Discrete Shearlets as a Sparsifying Transform in Low-Rank Plus Sparse Decomposition for Undersampled (k, t)-Space MR Data
Abstract
:1. Introduction
2. Theory
2.1. Focal Underdetermined System Solver (k-t FOCUSS)
2.2. Robust Principal Component Analysis (RPCA)
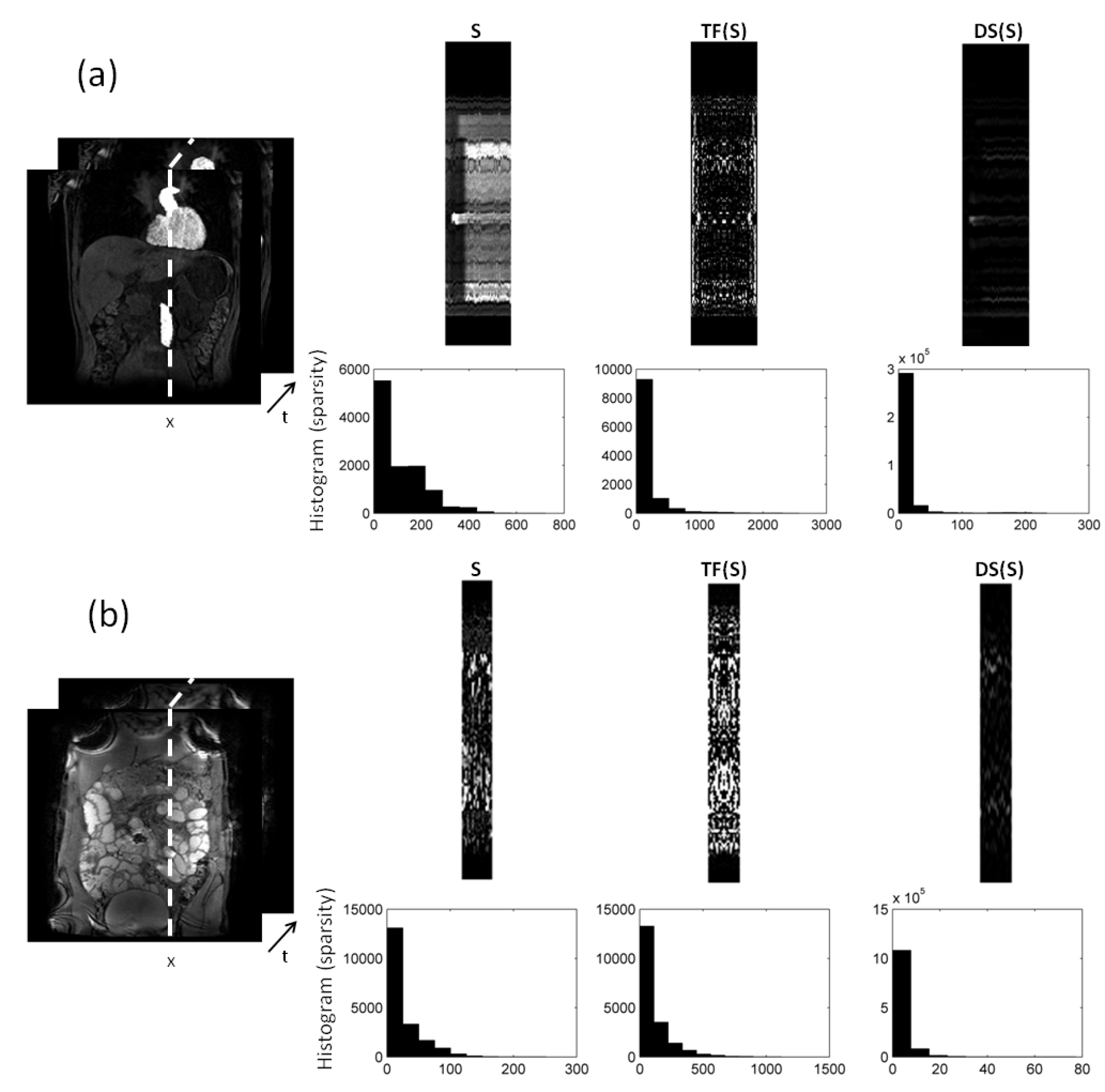
2.3. Sparsifying Transforms
2.4. Image Registration—Motility Metric
3. Materials and Methods
3.1. Small Bowel MR Acquisition
3.2. Simulated Abdominal DCE Data
3.3. Simulated Undersampled Acquisition
3.4. Quantitative Evaluation
3.5. Implementation Details
4. Results
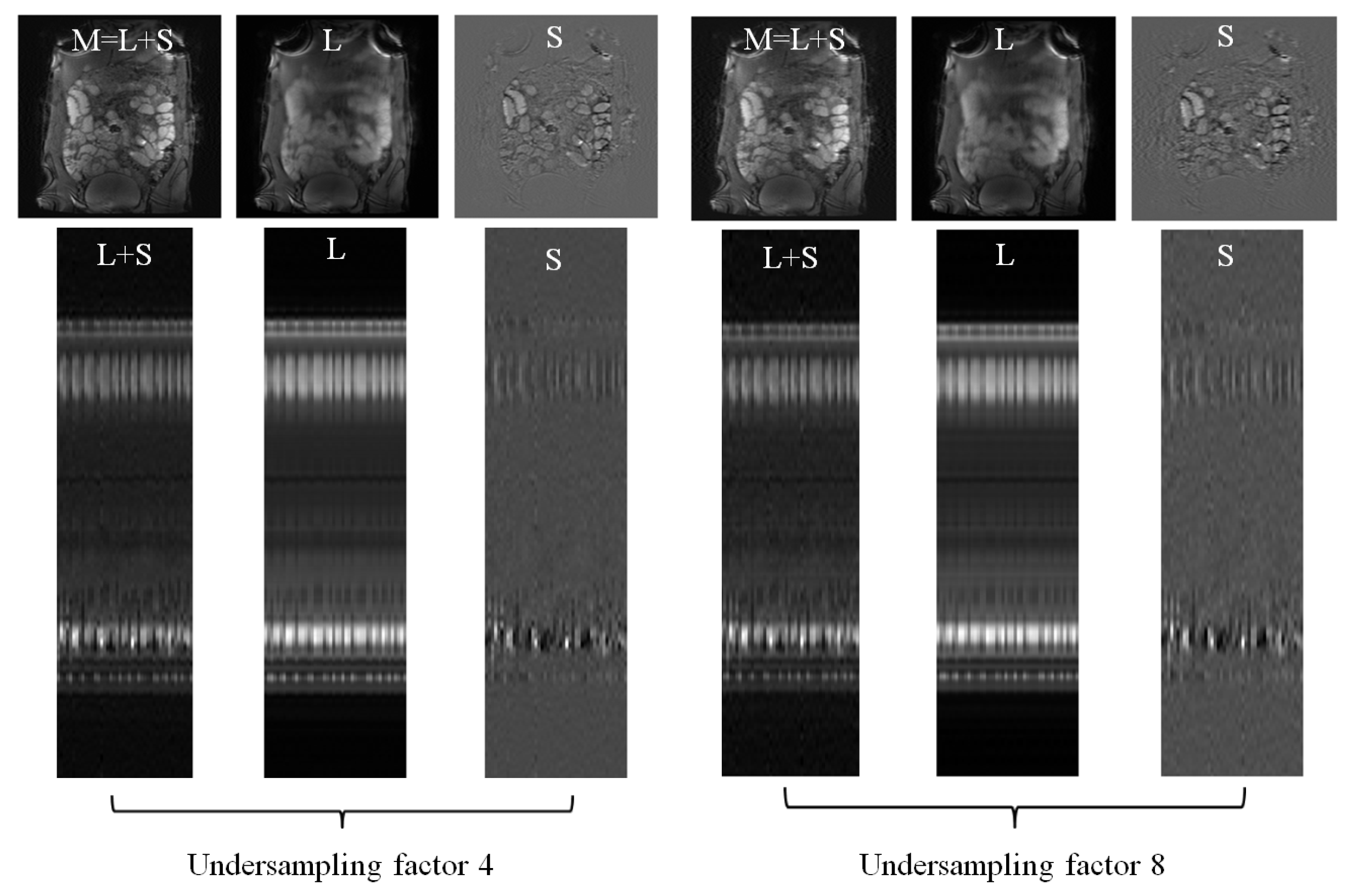
4.1. Low-Rank Plus Sparse Image Decomposition
4.2. Low-Rank Plus Sparse Image Decomposition from Undersampled -Space Data
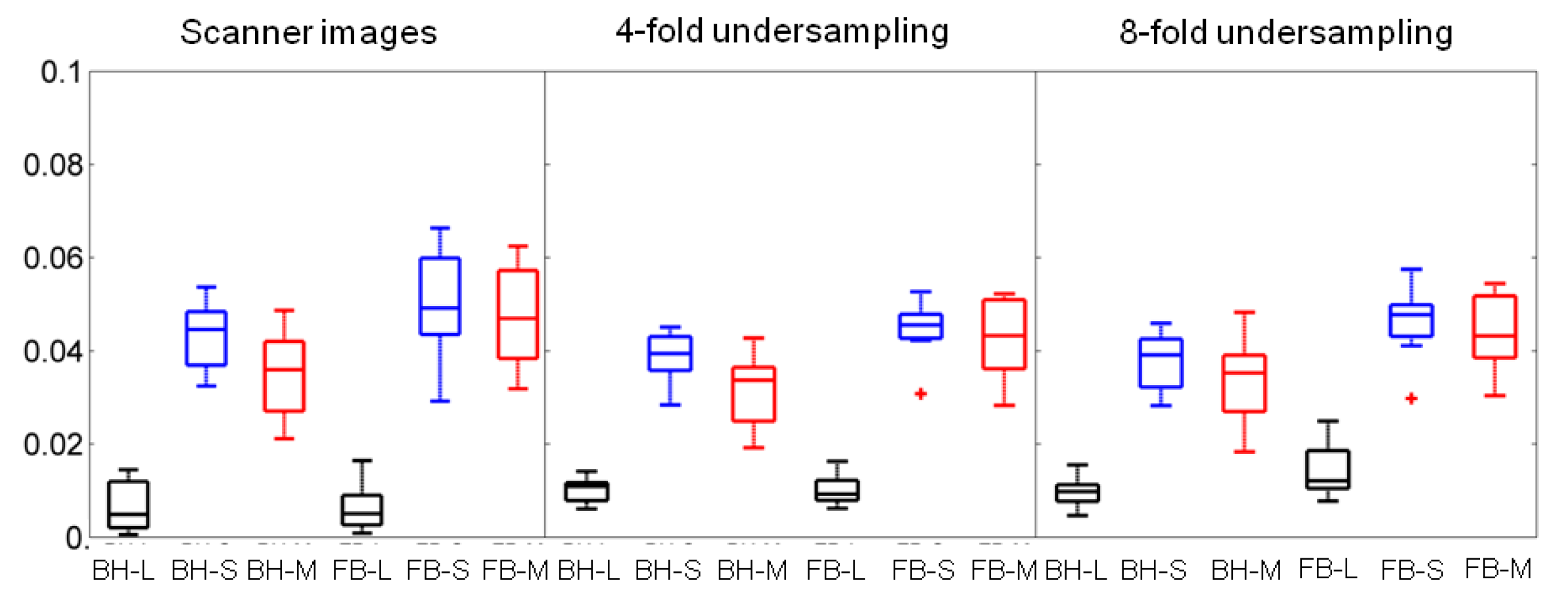
4.3. Quantification of Motility in Low-Rank Plus Sparse Decomposition Using Discrete Shearlets as Sparsifying Transforms
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Liu, Y.; Liu, T.; Liu, J.; Zhu, C. Smooth robust tensor principal component analysis for compressed sensing of dynamic MRI. Pattern Recognit. 2020, 102, 107252. [Google Scholar] [CrossRef]
- Candès, E.J.; Li, X.; Ma, Y.; Wright, J. Robust principal component analysis? J. ACM 2011, 58, 1–37. [Google Scholar] [CrossRef]
- Gao, H.; Cai, J.F.; Shen, Z.; Zhao, H. Robust principal component analysis-based four-dimensional computed tomography MRI. Phys. Med. Biol. 2011, 56, 3181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trémoulhéac, B.; Atkinson, D.; Arridge, S.R. Motion and contrast enhancement separation model reconstruction from partial measurements in dynamic MRI. In Proceedings of the MICCAI Workshop on Sparsity Techniques in Medical Imaging, Nice, France, 1–5 October 2012. [Google Scholar]
- Trémoulhéac, B.; Dikaios, N.; Atkinson, D.; Arridge, S.R. Dynamic MR Image reconstruction–separation from undersampled (k,t)-space via low-rank plus sparse prior. IEEE Trans. Med. Imaging 2014, 33, 1689–1701. [Google Scholar] [CrossRef] [PubMed]
