossRef]
- Li, X.; Desrosiers, C.; Liu, X. Deep Neural Forest for Out-of-Distribution Detection of Skin Lesion Images. IEEE J. Biomed. Health Inform. 2022, 27, 157–165. [Google Scholar] [CrossRef] [PubMed]
- Pfeifer, L.M.; Valdenegro-Toro, M. Automatic Detection and Classification of Tick-borne Skin Lesions using Deep Learning. arXiv 2020. [Google Scholar] [CrossRef]
- Hsu, R.L.; Abdel-Mottaleb, M.; Jain, A.K. Face detection in color images. IEEE Trans. Pattern Anal. Mach. Intell. 2002, 24, 696–706. [Google Scholar]
- Argyros, A.A.; Lourakis, M.I.A. Real-time tracking of multiple skin-colored objects with a possibly moving camera. In Computer Vision—ECCV 2004; Springer: Berlin/Heidelberg, Germany, 2004. [Google Scholar]
- Roy, K.; Mohanty, A.; Sahay, R.R. Deep Learning Based Hand Detection in Cluttered Environment Using Skin Segmentation. In Proceedings of the 2017 IEEE International Conference on Computer Vision Workshops (ICCVW), Venice, Italy, 22–29 October 2017; pp. 640–649. [Google Scholar]
- Sang, H.; Ma, Y.; Huang, J. Robust Palmprint Recognition Base on Touch-Less Color Palmprint Images Acquired. J. Signal Inf. Process. 2013, 4, 134–139. [Google Scholar] [CrossRef]
- De-La-Torre, M.; Granger, E.; Radtke, P.V.W.; Sabourin, R.; Gorodnichy, D.O. Partially-supervised learning from facial trajectories for face recognition in video surveillance. Inf. Fusion 2015, 24, 31–53. [Google Scholar] [CrossRef]
- Lee, J.-S.; Kuo, Y.-M.; Chung, P.-C.; Chen, E.-L. Naked image detection based on adaptive and extensible skin color model. Pattern Recognit. 2007, 40, 2261–2270. [Google Scholar] [CrossRef]
- Han, J.; Award, G.M.; Sutherland, A.; Wu, H. Automatic skin segmentation for gesture recognition combining region and support vector machine active learning. In Proceedings of the 7th International Conference on Automatic Face and Gesture Recognition, Southampton, UK, 10–12 April 2006; pp. 237–242. [Google Scholar]
- Stöttinger, J.; Hanbury, A.; Liensberger, C.; Khan, R. Skin paths for contextual flagging adult videos. In Proceedings of the Lecture Notes in Computer Science (Including Subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics), Las Vegas, NV, USA, 30 November–2 December 2009; Volume 5876 LNCS, pp. 303–314. [Google Scholar]
- Kong, S.G.; Heo, J.; Abidi, B.R.; Paik, J.; Abidi, M.A. Recent advances in visual and infrared face recognition-A review. Comput. Vis. Image Underst. 2005, 97, 103–135. [Google Scholar] [CrossRef]
- Healey, G.; Prasad, M.; Tromberg, B. Face recognition in hyperspectral images. IEEE Trans. Pattern Anal. Mach. Intell. 2003, 25, 1552–1560. [Google Scholar]
- Topiwala, A.; Al-Zogbi, L.; Fleiter, T.; Krieger, A. Adaptation and Evaluation of Deep Learning Techniques for Skin Segmentation on Novel Abdominal Dataset. In Proceedings of the 2019 IEEE 19th International Conference on Bioinformatics and Bioengineering (BIBE), Athens, Greece, 28–30 October 2019; pp. 752–759. [Google Scholar]
- Tsai, T.H.; Huang, S.A. Refined U-net: A new semantic technique on hand segmentation. Neurocomputing 2022, 495, 1–10. [Google Scholar] [CrossRef]
- Goceri, E. Automated Skin Cancer Detection: Where We Are and The Way to The Future. In Proceedings of the 2021 44th International Conference on Telecommunications and Signal Processing (TSP), Online, 26–28 July 2021; pp. 48–51. [Google Scholar]
- Rawat, V.; Singh, D.P.; Singh, N.; Kumar, P.; Goyal, T. A Comparative Study of various Skin Cancer using Deep Learning Techniques. In Proceedings of the 2022 International Conference on Computational Intelligence and Sustainable Engineering Solutions (CISES), Greater Noida, India, 20–21 May 2022; pp. 505–511. [Google Scholar]
