Automatic Classification of Simulated Breast Tomosynthesis Whole Images for the Presence of Microcalcification Clusters Using Deep CNNs
Abstract
:1. Introduction
2. Materials and Methods
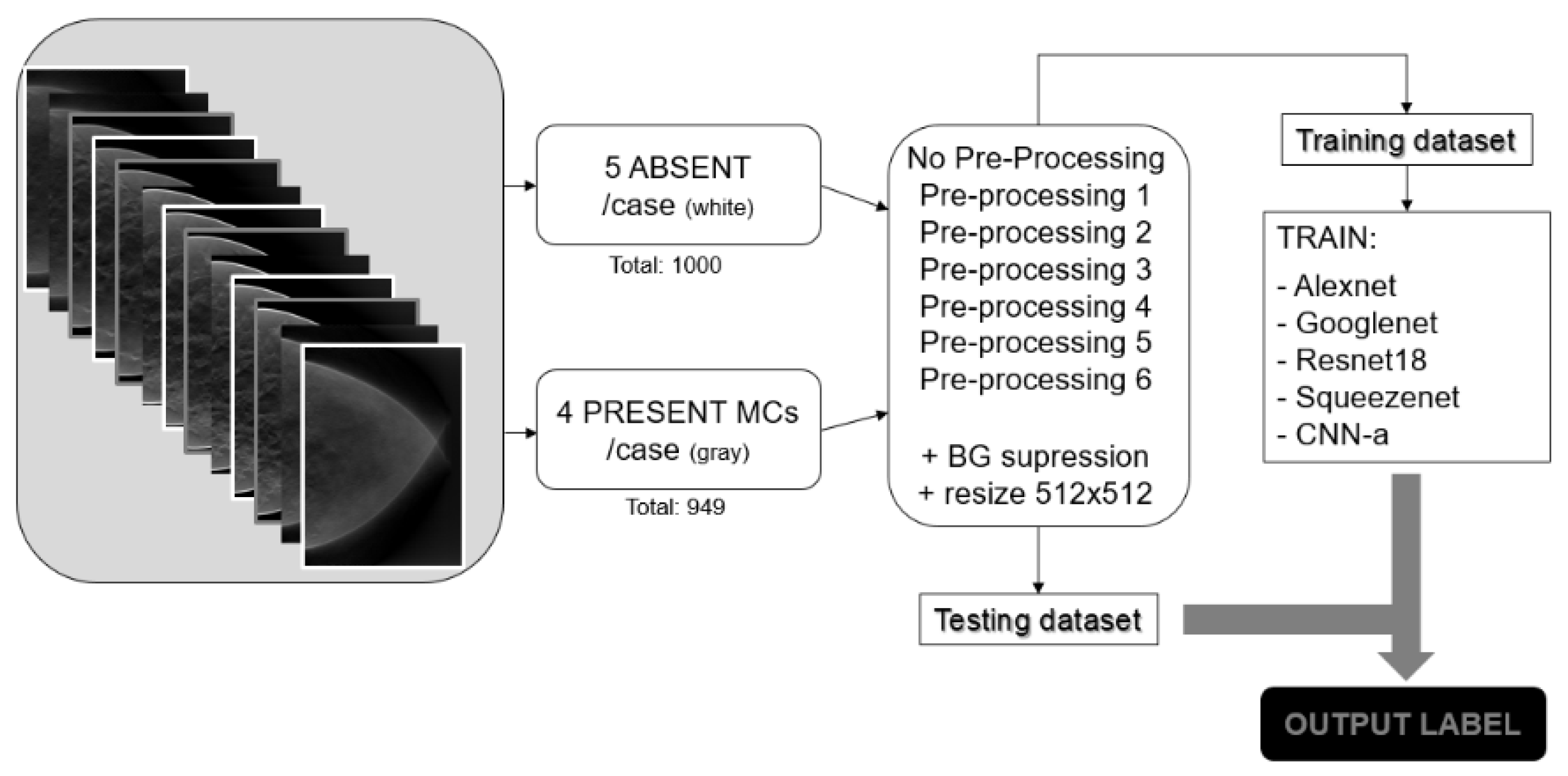
2.1. Database
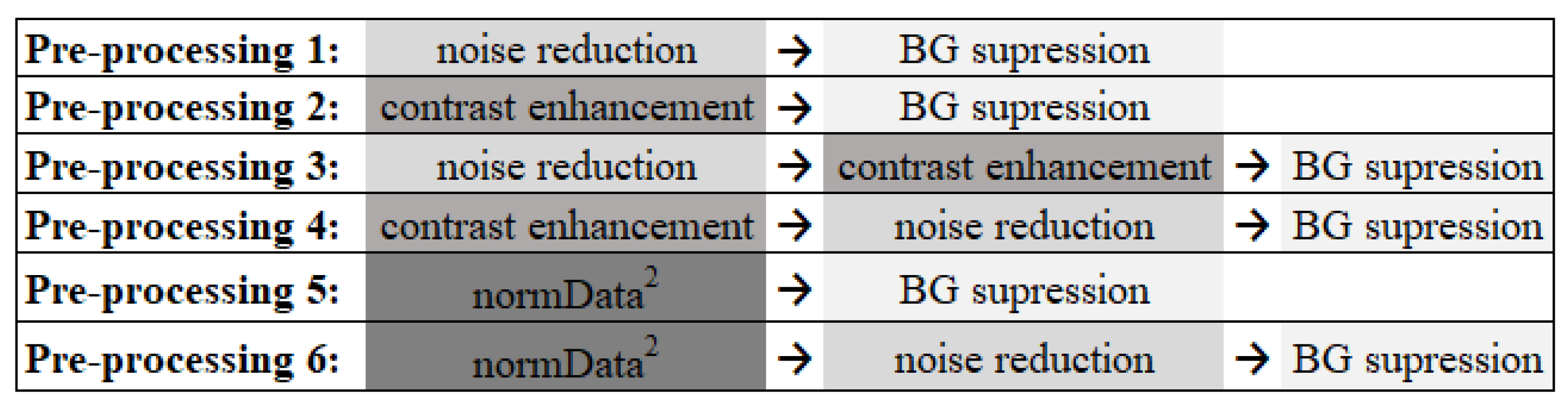
2.2. Data PreProcessing
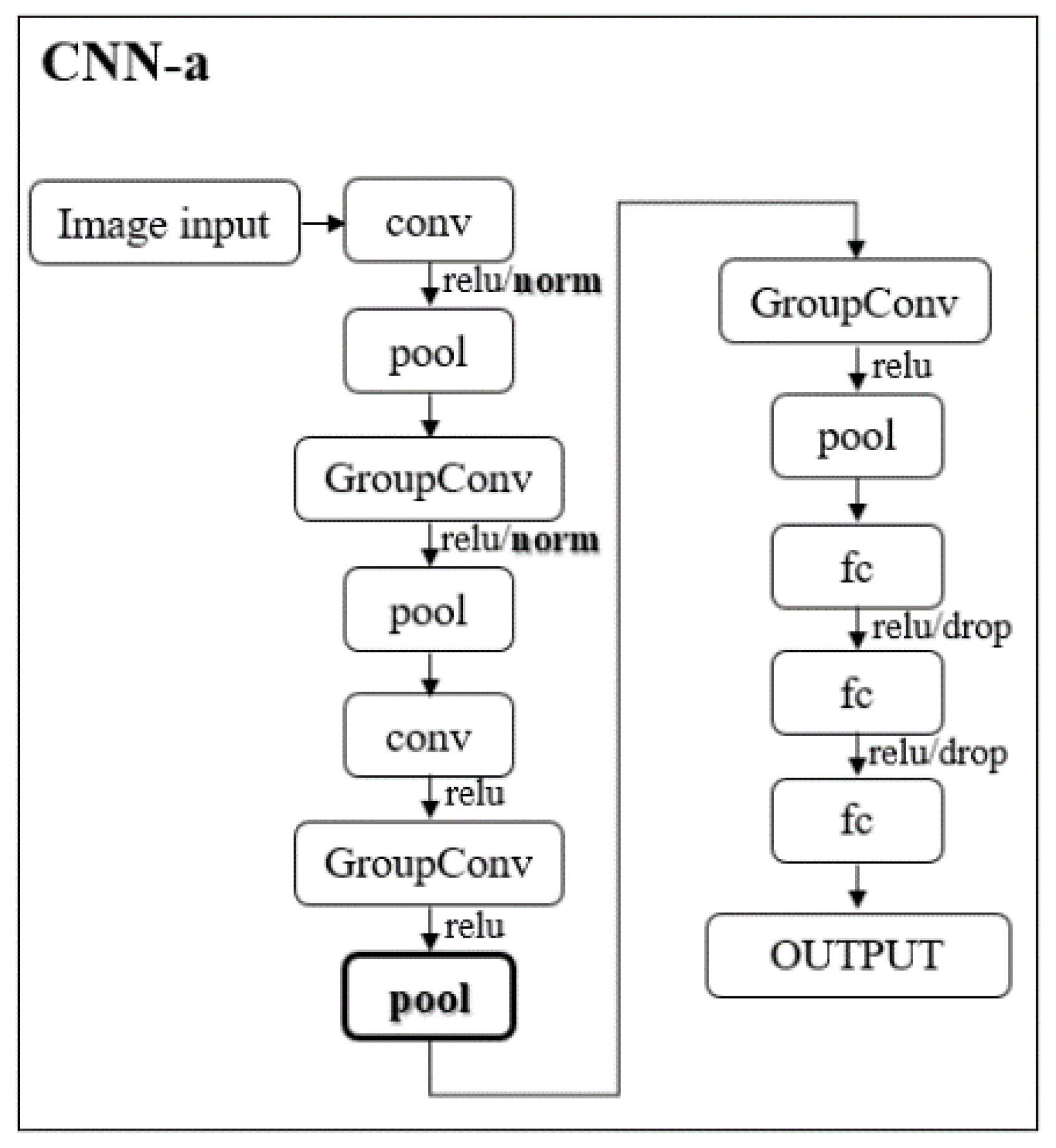
2.3. CNNs
2.4. Methodology Pipeline
2.5. Training Options
2.6. Evaluation Metrics
3. Results
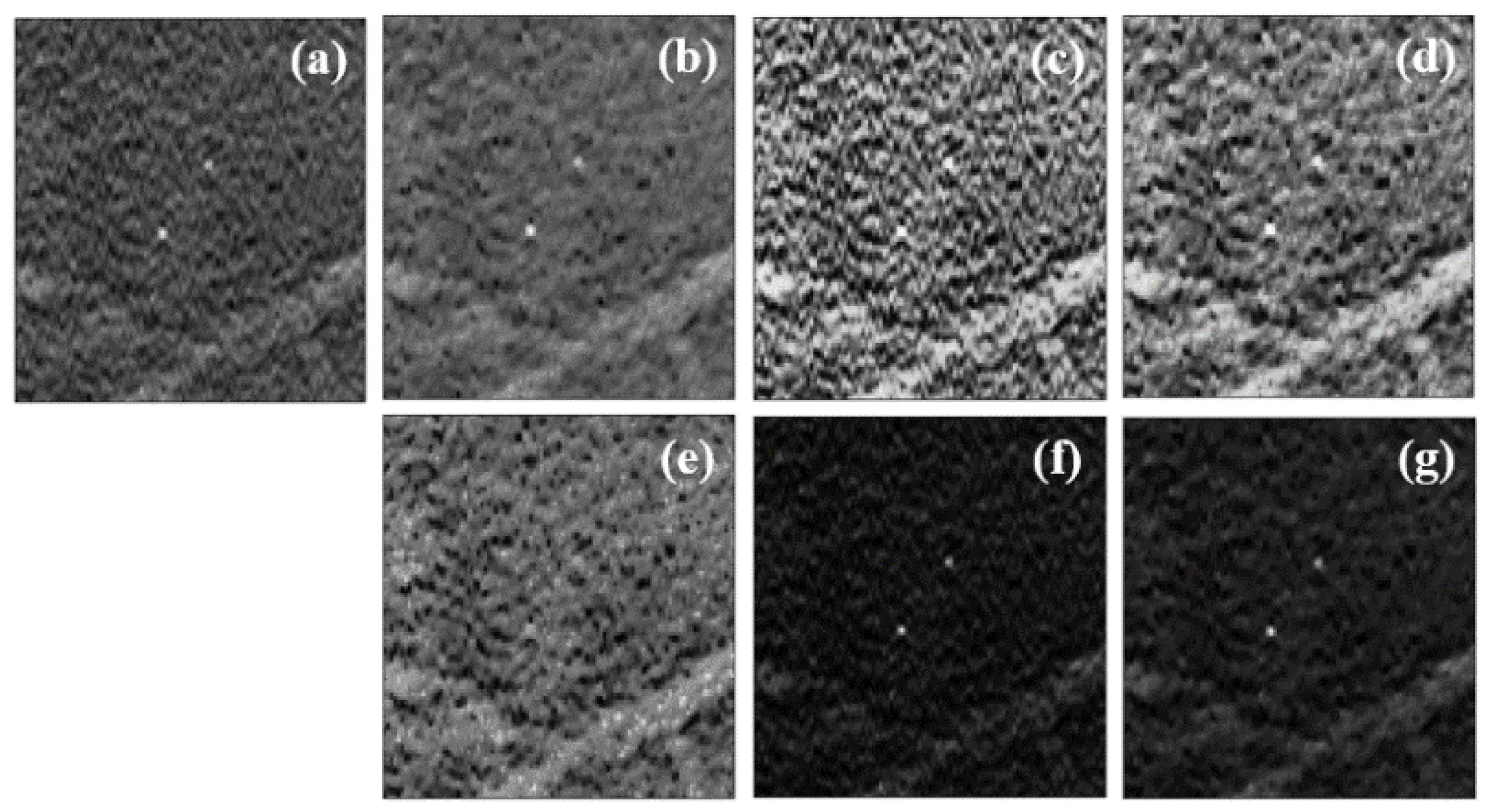
3.1. Data Preprocessing
3.2. Performance Analysis
3.3. Influence of Breast Density on Classification
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Tabár, L.; Dean, P.B.; Chen, T.H.H.; Yen, A.M.F.; Chen, S.L.S.; Fann, J.C.Y.; Chiu, S.Y.H.; Ku, M.M.S.; Wu, W.Y.Y.; Hsu, C.Y.; et al. The incidence of fatal breast cancer measures the increased effectiveness of therapy in women participating in mammography screening. Cancer 2019, 125, 515–523. [Google Scholar] [CrossRef] [PubMed]
- Skaane, P.; Sebuødegård, S.; Bandos, A.I.; Gur, D.; Østerås, B.H.; Gullien, R.; Hofvind, S. Performance of breast cancer screening using digital breast tomosynthesis: Results from the prospective population-based Oslo Tomosynthesis Screening Trial. Breast Cancer Res. Treat. 2018, 169, 489–496. [Google Scholar] [CrossRef] [PubMed]
- Ciatto, S.; Houssami, N.; Bernardi, D.; Caumo, F.; Pellegrini, M.; Brunelli, S.; Tuttobene, P.; Bricolo, P.; Fantò, C.; Valentini, M.; et al. Integration of 3D digital mammography with tomosynthesis for population breast-cancer screening (STORM): A prospective comparison study. Lancet Oncol. 2013, 14, 583–589. [Google Scholar] [CrossRef]
- Haas, B.M.; Kalra, V.; Geisel, J.; Raghu, M.; Durand, M.; Philpotts, L.E. Comparison of Tomosynthesis Plus Digital Mammography and Digital Mammography Alone for Breast Cancer Screening. Radiology 2013, 269, 694–700. [Google Scholar] [CrossRef]
- Rose, S.L.; Tidwell, A.L.; Bujnoch, L.J.; Kushwaha, A.C.; Nordmann, A.S.; Sexton Jr, R. Implementation of Breast Tomosynthesis in a Routine Screening Practice: An Observational Study. Am. J. Roentgenol. 2013, 200, 1401–1408. [Google Scholar] [CrossRef]
- Greenberg, J.S.; Javitt, M.C.; Katzen, J.; Michael, S.; Holland, A.E. Clinical Performance Metrics of 3D Digital Breast Tomosynthesis Compared With 2D Digital Mammography for Breast Cancer Screening in Community Practice. Am. J. Roentgenol. 2014, 203, 687–693. [Google Scholar] [CrossRef]
- McDonald, E.S.; Oustimov, A.; Weinstein, S.P.; Synnestvedt, M.B.; Schnall, M.; Conant, E.F. Effectiveness of Digital Breast Tomosynthesis Compared With Digital Mammography: Outcomes Analysis From 3 Years of Breast Cancer Screening. JAMA Oncol. 2016, 2, 737–743. [Google Scholar] [CrossRef]
- Zackrisson, S.; Lång, K.; Rosso, A.; Johnson, K.; Dustler, M.; Förnvik, D.; Förnvik, H.; Sartor, H.; Timberg, P.; Tingberg, A.; et al. One-view breast tomosynthesis versus two-view mammography in the Malmö Breast Tomosynthesis Screening Trial (MBTST): A prospective, population-based, diagnostic accuracy study. Lancet Oncol. 2018, 19, 1493–1503. [Google Scholar] [CrossRef]
- Bernardi, D.; Macaskill, P.; Pellegrini, M.; Valentini, M.; Fantò, C.; Ostillio, L.; Tuttobene, P.; Luparia, A.; Houssami, N. Breast cancer screening with tomosynthesis (3D mammography) with acquired or synthetic 2D mammography compared with 2D mammography alone (STORM-2): A population-based prospective study. Lancet Oncol. 2016, 17, 1105–1113. [Google Scholar] [CrossRef]