- Otazo, R.; Candes, E.; Sodickson, D.K. Low-rank plus sparse matrix decomposition for accelerated dynamic MRI with separation of background and dynamic components. Magn. Reson. Med. 2015, 73, 1125–1136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mahapatra, D.; Li, Z.; Vos, F.; Buhmann, J. Joint segmentation and groupwise registration of cardiac DCE MRI using sparse data representations. In Proceedings of the 2015 IEEE 12th International Symposium on Biomedical Imaging (ISBI), Brooklyn, NY, USA, 16–19 April 2015; pp. 1312–1315. [Google Scholar] [CrossRef]
- Hamy, V.; Dikaios, N.; Punwani, S.; Melbourne, A.; Latifoltojar, A.; Makanyanga, J.; Chouhan, M.; Helbren, E.; Menys, A.; Taylor, S.; et al. Respiratory motion correction in dynamic MRI using robust data decomposition registration—Application to DCE-MRI. Med. Image Anal. 2014, 18, 301–313. [Google Scholar] [CrossRef] [PubMed]
- Menys, A.; Hamy, V.; Makanyanga, J.; Hoad, C.; Gowland, P.; Odille, F.; Taylor, S.; Atkinson, D. Dual registration of abdominal motion for motility assessment in free-breathing data sets acquired using dynamic MRI. Phys. Med. Biol. 2014, 59, 4603. [Google Scholar] [CrossRef]
- Odille, F.; Menys, A.; Ahmed, A.; Punwani, S.; Taylor, S.A.; Atkinson, D. Quantitative assessment of small bowel motility by nonrigid registration of dynamic MR images. Magn. Reson. Med. 2012, 68, 783–793. [Google Scholar] [CrossRef]
- Menys, A.; Taylor, S.A.; Emmanuel, A.; Ahmed, A.; Plumb, A.A.; Odille, F.; Alam, A.; Halligan, S.; Atkinson, D. Global small bowel motility: Assessment with dynamic MR imaging. Radiology 2013, 269, 443–450. [Google Scholar] [CrossRef]
- Lustig, M.; Donoho, D.; Pauly, J.M. Sparse MRI: The application of compressed sensing for rapid MR imaging. Magn. Reson. Med. 2007, 58, 1182–1195. [Google Scholar] [CrossRef]