- Afroz, A.; Zia, R.; Garcia, A.O.; Khan, M.U.; Jilani, U.; Ahmed, K.M. Skin lesion classification using machine learning approach: A survey. In Proceedings of the 2022 Global Conference on Wireless and Optical Technologies (GCWOT), Malaga, Spain, 14–17 February 2022; pp. 1–8. [Google Scholar]
- Jones, O.T.; Matin, R.N.; van der Schaar, M.; Prathivadi Bhayankaram, K.; Ranmuthu, C.K.I.; Islam, M.S.; Behiyat, D.; Boscott, R.; Calanzani, N.; Emery, J.; et al. Artificial intelligence and machine learning algorithms for early detection of skin cancer in community and primary care settings: A systematic review. Lancet Digit. Health 2022, 4, e466–e476. [Google Scholar] [CrossRef] [PubMed]
- Wen, D.; Khan, S.M.; Xu, A.J.; Ibrahim, H.; Smith, L.; Caballero, J.; Zepeda, L.; de Blas Perez, C.; Denniston, A.K.; Liu, X.; et al. Characteristics of publicly available skin cancer image datasets: A systematic review. Lancet Digit. Health 2022, 4, e64–e74. [Google Scholar] [CrossRef]
- Kakumanu, P.; Makrogiannis, S.; Bourbakis, N. A survey of skin-color modeling and detection methods. Pattern Recognit. 2007, 40, 1106–1122. [Google Scholar] [CrossRef]
- Zarit, B.D.; Super, B.J.; Quek, F.K.H. Comparison of five color models in skin pixel classification. In Proceedings of the Proceedings International Workshop on Recognition, Analysis, and Tracking of Faces and Gestures in Real-Time Systems. In Conjunction with ICCV’99 (Cat. No.PR00378), Corfu, Greece, 26–27 September 1999; pp. 58–63. [Google Scholar]
- Ibrahim, N.B.; Selim, M.M.; Zayed, H.H. A Dynamic Skin Detector Based on Face Skin Tone Color. In Proceedings of the 8th International Conference on In Informatics and Systems (INFOS), Giza, Egypt, 14–16 May 2012; pp. 1–5. [Google Scholar]
- Naji, S.; Jalab, H.A.; Kareem, S.A. A survey on skin detection in colored images. Artif. Intell. Rev. 2018, 52, 1041–1087. [Google Scholar] [CrossRef]
- Xu, H.; Sarkar, A.; Abbott, A.L. Color Invariant Skin Segmentation. In Proceedings of the 2022 IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops (CVPRW), New Orleans, LA, USA, 21–24 June 2022; pp. 2906–2915. [Google Scholar]
- Nazari, K.; Mazaheri, S.; Bigham, B.S. Creating A New Color Space utilizing PSO and FCM to Perform Skin Detection by using Neural Network and ANFIS. arXiv 2021. [Google Scholar] [CrossRef]
- Chen, W.C.; Wang, M.S. Region-based and content adaptive skin detection in color images. Int. J. Pattern Recognit. Artif. Intell. 2007, 21, 831–853. [Google Scholar] [CrossRef]
- Poudel, R.P.K.; Zhang, J.J.; Liu, D.; Nait-Charif, H. Skin Color Detection Using Region-Based Approach. Int. J. Image Process. 2013, 7, 385. [Google Scholar]
- Kruppa, H.; Bauer, M.A.; Schiele, B. Skin Patch Detection in Real-World Images. In Proceedings of the Annual Symposium for Pattern Recognition of the DAGM, Zurich, Switzerland, 16–18 September 2002; p. 109f. [Google Scholar]
- Sebe, N.; Cohen, I.; Huang, T.S.; Gevers, T. Skin detection: A Bayesian network approach. In Proceedings of the 17th International Conference on Pattern Recognition, Cambridge, UK, 26–26 August 2004; Volume 2, pp. 2–5. [Google Scholar]
- Kim, Y.; Hwang, I.; Cho, N.I. Convolutional neural networks and training strategies for skin detection. In Proceedings of the IEEE International Conference on Image Processing (ICIP), Beijing, China, 17–20 September 2017; pp. 3919–3923. [Google Scholar]
- Zuo, H.; Fan, H.; Blasch, E.; Ling, H. Combining Convolutional and Recurrent Neural Networks for Human Skin Detection. IEEE Signal Process. Lett. 2017, 24, 289–293. [Google Scholar] [CrossRef]