- Lång, K.; Andersson, I.; Rosso, A.; Tingberg, A.; Timberg, P.; Zackrisson, S. Performance of one-view breast tomosynthesis as a stand-alone breast cancer screening modality: Results from the Malmö Breast Tomosynthesis Screening Trial, a population-based study. Eur. Radiol. 2016, 26, 184–190. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, F.J.; Tucker, L.; Gillan, M.G.; Willsher, P.; Cooke, J.; Duncan, K.A.; Michell, M.J.; Dobson, H.M.; Lim, Y.Y.; Suaris, T.; et al. Accuracy of Digital Breast Tomosynthesis for Depicting Breast Cancer Subgroups in a UK Retrospective Reading Study (TOMMY Trial). Radiology 2015, 277, 697–706. [Google Scholar] [CrossRef] [PubMed]
- Hofvind, S.; Hovda, T.; Holen, Å.S.; Lee, C.I.; Albertsen, J.; Bjørndal, H.; Brandal, S.H.; Gullien, R.; Lømo, J.; Park, D.; et al. Digital Breast Tomosynthesis and Synthetic 2D Mammography versus Digital Mammography: Evaluation in a Population-based Screening Program. Radiology 2018, 287, 787–794. [Google Scholar] [CrossRef]
- Freer, P.E.; Riegert, J.; Eisenmenger, L.; Ose, D.; Winkler, N.; Stein, M.A.; Stoddard, G.J.; Hess, R. Clinical implementation of synthesized mammography with digital breast tomosynthesis in a routine clinical practice. Breast Cancer Res. Treat. 2017, 166, 501–509. [Google Scholar] [CrossRef]
- Skaane, P.; Bandos, A.I.; Gullien, R.; Eben, E.B.; Ekseth, U.; Haakenaasen, U.; Izadi, M.; Jebsen, I.N.; Jahr, G.; Krager, M.; et al. Comparison of Digital Mammography Alone and Digital Mammography Plus Tomosynthesis in a Population-based Screening Program. Radiology 2013, 267, 47–56. [Google Scholar] [CrossRef]
- Tagliafico, A.S.; Calabrese, M.; Bignotti, B.; Signori, A.; Fisci, E.; Rossi, F.; Valdora, F.; Houssami, N. Accuracy and reading time for six strategies using digital breast tomosynthesis in women with mammographically negative dense breasts. Eur. Radiol. 2017, 27, 5179–5184. [Google Scholar] [CrossRef]
- Balleyguier, C.; Arfi-Rouche, J.; Levy, L.; Toubiana, P.R.; Cohen-Scali, F.; Toledano, A.Y.; Boyer, B. Improving digital breast tomosynthesis reading time: A pilot multi-reader, multi-case study using concurrent Computer-Aided Detection (CAD). Eur. J. Radiol. 2017, 97, 83–89. [Google Scholar] [CrossRef]
- Benedikt, R.A.; Boatsman, J.E.; Swann, C.A.; Kirkpatrick, A.D.; Toledano, A.Y. Concurrent Computer-Aided Detection Improves Reading Time of Digital Breast Tomosynthesis and Maintains Interpretation Performance in a Multireader Multicase Study. Am. J. Roentgenol. 2017, 210, 685–694. [Google Scholar] [CrossRef]
- Chae, E.Y.; Kim, H.H.; Jeong, J.W.; Chae, S.H.; Lee, S.; Choi, Y.W. Decrease in interpretation time for both novice and experienced readers using a concurrent computer-aided detection system for digital breast tomosynthesis. Eur. Radiol. 2019, 29, 2518–2525. [Google Scholar] [CrossRef]
- Poplack, S.P.; Tosteson, T.D.; Kogel, C.A.; Nagy, H.M. Digital breast tomosynthesis: Initial experience in 98 women with abnormal digital screening mammography. AJR Am. J. Roentgenol. 2007, 189, 616–623. [Google Scholar] [CrossRef] [PubMed]
- Andersson, I.; Ikeda, D.M.; Zackrisson, S.; Ruschin, M.; Svahn, T.; Timberg, P.; Tingberg, A. Breast tomosynthesis and digital mammography: A comparison of breast cancer visibility and BIRADS classification in a population of cancers with subtle mammographic findings. Eur. Radiol. 2008, 18, 2817–2825. [Google Scholar] [CrossRef] [PubMed]
- Spangler, M.L.; Zuley, M.L.; Sumkin, J.H.; Abrams, G.; Ganott, M.A.; Hakim, C.; Perrin, R.; Chough, D.M.; Shah, R.; Gur, D. Detection and Classification of Calcifications on Digital Breast Tomosynthesis and 2D Digital Mammography: A Comparison. Am. J. Roentgenol. 2011, 196, 320–324. [Google Scholar] [CrossRef] [PubMed]
- Kopans, D.; Gavenonis, S.; Halpern, E.; Moore, R. Calcifications in the breast and digital breast tomosynthesis. Breast. J. 2011, 17, 638–644. [Google Scholar] [CrossRef] [PubMed]