- Donoho, D.L. Compressed sensing. IEEE Trans. Inf. Theory 2006, 52, 1289–1306. [Google Scholar] [CrossRef]
- Huizinga, J.D.; Chen, J.H.; Zhu, Y.F.; Pawelka, A.; McGinn, R.J.; Bardakjian, B.L.; Parsons, S.P.; Kunze, W.A.; Wu, R.Y.; Bercik, P.; et al. The origin of segmentation motor activity in the intestine. Nat. Commun. 2014, 5, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, K.; Kutyniok, G.; Labate, D. Sparse multidimensional representations using anisotropic dilation and shear operators. In Wavelets and Splines; Chen, G., Lai, M., Eds.; Nashboro Press: Nashville, TN, USA, 2006; pp. 189–201. [Google Scholar]
- Yi, S.; Labate, D.; Easley, G.R.; Krim, H. A shearlet approach to edge analysis and detection. IEEE Trans. Image Process. 2009, 18, 929–941. [Google Scholar] [CrossRef] [PubMed]
- Häuser, S.; Steidl, G. Fast Finite Shearlet Transform. arXiv 2014, arXiv:1202.1773. [Google Scholar]
- Yuan, M.; Yang, B.; Ma, Y.; Zhang, J.; Zhang, R.; Zhang, C. Compressed sensing MRI reconstruction from highly undersampled-space data using nonsubsampled shearlet transform sparsity prior. Math. Probl. Eng. 2015, 2015. [Google Scholar] [CrossRef] [Green Version]
- Liu, R.W.; Shi, L.; Simon, C.; Wang, D. Hybrid regularization for compressed sensing MRI: Exploiting shearlet transform and group-sparsity total variation. In Proceedings of the 2017 20th International Conference on Information Fusion (Fusion), Xi’an, China, 10–13 July 2017; pp. 1–8. [Google Scholar] [CrossRef]
- Ma, J.; März, M.; Funk, S.; Schulz-Menger, J.; Kutyniok, G.; Schaeffter, T.; Kolbitsch, C. Shearlet-based compressed sensing for fast 3D cardiac MR imaging using iterative reweighting. Phys. Med. Biol. 2018, 63, 235004. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yazdanpanah, A.P.; Regentova, E.E. Compressed sensing magnetic resonance imaging based on shearlet sparsity and nonlocal total variation. J. Med. Imaging 2017, 4, 026003. [Google Scholar] [CrossRef]