- Kumar, A.; Malhotra, S. Pixel-Based Skin Color Classifier: A Review. Int. J. Signal Process. Image Process. Pattern Recognit. 2015, 8, 283–290. [Google Scholar] [CrossRef]
- Mahmoodi, M.R.; Sayedi, S.M. A Comprehensive Survey on Human Skin Detection. Int. J. Image Graph. Signal Process. 2016, 8, 1–35. [Google Scholar] [CrossRef]
- Jones, M.J.; Rehg, J.M. Statistical color models with application to skin detection. Int. J. Comput. Vis. 2002, 46, 81–96. [Google Scholar] [CrossRef]
- Mahmoodi, M.R.; Sayedi, S.M. Leveraging spatial analysis on homogonous regions of color images for skin classification. In Proceedings of the 4th International Conference on Computer and Knowledge Engineering (ICCKE), Ferdowsi, Iran, 29–30 October 2014; pp. 209–214. [Google Scholar]
- Nidhu, R.; Thomas, M.G. Real Time Segmentation Algorithm for Complex Outdoor Conditions. Int. J. Sci. Technoledge 2014, 2, 71. [Google Scholar]
- Chen, L.; Zhou, J.; Liu, Z.; Chen, W.; Xiong, G. A skin detector based on neural network. In Proceedings of the Communications, Circuits and Systems and West Sino Expositions, Chengdu, China, 29 June–1 July 2002; Volume 1, pp. 615–619. [Google Scholar]
- Chen, Y.H.; Hu, K.T.; Ruan, S.J. Statistical skin color detection method without color transformation for real-time surveillance systems. Eng. Appl. Artif. Intell. 2012, 25, 1331–1337. [Google Scholar] [CrossRef]
- Kawulok, M.; Kawulok, J.; Nalepa, J. Spatial-based skin detection using discriminative skin-presence features. Pattern Recognit. Lett. 2014, 41, 3–13. [Google Scholar] [CrossRef]
- Jiang, Z.; Yao, M.; Jiang, W. Skin Detection Using Color, Texture and Space Information. In Proceedings of the Fourth International Conference on Fuzzy Systems and Knowledge Discovery, Hainan, China, 24–27 August 2007; pp. 366–370. [Google Scholar]
- Nunez, A.S.; Mendenhall, M.J. Detection of Human Skin in Near Infrared Hyperspectral Imagery. Int. Geosci. Remote Sens. Symp. 2008, 2, 621–624. [Google Scholar]
- Sandnes, F.E.; Neyse, L.; Huang, Y.-P. Simple and practical skin detection with static RGB-color lookup tables: A visualization-based study. In Proceedings of the IEEE International Conference on Systems, Man, and Cybernetics (SMC), Budapest, Hungary, 9–12 October 2016; pp. 2370–2375. [Google Scholar]
- Song, W.; Wu, D.; Xi, Y.; Park, Y.W.; Cho, K. Motion-based skin region of interest detection with a real-time connected component labeling algorithm. Multimed. Tools Appl. 2016, 76, 11199–11214. [Google Scholar] [CrossRef]
- Jairath, S.; Bharadwaj, S.; Vatsa, M.; Singh, R. Adaptive Skin Color Model to Improve Video Face Detection. In Machine Intelligence and Signal Processing; Springer: Berlin/Heidelberg, Germany, 2016; pp. 131–142. [Google Scholar]
- Gupta, A.; Chaudhary, A. Robust skin segmentation using color space switching. Pattern Recognit. Image Anal. 2016, 26, 61–68. [Google Scholar] [CrossRef]
- Oghaz, M.M.; Maarof, M.A.; Zainal, A.; Rohani, M.F.; Yaghoubyan, S.H. A hybrid Color space for skin detection using genetic algorithm heuristic search and principal component analysis technique. PLoS ONE 2015, 10, e0134828. [Google Scholar]
- Xu, T.; Zhang, Z.; Wang, Y. Patch-wise skin segmentation of human body parts via deep neural networks. J. Electron. Imaging 2015, 24, 043009. [Google Scholar] [CrossRef]
- Ma, C.; Shih, H. Human Skin Segmentation Using Fully Convolutional Neural Networks. In Proceedings of the IEEE 7th Global Conference on Consumer Electronics (GCCE), Nara, Japan, 9–12 October 2018; pp. 168–170. [Google Scholar]
- Dourado, A.; Guth, F.; de Campos, T.E.; Li, W. Domain adaptation for holistic skin detection. arXiv 2019. [Google Scholar] [CrossRef]