- Svane, G.; Azavedo, E.; Lindman, K.; Urech, M.; Nilsson, J.; Weber, N.; Lindqvist, L.; Ullberg, C. Clinical experience of photon counting breast tomosynthesis: Comparison with traditional mammography. Acta Radiol. 2011, 52, 134–142. [Google Scholar] [CrossRef] [PubMed]
- Wallis, M.G.; Moa, E.; Zanca, F.; Leifland, K.; Danielsson, M. Two-View and Single-View Tomosynthesis versus Full-Field Digital Mammography: High-Resolution X-Ray Imaging Observer Study. Radiology 2012, 262, 788–796. [Google Scholar] [CrossRef]
- Nyante, S.J.; Lee, S.S.; Benefield, T.S.; Hoots, T.N.; Henderson, L.M. The association between mammographic calcifications and breast cancer prognostic factors in a population-based registry cohort. Cancer 2017, 123, 219–227. [Google Scholar] [CrossRef]
- D’Orsi, C.J. ACR BI-RADS Atlas: Breast Imaging Reporting and Data System; American College of Radiology: Reston, VA, USA, 2013. [Google Scholar]
- Samala, R.K.; Chan, H.P.; Lu, Y.; Hadjiiski, L.M.; Wei, J.; Helvie, M.A. Digital breast tomosynthesis: Computer-aided detection of clustered microcalcifications on planar projection images. Phys. Med. Biol. 2014, 59, 7457–7477. [Google Scholar] [CrossRef]
- Samala, R.K.; Chan, H.P.; Hadjiiski, L.M.; Helvie, M.A. Analysis of computer-aided detection techniques and signal characteristics for clustered microcalcifications on digital mammography and digital breast tomosynthesis. Phys. Med. Biol. 2016, 61, 7092–7112. [Google Scholar] [CrossRef]
- Park, S.C.; Zheng, B.; Wang, X.H.; Gur, D. Applying a 2D Based CAD Scheme for Detecting Micro-Calcification Clusters Using Digital Breast Tomosynthesis Images: An Assessment. In Medical Imaging 2008: Computer-Aided Diagnosis; Medical Imaging: San Diego, CA, USA, 2008; Volume 6915, pp. 70–77. [Google Scholar]
- Reiser, I.; Nishikawa, R.M.; Edwards, A.V.; Kopans, D.B.; Schmidt, R.A.; Papaioannou, J.; Moore, R.H. Automated detection of microcalcification clusters for digital breast tomosynthesis using projection data only: A preliminary study. Med. Phys. 2008, 35, 1486–1493. [Google Scholar] [CrossRef]
- Bernard, S.; Muller, S.; Onativia, J. Computer-Aided Microcalcification Detection on Digital Breast Tomosynthesis Data: A Preliminary Evaluation. In Digital Mammography: 9th International Workshop; Krupinski, E.A., Ed.; Springer: Berlin/Heidelberg, Germany, 2008; pp. 151–157. [Google Scholar]
- Sahiner, B.; Chan, H.P.; Hadjiiski, L.M.; Helvie, M.A.; Wei, J.; Zhou, C.; Lu, Y. Computer-aided detection of clustered microcalcifications in digital breast tomosynthesis: A 3D approach. Med. Phys. 2012, 39, 28–39. [Google Scholar] [CrossRef] [PubMed]
- Samala, R.K.; Chan, H.P.; Lu, Y.; Hadjiiski, L.; Wei, J.; Sahiner, B.; Helvie, M.A. Computer-aided detection of clustered microcalcifications in multiscale bilateral filtering regularized reconstructed digital breast tomosynthesis volume. Med. Phys. 2014, 41, 021901. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Chan, H.P.; Hadjiiski, L.M.; Helvie, M.A.; Lu, Y.; Zhou, C.; Samala, R. Multichannel response analysis on 2D projection views for detection of clustered microcalcifications in digital breast tomosynthesis. Med. Phys. 2014, 41, 041913. [Google Scholar] [CrossRef] [PubMed]
- Samala, R.K.; Chan, H.P.; Lu, Y.; Hadjiiski, L.M.; Wei, J.; Helvie, M.A. Computer-aided detection system for clustered microcalcifications in digital breast tomosynthesis using joint information from volumetric and planar projection images. Phys. Med. Biol. 2015, 60, 8457–8479. [Google Scholar] [CrossRef]
- Fenton, J.J.; Taplin, S.H.; Carney, P.A.; Abraham, L.; Sickles, E.A.; D’Orsi, C.; Berns, E.A.; Cutter, G.; Hendrick, R.E.; Barlow, W.E.; et al. Influence of Computer-Aided Detection on Performance of Screening Mammography. N. Engl. J. Med. 2007, 356, 1399–1409. [Google Scholar] [CrossRef]