- Jung, H.; Ye, J.C.; Kim, E.Y. Improved k-t BLAST and k-t SENSE using FOCUSS. Phys. Med. Biol. 2007, 52, 3201–3226. [Google Scholar] [CrossRef]
- Tsao, J.; Boesiger, P.; Pruessmann, K.P. k-t BLAST and k-t SENSE: Dynamic MRI with high frame rate exploiting spatiotemporal correlations. Magn. Reson. Med. 2003, 50, 1031–1042. [Google Scholar] [CrossRef]
- Gorodnitsky, I.F.; George, J.S.; Rao, B.D. Neuromagnetic source imaging with FOCUSS: A recursive weighted minimum norm algorithm. Electroencephalogr. Clin. Neurophysiol. 1995, 95, 231–251. [Google Scholar] [CrossRef]
- Jung, H.; Sung, K.; Nayak, K.S.; Kim, E.Y.; Ye, J.C. k-t FOCUSS: A general compressed sensing framework for high resolution dynamic MRI. Magn. Reson. Med. 2009, 61, 103–116. [Google Scholar] [CrossRef] [PubMed]
- Lingala, S.G.; Hu, Y.; DiBella, E.; Jacob, M. Accelerated dynamic MRI exploiting sparsity and low-rank structure: k-t SLR. IEEE Trans. Med. Imag. 2011, 30, 1042–1054. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dikaios, N.; Tremoulheac, B.; Menys, A.; Hamy, V.; Arridge, S.; Atkinson, D. Joint reconstruction of low-rank and sparse components from undersampled (k, t)-space small bowel data. In Proceedings of the 2013 IEEE Nuclear Science Symposium and Medical Imaging Conference (2013 NSS/MIC), Seoul, Korea, 27 October–2 November 2013; pp. 1–5. [Google Scholar] [CrossRef]
- Qu, X.; Guo, D.; Ning, B.; Hou, Y.; Lin, Y.; Cai, S.; Chen, Z. Undersampled MRI reconstruction with patch-based directional wavelets. Magn. Reson. Med. 2012, 30, 964–977. [Google Scholar] [CrossRef] [PubMed]
- Lai, Z.; Qu, X.; Liu, Y.; Guo, D.; Ye, J.; Zhan, Z.; Chen, Z. Image reconstruction of compressed sensing MRI using graph-based redundant wavelet transform. Med. Image Anal. 2016, 27, 93–104. [Google Scholar] [CrossRef] [PubMed]