- Conaire, C.Ó.; O’Connor, N.E.; Smeaton, A.F. Detector adaptation by maximising agreement between independent data sources. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, Minneapolis, MN, USA, 17–22 June 2007. [Google Scholar]
- Cheddad, A.; Condell, J.; Curran, K.; Mc Kevitt, P. A skin tone detection algorithm for an adaptive approach to steganography. Signal Process. 2009, 89, 2465–2478. [Google Scholar] [CrossRef]
- Kawulok, M. Fast propagation-based skin regions segmentation in color images. In Proceedings of the 10th IEEE International Conference and Workshops on Automatic Face and Gesture Recognition, Shanghai, China, 22–26 April 2013. [Google Scholar]
- Kawulok, M.; Kawulok, J.; Nalepa, J.; Smolka, B. Self-adaptive algorithm for segmenting skin regions. EURASIP J. Adv. Signal Process. 2014, 2014, 170. [Google Scholar] [CrossRef]
- Brancati, N.; De Pietro, G.; Frucci, M.; Gallo, L. Human skin detection through correlation rules between the YCb and YCr subspaces based on dynamic color clustering. Comput. Vis. Image Underst. 2017, 155, 33–42. [Google Scholar] [CrossRef]
- Nanni, L.; Lumini, A.; Loreggia, A.; Formaggio, A.; Cuza, D. An Empirical Study on Ensemble of Segmentation Approaches. Signals 2022, 3, 341–358. [Google Scholar] [CrossRef]
- Huang, C.-H.; Wu, H.-Y.; Lin, Y.-L. HarDNet-MSEG: A Simple Encoder-Decoder Polyp Segmentation Neural Network that Achieves over 0.9 Mean Dice and 86 FPS. arXiv 2021. [Google Scholar] [CrossRef]
- Dong, B.; Wang, W.; Li, J.; Fan, D.-P. Polyp-PVT: Polyp Segmentation with Pyramid Vision Transformers. arXiv 2021. [Google Scholar] [CrossRef]
- Farooq, M.A.; Azhar, M.A.M.; Raza, R.H. Automatic Lesion Detection System (ALDS) for Skin Cancer Classification Using SVM and Neural Classifiers. In Proceedings of the 2016 IEEE 16th International Conference on Bioinformatics and Bioengineering (BIBE), Taichung, Taiwan, 31 October–2 November 2016; pp. 301–308. [Google Scholar]
- He, X.; Lei, B.; Wang, T. SANet:Superpixel Attention Network for Skin Lesion Attributes Detection. arXiv 2019. [Google Scholar] [CrossRef]
- Arsalan, M.; Kim, D.S.; Owais, M.; Park, K.R. OR-Skip-Net: Outer residual skip network for skin segmentation in non-ideal situations. Expert Syst. Appl. 2020, 141, 112922. [Google Scholar] [CrossRef]
- Minhas, K.; Khan, T.M.; Arsalan, M.; Naqvi, S.S.; Ahmed, M.; Khan, H.A.; Haider, M.A.; Haseeb, A. Accurate Pixel-Wise Skin Segmentation Using Shallow Fully Convolutional Neural Network. IEEE Access 2020, 8, 156314–156327. [Google Scholar] [CrossRef]
- Zhang, K.; Wang, Y.; Li, W.; Li, C.; Lei, Z. Real-time adaptive skin detection using skin color model updating unit in videos. J. Real-Time Image Process. 2022, 19, 303–315. [Google Scholar] [CrossRef]
- Tarasiewicz, T.; Nalepa, J.; Kawulok, M. Skinny: A Lightweight U-Net for Skin Detection and Segmentation. In Proceedings of the 2020 IEEE International Conference on Image Processing (ICIP), Abu Dhabi, United Arab Emirates, 25–28 October 2020; pp. 2386–2390. [Google Scholar]
- Xie, Z.; Wang, S.; Zhao, W.; Guo, Z. A robust context attention network for human hand detection. Expert Syst. Appl. 2022, 208, 118132. [Google Scholar] [CrossRef]
- Khan, A.U.; Borji, A. Analysis of Hand Segmentation in the Wild. In Proceedings of the 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018; pp. 4710–4719. [Google Scholar]
- Shelhamer, E.; Long, J.; Darrell, T. Fully Convolutional Networks for Semantic Segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 640–651. [Google Scholar] [CrossRef]