- Lehman, C.D.; Wellman, R.D.; Buist, D.S.M.; Kerlikowske, K.; Tosteson, A.N.A.; Miglioretti, D.L.; Breast Cancer Surveillance Consortium. Diagnostic Accuracy of Digital Screening Mammography With and Without Computer-Aided Detection. JAMA Intern. Med. 2015, 175, 1828–1837. [Google Scholar] [CrossRef]
- Katzen, J.; Dodelzon, K. A review of computer aided detection in mammography. Clin. Imaging. 2018, 52, 305–309. [Google Scholar] [CrossRef]
- Sechopoulos, I.; Teuwen, J.; Mann, R. Artificial intelligence for breast cancer detection in mammography and digital breast tomosynthesis: State of the art. Semin. Cancer Biol. 2020, 72, 214–225. [Google Scholar] [CrossRef]
- Rodriguez-Ruiz, A.; Wellman, R.D.; Buist, D.S.; Kerlikowske, K.; Tosteson, A.N.; Miglioretti, D.L.; Breast Cancer Surveillance Consortium. Stand-Alone Artificial Intelligence for Breast Cancer Detection in Mammography: Comparison With 101 Radiologists. J. Natl. Cancer Inst. 2019, 111, 916–922. [Google Scholar] [CrossRef]
- McKinney, S.M.; Sieniek, M.; Godbole, V.; Godwin, J.; Antropova, N.; Ashrafian, H.; Back, T.; Chesus, M.; Corrado, G.S.; Darzi, A.; et al. International evaluation of an AI system for breast cancer screening. Nature 2020, 577, 89–94. [Google Scholar] [CrossRef]
- Kim, H.-E.; Kim, H.H.; Han, B.K.; Kim, K.H.; Han, K.; Nam, H.; Lee, E.H.; Kim, E.K. Changes in cancer detection and false-positive recall in mammography using artificial intelligence: A retrospective, multireader study. Lancet Digit. Health 2020, 2, e138–e148. [Google Scholar] [CrossRef]
- Wang, X.; Liang, G.; Zhang, Y.; Blanton, H.; Bessinger, Z.; Jacobs, N. Inconsistent Performance of Deep Learning Models on Mammogram Classification. J. Am. Coll. Radiol. 2020, 17, 796–803. [Google Scholar] [CrossRef]
- Schaffter, T.; Buist, D.S.; Lee, C.I.; Nikulin, Y.; Ribli, D.; Guan, Y.; Lotter, W.; Jie, Z.; Du, H.; Wang, S.; et al. Evaluation of Combined Artificial Intelligence and Radiologist Assessment to Interpret Screening Mammograms. JAMA Netw. Open 2020, 3, e200265. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Ruiz, A.; Krupinski, E.; Mordang, J.J.; Schilling, K.; Heywang-Köbrunner, S.H.; Sechopoulos, I.; Mann, R.M. Detection of Breast Cancer with Mammography: Effect of an Artificial Intelligence Support System. Radiology 2019, 290, 305–314. [Google Scholar] [CrossRef] [PubMed]
- Conant, E.F.; Toledano, A.Y.; Periaswamy, S.; Fotin, S.V.; Go, J.; Boatsman, J.E.; Hoffmeister, J.W. Improving Accuracy and Efficiency with Concurrent Use of Artificial Intelligence for Digital Breast Tomosynthesis. Radiol. Artif. Intell. 2019, 1, e180096. [Google Scholar] [CrossRef]
- van Winkel, S.L.; Rodríguez-Ruiz, A.; Appelman, L.; Gubern-Mérida, A.; Karssemeijer, N.; Teuwen, J.; Wanders, A.J.; Sechopoulos, I.; Mann, R.M. Impact of artificial intelligence support on accuracy and reading time in breast tomosynthesis image interpretation: A multi-reader multi-case study. Eur. Radiol. 2021, 31, 8682–8691. [Google Scholar] [CrossRef]
- Samala, R.; Chan, H.P.; Hadjiiski, L.M.; Cha, K.; Helvie, M.A. Deep-learning convolution neural network for computer-aided detection of microcalcifications in digital breast tomosynthesis. In Medical Imaging 2016: Computer-Aided Diagnosis; SPIE Medical Imaging: San Diego, CA, USA, 2016; Volume 9785. [Google Scholar]
- Fotin, S.; Yin, Y.; Haldankar, H.; Hoffmeister, J.W.; Periaswamy, S. Detection of soft tissue densities from digital breast tomosynthesis: Comparison of conventional and deep learning approaches. In Medical Imaging 2016: Computer-Aided Diagnosis; SPIE Medical Imaging: San Diego, CA, USA, 2016; Volume 9785. [Google Scholar]