- Deka, B.; Datta, S.; Handique, S. Wavelet tree support detection for compressed sensing MRI reconstruction. IEEE Signal Process. Lett. 2018, 25, 730–734. [Google Scholar] [CrossRef]
- Liu, R.W.; Ma, Q.; Yu, S.C.H.; Chui, K.T.; Xiong, N. Variational regularized tree-structured wavelet sparsity for CS-SENSE parallel imaging. IEEE Access 2018, 6, 61050–61064. [Google Scholar] [CrossRef]
- Islam, M.S.; Islam, R. Multiscale wavelet-based regularized reconstruction algorithm for three-dimensional compressed sensing magnetic resonance imaging. Signal Image Video Process. 2021, 1–9. [Google Scholar] [CrossRef]
- Chen, Y.; Firmin, D.; Yang, G. Wavelet improved GAN for MRI reconstruction. Medical Imaging 2021: Physics of Medical Imaging. Int. Soc. Opt. Photonics 2021, 11595, 1159513. [Google Scholar] [CrossRef]
- Wang, D.; Arlinghaus, L.R.; Yankeelov, T.E.; Yang, X.; Smith, D.S. Quantitative evaluation of temporal regularizers in compressed sensing dynamic contrast enhanced MRI of the breast. Int. J. Biomed. Imaging 2017, 2017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Li, B.; Tian, L. Multimodal medical volumetric data fusion using 3-D discrete shearlet transform and global-to-local rule. IEEE Trans. Biomed. Eng. 2013, 61, 197–206. [Google Scholar] [CrossRef]
- Alinsaif, S.; Lang, J.; Alzheimer’s Disease Neuroimaging Initiative. 3D shearlet-based descriptors combined with deep features for the classification of Alzheimer’s disease based on MRI data. Comput. Biol. Med. 2021, 138, 104879. [Google Scholar] [CrossRef] [PubMed]
- Gollifer, R.M.; Menys, A.; Makanyanga, J.; Puylaert, C.A.; Vos, F.M.; Stoker, J.; Atkinson, D.; Taylor, S.A. Relationship between MRI quantified small bowel motility and abdominal symptoms in Crohn’s disease patients—A validation study. Br. J. Radiol. 2018, 91, 20170914. [Google Scholar] [CrossRef] [PubMed]