- Badrinarayanan, V.; Kendall, A.; Cipolla, R. SegNet: A Deep Convolutional Encoder-Decoder Architecture for Image Segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 2481–2495. [Google Scholar] [CrossRef]
- Chen, L.C.; Papandreou, G.; Kokkinos, I.; Murphy, K.; Yuille, A.L. DeepLab: Semantic Image Segmentation with Deep Convolutional Nets, Atrous Convolution, and Fully Connected CRFs. IEEE Trans. Pattern Anal. Mach. Intell. 2018, 40, 834–848. [Google Scholar] [CrossRef]
- Chen, L.-C.; Papandreou, G.; Schroff, F.; Adam, H. Rethinking atrous convolution for semantic image segmentation. arXiv 2017, arXiv:1706.05587. [Google Scholar]
- Chen, L.C.; Zhu, Y.; Papandreou, G.; Schroff, F.; Adam, H. Encoder-decoder with atrous separable convolution for semantic image segmentation. In Proceedings of the Lecture Notes in Computer Science (Including Subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics), Munich, Germany, 8–14 September 2018, ISSN 1611-3349. [Google Scholar]
- Minaee, S.; Boykov, Y.Y.; Porikli, F.; Plaza, A.J.; Kehtarnavaz, N.; Terzopoulos, D. Image Segmentation Using Deep Learning: A Survey. IEEE Trans. Pattern Anal. Mach. Intell. 2021, 44, 3523–3542. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 26 June–1 July 2016; pp. 770–778. [Google Scholar]
- Khan, A.; Sohail, A.; Zahoora, U.; Qureshi, A.S. A survey of the recent architectures of deep convolutional neural networks. Artif. Intell. Rev. 2020, 53, 5455–5516. [Google Scholar] [CrossRef]
- Zhang, W.; Fu, C.; Zheng, Y.; Zhang, F.; Zhao, Y.; Sham, C.W. HSNet: A hybrid semantic network for polyp segmentation. Comput. Biol. Med. 2022, 150, 106173. [Google Scholar] [CrossRef] [PubMed]
- Jadon, S. A survey of loss functions for semantic segmentation. In Proceedings of the 2020 IEEE Conference on Computational Intelligence in Bioinformatics and Computational Biology, CIBCB 2020, Virtual, 27–29 October 2020; pp. 1–7. [Google Scholar]
- Salehi, S.S.M.; Erdogmus, D.; Gholipour, A. Tversky loss function for image segmentation using 3D fully convolutional deep networks. In Proceedings of the Lecture Notes in Computer Science (Including Subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics), Quebec City, QC, Canada, 10 September 2017; Volume 10541. [Google Scholar]
- Lin, T.-Y.; Goyal, P.; Girshick, R.; He, K.; Dollar, P. Focal Loss for Dense Object Detection. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 42, 2980–2988. [Google Scholar]
- Qin, X.; Zhang, Z.; Huang, C.; Gao, C.; Dehghan, M.; Jagersand, M. Basnet: Boundary-aware salient object detection. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 16–20 June 2019; pp. 7479–7489. [Google Scholar]
- Wang, Z.; Bovik, A.C.; Sheikh, H.R.; Simoncelli, E.P. Image quality assessment: From error visibility to structural similarity. IEEE Trans. Image Process. 2004, 13, 600–612. [Google Scholar] [CrossRef] [PubMed]
- Aurelio, Y.S.; de Almeida, G.M.; de Castro, C.L.; Braga, A.P. Learning from Imbalanced Data Sets with Weighted Cross-Entropy Function. Neural Process. Lett. 2019, 50, 1937–1949. [Google Scholar] [CrossRef]
- Rahman, M.A.; Wang, Y. Optimizing intersection-over-union in deep neural networks for image segmentation. In Proceedings of the Lecture Notes in Computer Science (Including Subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics), Las Vegas, NV, USA, 12–14 December 2016; Volume 10072 LNCS. [Google Scholar]
- Yang, D.; Roth, H.; Wang, X.; Xu, Z.; Myronenko, A.; Xu, D. Enhancing Foreground Boundaries for Medical Image Segmentation. arXiv 2020, arXiv:2005.14355. [Google Scholar]
- Chen, Z.; Zhou, H.; Lai, J.; Yang, L.; Xie, X. Contour-Aware Loss: Boundary-Aware Learning for Salient Object Segmentation. IEEE Trans. Image Process. 2021, 30, 431–443. [Google Scholar] [CrossRef]