- Kim, D.H.; Kim, S.T.; Ro, Y.M. Latent feature representation with 3-D multi-view deep convolutional neural network for bilateral analysis in digital breast tomosynthesis. In Proceedings of the 2016 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Shanghai, China, 20–25 March 2016. [Google Scholar]
- Samala, R.K.; Chan, H.P.; Hadjiiski, L.; Helvie, M.A.; Wei, J.; Cha, K. Mass detection in digital breast tomosynthesis: Deep convolutional neural network with transfer learning from mammography. Med. Phys. 2016, 43, 6654. [Google Scholar] [CrossRef]
- Kim, D.H.; Kim, S.T.; Chang, J.M.; Ro, Y.M. Latent feature representation with depth directional long-term recurrent learning for breast masses in digital breast tomosynthesis. Phys. Med Biol. 2017, 62, 1009–1031. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, Y.; Han, E.Y.; Jacobs, N.; Han, Q.; Wang, X.; Liu, J. Classification of Whole Mammogram and Tomosynthesis Images Using Deep Convolutional Neural Networks. IEEE Trans. NanoBioscience 2018, 17, 237–242. [Google Scholar] [CrossRef]
- Samala, R.K.; Chan, H.P.; Hadjiiski, L.M.; Helvie, M.A.; Richter, C.; Cha, K. Evolutionary pruning of transfer learned deep convolutional neural network for breast cancer diagnosis in digital breast tomosynthesis. Phys. Med. Biol. 2018, 63, 095005. [Google Scholar] [CrossRef]
- Yousefi, M.; Krzyżak, A.; Suen, C.Y. Mass detection in digital breast tomosynthesis data using convolutional neural networks and multiple instance learning. Comput. Biol. Med. 2018, 96, 283–293. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Ruiz, A.; Teuwen, J.; Vreemann, S.; Bouwman, R.W.; van Engen, R.E.; Karssemeijer, N.; Mann, R.M.; Gubern-Merida, A.; Sechopoulos, I. New reconstruction algorithm for digital breast tomosynthesis: Better image quality for humans and computers. Acta Radiol. 2018, 59, 1051–1059. [Google Scholar] [CrossRef] [PubMed]
- Mordang, J.J.; Janssen, T.; Bria, A.; Kooi, T.; Gubern-Mérida, A.; Karssemeijer, N. Automatic Microcalcification Detection in Multi-vendor Mammography Using Convolutional Neural Networks. In International Workshop on Breast Imaging; Springer International Publishing: Berlin/Heidelberg, Germany, 2016. [Google Scholar]
- Zhang, Y.; Wang, X.; Blanton, H.; Liang, G.; Xing, X.; Jacobs, N. 2D Convolutional Neural Networks for 3D Digital Breast Tomosynthesis Classification. In Proceedings of the 2019 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), San Diego, CA, USA, 18–21 November 2019. [Google Scholar]
- Liang, G.; Wang, X.; Zhang, Y.; Xing, X.; Blanton, H.; Salem, T.; Jacobs, N. Joint 2D-3D Breast Cancer Classification. In Proceedings of the 2019 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), San Diego, CA, USA, 18–21 November 2019. [Google Scholar]
- Mendel, K.; Li, H.; Sheth, D.; Giger, M. Transfer Learning From Convolutional Neural Networks for Computer-Aided Diagnosis: A Comparison of Digital Breast Tomosynthesis and Full-Field Digital Mammography. Acad. Radiol. 2019, 26, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Matthews, T.P.; Shah, M.; Mombourquette, B.; Tsue, T.; Long, A.; Almohsen, R.; Pedemonte, S.; Su, J. Adaptation of a deep learning malignancy model from full-field digital mammography to digital breast tomosynthesis. arXiv 2020, arXiv:2001.08381. [Google Scholar]
- Li, X.; Qin, G.; He, Q.; Sun, L.; Zeng, H.; He, Z.; Chen, W.; Zhen, X.; Zhou, L. Digital breast tomosynthesis versus digital mammography: Integration of image modalities enhances deep learning-based breast mass classification. Eur. Radiol. 2020, 30, 778–788. [Google Scholar] [CrossRef]