- Menys, A.; Plumb, A.; Atkinson, D.; Taylor, S. The challenge of segmental small bowel motility quantitation using MR enterography. Br. J. Radiol. 2014, 87, 20140330. [Google Scholar] [CrossRef] [PubMed]
- Dikaios, N.; Punwani, S.; Hamy, V.; Purpura, P.; Rice, S.; Forster, M.; Mendes, R.; Taylor, S.; Atkinson, D. Noise estimation from averaged diffusion weighted images: Can unbiased quantitative decay parameters assist cancer evaluation? Magn. Reson. Med. 2014, 71, 2105–2117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dikaios, N.; Arridge, S.; Hamy, V.; Punwani, S.; Atkinson, D. Direct parametric reconstruction from undersampled (k, t)-space data in dynamic contrast enhanced MRI. Med. Image Anal. 2014, 18, 989–1001. [Google Scholar] [CrossRef]
- Mann, H.B.; Whitney, D.R. On a test of whether one of two random variables is stochastically larger than the other. Ann. Math. Stat. 1947, 18, 50–60. [Google Scholar] [CrossRef]




| decomposition and reconstruction algorithm |
|---|
| Input: -space samples y, decomposition parameters , |
| Initialize: and , iteration k=1; |
| while stopping criterion is not met, do |
| Singular value thresholding |
| Shrinkage operator |
| Subtract aliasing artifacts from |
| Stopping criterion |
| or |
| end while |
| Output: , |
| Unders. Factor 4 | Unders. Factor 8 | |
|---|---|---|
| k-t FOCUSS | 0.162 | 0.203 |
| using I as sparsifying transform | 0.149 | 0.194 |
| using TF as sparsifying transform | 0.147 | 0.189 |
| using WT as sparsifying transform | 0.147 | 0.188 |
| using DS as sparsifying transform | 0.144 | 0.187 |
| Undersampling Factor 4 | Median | iQR |
| k-t FOCUSS | 0.077 | 0.013 |
| using I as sparsifying transform | 0.080 | 0.012 |
| using TF as sparsifying transform | 0.079 | 0.012 |
| using WT as sparsifying transform | 0.075 | 0.011 |
| using DS as sparsifying transform | 0.064 * | 0.006 |
| Undersampling Factor 8 | Median | iQR |