- Nanni, L.; Cuza, D.; Lumini, A.; Loreggia, A.; Brahnam, S. Deep ensembles in bioimage segmentation. arXiv 2021, arXiv:2112.12955. [Google Scholar]
- Nanni, L.; Brahnam, S.; Paci, M.; Ghidoni, S. Comparison of Different Convolutional Neural Network Activation Functions and Methods for Building Ensembles for Small to Midsize Medical Data Sets. Sensors 2022, 22, 6129. [Google Scholar] [CrossRef]
- Nanni, L.; Cuza, D.; Lumini, A.; Loreggia, A.; Brahman, S. Polyp Segmentation with Deep Ensembles and Data Augmentation. In Artificial Intelligence and Machine Learning for Healthcare; Springer: Cham, Switzerland, 2023; pp. 133–153. [Google Scholar]
- Powers, D.M.W. Evaluation: From Precision, Recall and F-Measure To Roc, Informedness, Markedness & Correlation. J. Mach. Learn. Technol. 2011, 2, 37–63. [Google Scholar]
- Zhu, Q.; Wu, C.-T.; Cheng, K.; Wu, Y. An adaptive skin model and its application to objectionable image filtering. In Proceedings of the 12th Annual ACM International Conference on Multimedia, New York, NY, USA, 10–16 October 2004; p. 56. [Google Scholar]
- Ruiz-Del-Solar, J.; Verschae, R. Skin detection using neighborhood information. In Proceedings of the Sixth IEEE International Conference on Automatic Face and Gesture Recognition, Seoul, Republic of Korea, 17–19 May 2004; pp. 463–468. [Google Scholar]
- Phung, S.L.; Bouzerdoum, A.; Chai, D. Skin segmentation using color pixel classification: Analysis and comparison. IEEE Trans. Pattern Anal. Mach. Intell. 2005, 27, 148–154. [Google Scholar] [CrossRef] [PubMed]
- Abdallah, A.S.; El-Nasr, M.A.; Abbott, A.L. A new color image database for benchmarking of automatic face detection and human skin segmentation techniques. Int. J. Comput. Inf. Eng. 2007, 20, 353–357. [Google Scholar]
- Schmugge, S.J.; Jayaram, S.; Shin, M.C.; Tsap, L.V. Objective evaluation of approaches of skin detection using ROC analysis. Comput. Vis. Image Underst. 2007, 108, 41–51. [Google Scholar] [CrossRef]
- Huang, L.; Xia, T.; Zhang, Y.; Lin, S. Human skin detection in images by MSER analysis. In Proceedings of the 2011 18th IEEE International Conference on Image Processing, Brussels, Belgium, 11–14 September 2011; pp. 1257–1260. [Google Scholar]
- Sanmiguel, J.C.; Suja, S. Skin detection by dual maximization of detectors agreement for video monitoring. Pattern Recognit. Lett. 2013, 34, 2102–2109. [Google Scholar] [CrossRef]
- Casati, J.P.B.; Moraes, D.R.; Rodrigues, E.L.L. SFA: A human skin image database based on FERET and AR facial images. In Proceedings of the IX workshop de Visao Computational, Rio de Janeiro, Brazil, 3–5 June 2013. [Google Scholar]
- Tan, W.R.; Chan, C.S.; Yogarajah, P.; Condell, J. A Fusion Approach for Efficient Human Skin Detection. Ind. Inform. IEEE Trans. 2012, 8, 138–147. [Google Scholar] [CrossRef]
- Mahmoodi, M.R.; Sayedi, S.M.; Karimi, F.; Fahimi, Z.; Rezai, V.; Mannani, Z. SDD: A skin detection dataset for training and assessment of human skin classifiers. In Proceedings of the Knowledge-Based Engineering and Innovation (KBEI), Tehran, Iran, 5–6 November 2015; pp. 71–77. [Google Scholar]
- Li, Y.; Ye, Z.; Rehg, J.M. Delving Into Egocentric Actions. In Proceedings of the 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015; pp. 287–295. [Google Scholar]
- Wang, W.; Yu, K.; Hugonot, J.; Fua, P.; Salzmann, M. Recurrent U-Net for Resource-Constrained Segmentation. In Proceedings of the 2019 IEEE/CVF International Conference on Computer Vision (ICCV), Seoul, Republic of Korea, 27–28 October 2019; pp. 2142–2151. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).