- Wang, L.; Zheng, C.; Chen, W.; He, Q.; Li, X.; Zhang, S.; Qin, G.; Chen, W.; Wei, J.; Xie, P.; et al. Multi-path synergic fusion deep neural network framework for breast mass classification using digital breast tomosynthesis. Phys. Med. Biol. 2020, 65, 235045. [Google Scholar] [CrossRef]
- Seyyedi, S.; Wong, M.J.; Ikeda, D.M.; Langlotz, C.P. SCREENet: A Multi-view Deep Convolutional Neural Network for Classification of High-resolution Synthetic Mammographic Screening Scans. arXiv 2020, arXiv:abs/2009.08563. [Google Scholar]
- Matthews, T.P.; Singh, S.; Mombourquette, B.; Su, J.; Shah, M.P.; Pedemonte, S.; Long, A.; Maffit, D.; Gurney, J.; Hoil, R.M.; et al. A Multisite Study of a Breast Density Deep Learning Model for Full-Field Digital Mammography and Synthetic Mammography. Radiol. Artif. Intell. 2021, 3, e200015. [Google Scholar] [CrossRef]
- Zheng, J.; Sun, H.; Wu, S.; Jiang, K.; Peng, Y.; Yang, X.; Zhang, F.; Li, M. 3D Context-Aware Convolutional Neural Network for False Positive Reduction in Clustered Microcalcifications Detection. IEEE J. Biomed. Health Inform. 2021, 25, 764–773. [Google Scholar] [CrossRef]
- Aswiga, R.V.; Shanthi, A.P. Augmenting Transfer Learning with Feature Extraction Techniques for Limited Breast Imaging Datasets. J. Digit. Imaging 2021, 34, 618–629. [Google Scholar]
- Xiao, B.; Sun, H.; Meng, Y.; Peng, Y.; Yang, X.; Chen, S.; Yan, Z.; Zheng, J. Classification of microcalcification clusters in digital breast tomosynthesis using ensemble convolutional neural network. Biomed. Eng. Online 2021, 20, 71. [Google Scholar] [CrossRef] [PubMed]
- El-Shazli, A.M.A.; Youssef, S.M.; Soliman, A.H. Intelligent Computer-Aided Model for Efficient Diagnosis of Digital Breast Tomosynthesis 3D Imaging Using Deep Learning. Appl. Sci. 2022, 12, 5736. [Google Scholar] [CrossRef]
- Bai, J.; Posner, R.; Wang, T.; Yang, C.; Nabavi, S. Applying deep learning in digital breast tomosynthesis for automatic breast cancer detection: A review. Med. Image Anal. 2021, 71, 102049. [Google Scholar] [CrossRef] [PubMed]
- Buda, M.; Saha, A.; Walsh, R.; Ghate, S.; Li, N.; Święcicki, A.; Lo, J.Y.; Mazurowski, M.A. Detection of masses and architectural distortions in digital breast tomosynthesis: A publicly available dataset of 5060 patients and a deep learning model. arXiv 2020, arXiv:2011.07995. [Google Scholar]
- Badano, A.; Graff, C.G.; Badal, A.; Sharma, D.; Zeng, R.; Samuelson, F.W.; Glick, S.J.; Myers, K.J. Evaluation of Digital Breast Tomosynthesis as Replacement of Full-Field Digital Mammography Using an In Silico Imaging Trial. JAMA Netw. Open 2018, 1, e185474. [Google Scholar] [CrossRef]
- VICTRE. The VICTRE Trial: Open-Source, In-Silico Clinical Trial For Evaluating Digital Breast Tomosynthesis. 2018. Available online: https://wiki.cancerimagingarchive.net/display/Public/The+VICTRE+Trial%3A+Open-Source%2C+In-Silico+Clinical+Trial+For+Evaluating+Digital+Breast+Tomosynthesis (accessed on 1 November 2021).
- Sidky, E.Y.; Pan, X.; Reiser, I.S.; Nishikawa, R.M.; Moore, R.H.; Kopans, D.B. Enhanced imaging of microcalcifications in digital breast tomosynthesis through improved image-reconstruction algorithms. Med. Phys. 2009, 36, 4920–4932. [Google Scholar] [CrossRef]
- Lu, Y.; Chan, H.P.; Wei, J.; Hadjiiski, L.M. Selective-diffusion regularization for enhancement of microcalcifications in digital breast tomosynthesis reconstruction. Med. Phys. 2010, 37, 6003–6014. [Google Scholar] [CrossRef]
- Mota, A.M.; Matela, N.; Oliveira, N.; Almeida, P. Total variation minimization filter for DBT imaging. Med. Phys. 2015, 42, 2827–2836. [Google Scholar] [CrossRef]