| k-t FOCUSS | 0.120 | 0.025 |
| using I as sparsifying transform | 0.119 | 0.018 |
| using TF as sparsifying transform | 0.115 | 0.017 |
| using WT as sparsifying transform | 0.112 | 0.021 |
| using DS as sparsifying transform | 0.106 | 0.029 |
| L | ||
|---|---|---|
| DCE images | 0.016 | 0.031 |
| US4 | 0.021 | 0.025 |
| US8 | 0.024 | 0.029 |
| Scanner images | BH | FB | p-value |
| S | 0.044 | 0.049 | 0.20 |
| 0.036 | 0.047 | 0.02 | |
| Four-fold | BH | FB | p-value |
| S | 0.039 | 0.045 | 0.08 |
| 0.034 | 0.043 | 0.02 | |
| Eight-fold | BH | FB | p-value |
| S | 0.039 | 0.047 | 0.04 |
| 0.035 | 0.043 | 0.01 |
| L | S | ||
|---|---|---|---|
| Four-fold | 0.10 | 0.27 | 0.35 |
| Eight-fold | 0.01 | 0.45 | 0.49 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Protonotarios, N.E.; Tzampazidou, E.; Kastis, G.A.; Dikaios, N. Discrete Shearlets as a Sparsifying Transform in Low-Rank Plus Sparse Decomposition for Undersampled (k, t)-Space MR Data. J. Imaging 2022, 8, 29. https://doi.org/10.3390/jimaging8020029
Protonotarios NE, Tzampazidou E, Kastis GA, Dikaios N. Discrete Shearlets as a Sparsifying Transform in Low-Rank Plus Sparse Decomposition for Undersampled (k, t)-Space MR Data. Journal of Imaging. 2022; 8(2):29. https://doi.org/10.3390/jimaging8020029
Chicago/Turabian StyleProtonotarios, Nicholas E., Evangelia Tzampazidou, George A. Kastis, and Nikolaos Dikaios. 2022. "Discrete Shearlets as a Sparsifying Transform in Low-Rank Plus Sparse Decomposition for Undersampled (k, t)-Space MR Data" Journal of Imaging 8, no. 2: 29. https://doi.org/10.3390/jimaging8020029
APA StyleProtonotarios, N. E., Tzampazidou, E., Kastis, G. A., & Dikaios, N. (2022). Discrete Shearlets as a Sparsifying Transform in Low-Rank Plus Sparse Decomposition for Undersampled (k, t)-Space MR Data. Journal of Imaging, 8(2), 29. https://doi.org/10.3390/jimaging8020029