- Michielsen, K.; Nuyts, J.; Cockmartin, L.; Marshall, N.; Bosmans, H. Design of a model observer to evaluate calcification detectability in breast tomosynthesis and application to smoothing prior optimization. Med. Phys. 2016, 43, 6577–6587. [Google Scholar] [CrossRef]
- Mota, A.M.; Clarkson, M.J.; Almeida, P.; Matela, N. An Enhanced Visualization of DBT Imaging Using Blind Deconvolution and Total Variation Minimization Regularization. IEEE Trans. Med. Imaging 2020, 39, 4094–4101. [Google Scholar] [CrossRef]
- Zuiderveld, K. Contrast limited adaptive histogram equalization. In Graphics Gems IV1994; Academic Press Professional, Inc.: San Diego, CA, USA, 1994; pp. 474–485. [Google Scholar]
- MathWorks. MATLAB Adapthisteq Function. [Cited 2021 May]. 2021. Available online: https://www.mathworks.com/help/images/ref/adapthisteq.html (accessed on 1 November 2021).
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. Adv. Neural Inf. Processing Syst. 2012, 25, 1097–1105. [Google Scholar] [CrossRef]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016. [Google Scholar]
- Iandola, F.N.; Han, S.; Moskewicz, M.W.; Ashraf, K.; Dally, W.J.; Keutzer, K. SqueezeNet: AlexNet-level accuracy with 50x fewer parameters and <0.5 MB model size. arXiv 2016, arXiv:1602.07360. [Google Scholar]
- MathWorks. Tranfer Learning. [Cited May 2021]. 2021. Available online: https://www.mathworks.com/discovery/transfer-learning.html (accessed on 1 November 2021).
- Vourtsis, A.; Berg, W.A. Breast density implications and supplemental screening. Eur. Radiol. 2019, 29, 1762–1777. [Google Scholar] [CrossRef] [PubMed]
- Zeng, R.; Samuelson, F.W.; Sharma, D.; Badal, A.; Christian, G.G.; Glick, S.J.; Myers, K.J.; Badano, A.G. Computational reader design and statistical performance evaluation of an in-silico imaging clinical trial comparing digital breast tomosynthesis with full-field digital mammography. J. Med. Imaging 2020, 7, 042802. [Google Scholar] [CrossRef] [PubMed]


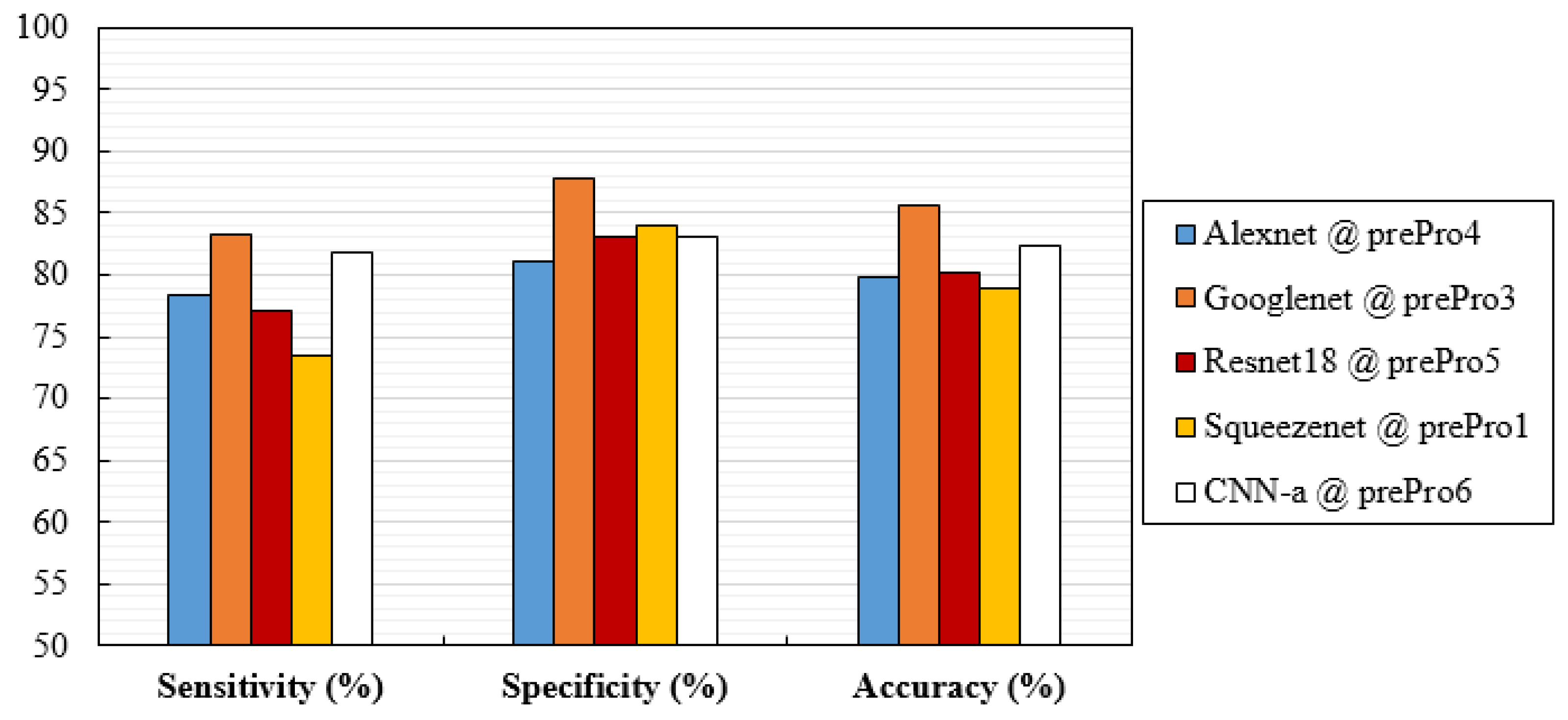
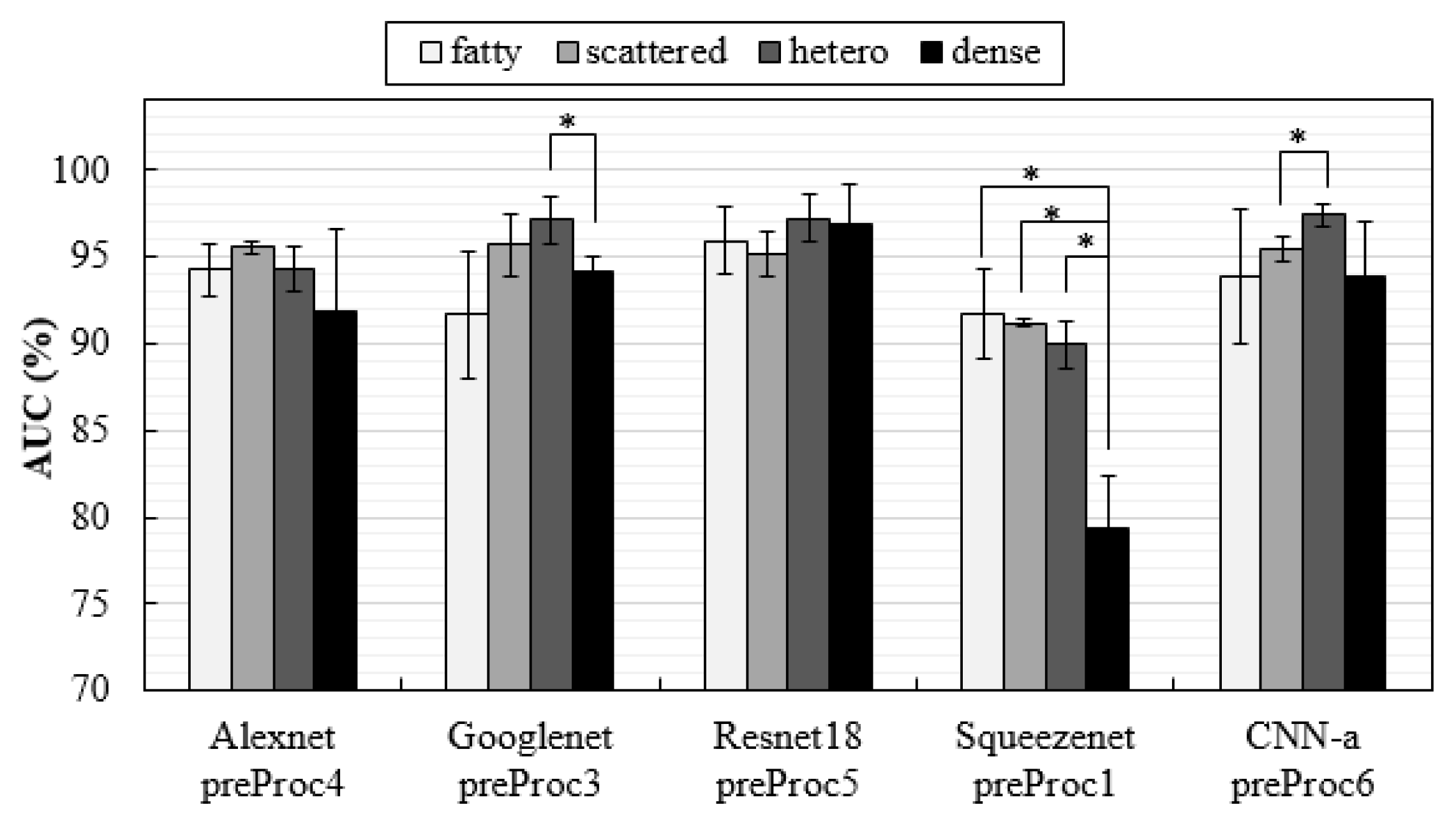
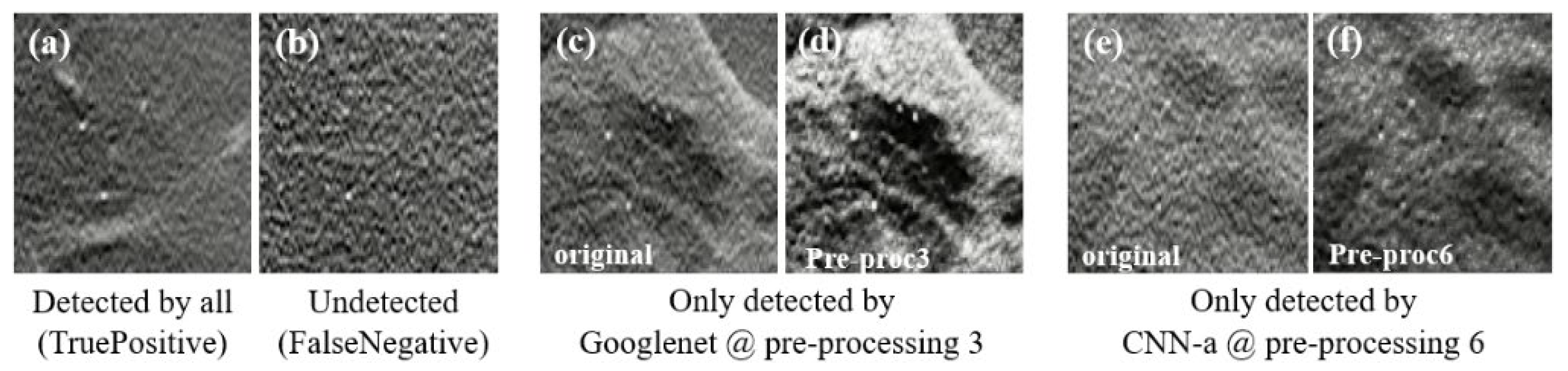
| Ref. | Classification Task | ROI/Patch/Image | Model | Best Metric |
|---|---|---|---|---|
| [50] | True MCs vs. false positives | ROI (16 × 16) | Own | AUC: 0.93 |
| [51] | Presence/absence of masses and architectural distortions | Patch (256 × 256) | Based on AlexNet | Accuracy: 0.8640 |
| [52] | Presence/absence of masses | ROI (32 × 32 × 25) | Own | AUC: 0.847 |
| [53] | True masses vs. false positives | ROI (128 × 128) | Own | AUC: 0.90 |
| [54] | True masses vs. false positives | ROI (64 × 64) | Based on VGG16 | AUC: 0.919 |
| [55] | Positive (malignant, benign masses) vs. negative images | Image (224 × 224) | Based on AlexNet | AUC: 0.6632 |
| [56] | Malignant vs. benign masses | ROI (128 × 128) | Based on AlexNet | AUC: 0.90 |
| [57] | Malignant vs. benign masses | Image (256 × 256) | Own | AUC: 0.87 |
| [58] | Presence/absence of MCs | Patch (29 × 29 × 9) | Based on [59] | pAUC: 0.880 |
| [60] | Positive vs. negative volumes | Image (1024 × 1024) | Based on AlexNet, ResNet50, Xception | AUC: 0.854 (AlexNet) |
| [61] | Positive vs. negative volumes | Image (832 × 832) | Based on AlexNet, ResNet, DenseNet and SqueezeNet | AUC: 0.91 (DenseNet) |
| [62] | Benign vs. malignant lesions | ROI (224 × 224) | Based on VGG19 | AUC (MCs): 0.97 |
| [63] | Positive vs. negative patches | Patch (512 × 512) | Based on ResNet | AUC: 0.847 |
| [64] | Malignant vs. benign vs. normal masses | ROI (256 × 256) | Based on VGG16 | AUC: 0.917, 0.951, 0.993 (malignant, benign, normal) |
| [65] | Malignant vs. benign masses | ROI (224 × 224) | Based on DenseNet121 | AUC: 0.8703 |
| [66] | BIRADS 0 vs. BIRADS 1 vs. BIRADS 2 | Image (2200 × 1600) | Based on ResNet50 | AUC: 0.912 (BIRADS 0 vs. non-0) |
| [67] | Predict breast density | Image | Based on ResNet34 | AUC: 0.952 |
| [68] | True MCs vs. false positives | ROI (128 × 128) | Based on ResNet18 | AUC: 0.9765 |
| [69] | Malignant vs. benign vs. normal images | Image (150 × 150) | Own | AUC: 0.89 |
| [70] | Malignant vs. benign MCs | Patch (224 × 224) | Ensemble CNN (2D ResNet34 and anisotropic 3D Resnet) | AUC: 0.8837 |
| [71] | Malignant vs. benign vs. normal slices based on masses and architectural distortions | Image (input size of each CNN: 224 × 224, 227 × 227) | ResNet18, AlexNet, GoogLeNet, VGG16, MobileNetV2, DenseNet201, Mod_AlexNet | Accuracy: 0.9161 (Mod_AlexNet) |
| Absent | Present MCs | |||
|---|---|---|---|---|
| Density | Number of Cases | Number of Slices | Number of Cases | Number of Slices |
| Fatty | 20 | 100 | 25 | 99 |
| Scattered | 80 | 400 | 100 | 386 |
| Heterogeneous | 80 | 400 | 100 | 371 |
| Dense | 20 | 100 | 25 | 93 |
| Total | 1000 | 949 | ||
| AUC (%): Mean ± SD | |||||
|---|---|---|---|---|---|
| AlexNet | GoogLeNet | ResNet18 | SqueezeNet | CNN-a | |
| Original data | 87.92 ± 2.01 | 90.14 ± 0.38 | 86.84 ± 2.62 | 87.43 ± 0.78 | 89.79 ± 1.23 |
| Preprocessing 1 | 87.35 ± 1.63 | 88.38 ± 1.12 | 87.96 ± 0.96 | 88.78 ± 0.99 | 90.66 ± 0.15 |
| Preprocessing 2 | 87.29 ± 0.78 | 93.02 ± 3.59 | 86.42 ± 3.26 | 86.84 ± 3.82 | 86.95 ± 0.97 |
| Preprocessing 3 | 88.61 ± 0.43 | 94.19 ± 1.12 | 86.33 ± 1.46 | 82.15 ± 1.51 | 85.80 ± 1.73 |
| Preprocessing 4 | 90.82 ± 1.29 | 94.15 ± 1.54 | 90.13 ± 0.32 | 86.33 ± 6.31 | 89.07 ± 1.62 |
| Preprocessing 5 | 87.62 ± 0.35 | 88.65 ± 4.27 | 90.44 ± 0.41 | 85.18 ± 2.78 | 89.54 ± 2.63 |
| Preprocessing 6 | 87.47 ± 1.13 | 89.76 ± 1.76 | 89.00 ± 1.33 | 84.09 ± 3.13 | 91.17 ± 0.07 |
| p-Value | GoogLeNet PreProc3 | ResNet18 PreProc5 | SqueezeNet PreProc1 | CNN-a PreProc6 |
|---|---|---|---|---|
| (94.19 ± 1.12) | (90.44 ± 0.41) | (88.78 ± 0.99) | (91.17 ± 0.07) | |
| AlexNet preProc4 | 0.027 | 0.654 | 0.095 | 0.662 |
| (90.82 ± 1.29) | (AlexNet < GoogLeNet) | |||
| GoogLeNet preProc3 | 0.006 | 0.003 | 0.010 | |
| (94.19 ± 1.12) | (GoogLeNet > ResNet18) | (GoogLeNet > SqueezeNet) | (GoogLeNet > CNN-a) | |
| ResNet18 preProc5 | 0.055 | 0.038 | ||
| (90.44 ± 0.41) | (ResNet18 < CNN-a) | |||
| SqueezeNet preProc1 | 0.014 | |||
| (88.78 ± 0.99) | (SqueezeNet < CNN-a) |
| Training Time (h) | Inference Time/Slice (s) | |
|---|---|---|
| CNN-a | 2.4 | 0.0057 |
| AlexNet | 4.1 | 0.0062 |
| SqueezeNet | 4.4 | 0.0083 |
| ResNet18 | 7.8 | 0.0143 |
| GoogLeNet | 8.9 | 0.0158 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mota, A.M.; Clarkson, M.J.; Almeida, P.; Matela, N. Automatic Classification of Simulated Breast Tomosynthesis Whole Images for the Presence of Microcalcification Clusters Using Deep CNNs. J. Imaging 2022, 8, 231. https://doi.org/10.3390/jimaging8090231
Mota AM, Clarkson MJ, Almeida P, Matela N. Automatic Classification of Simulated Breast Tomosynthesis Whole Images for the Presence of Microcalcification Clusters Using Deep CNNs. Journal of Imaging. 2022; 8(9):231. https://doi.org/10.3390/jimaging8090231
Chicago/Turabian StyleMota, Ana M., Matthew J. Clarkson, Pedro Almeida, and Nuno Matela. 2022. "Automatic Classification of Simulated Breast Tomosynthesis Whole Images for the Presence of Microcalcification Clusters Using Deep CNNs" Journal of Imaging 8, no. 9: 231. https://doi.org/10.3390/jimaging8090231
APA StyleMota, A. M., Clarkson, M. J., Almeida, P., & Matela, N. (2022). Automatic Classification of Simulated Breast Tomosynthesis Whole Images for the Presence of Microcalcification Clusters Using Deep CNNs. Journal of Imaging, 8(9), 231. https://doi.org/10.3390/jimaging8090231














