Neutron Imaging and Learning Algorithms: New Perspectives in Cultural Heritage Applications
Abstract
1. State-of-the-Art in Imaging and in Machine and Deep Learning
1.1. Imaging Techniques
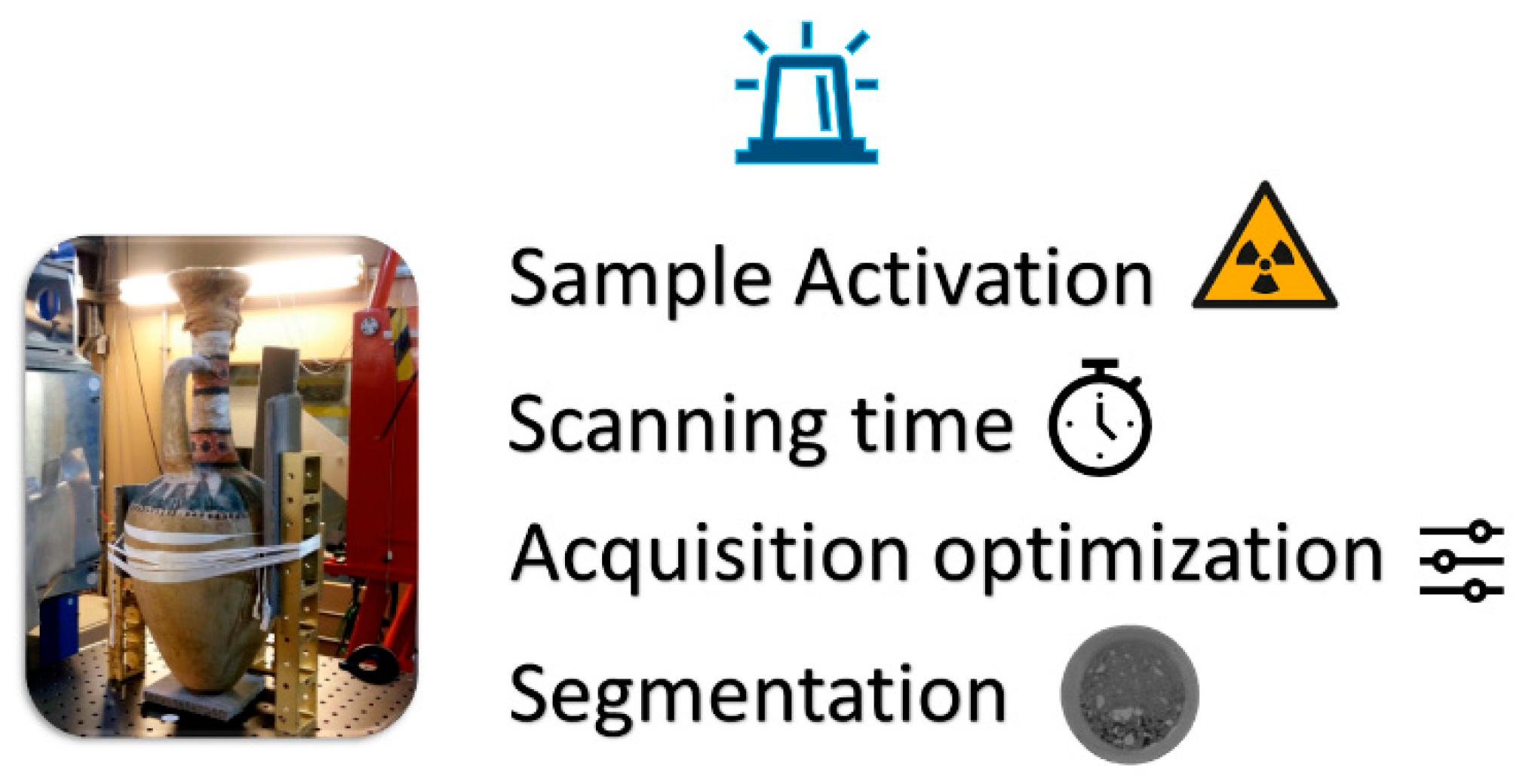
1.2. Neutron Imaging and Challenges
1.3. Neutron Imaging Optimization
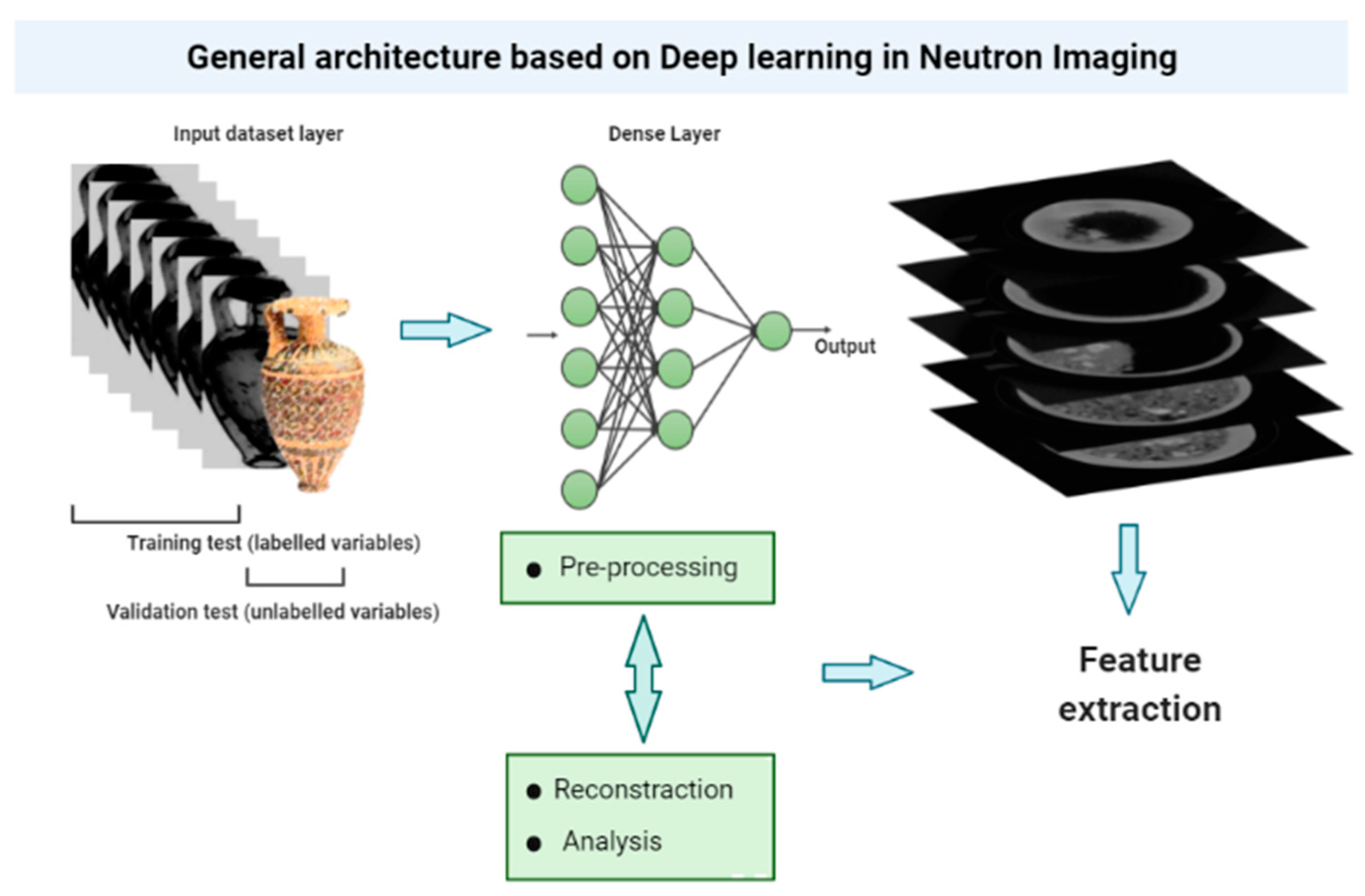
1.4. Neutron Imaging and Deep Learning
2. Deep Learning Applications
2.1. Learning Algorithms in X-ray Tomography (Biomedicine, Materials Science, and Cultural Heritage)
2.2. Learning Algorithms in Cultural Heritage Imaging
2.3. Deep Learning in Neutron Imaging and Future Applications in Cultural Heritage
3. Conclusions: Future Insights and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chitradevi, B.; Srimathi, P. An overview on image processing techniques. Int. J. Innov. Res. Comput. Commun. Eng. 2014, 2, 6466–6472. [Google Scholar]
- Danan, E.; Shabairou, N.; Danan, Y.; Zalevsky, Z. Signal-to-Noise Ratio Improvement for Multiple-Pinhole Imaging Using Supervised Encoder–Decoder Convolutional Neural Network Architecture. Photonics 2022, 9, 69. [Google Scholar] [CrossRef]
- Kardjilov, N.; Festa, G. (Eds.) Neutron Methods for Archaeology and Cultural Heritage; Springer: Berlin, Germany, 2017. [Google Scholar]
- Festa, G.; Romanelli, G.; Senesi, R.; Arcidiacono, L.; Scatigno, C.; Parker, S.F.; Marques, M.P.M.; Andreani, C. Neutrons for Cultural Heritage—Techniques, Sensors, and Detection. Sensors 2020, 20, 502. [Google Scholar] [CrossRef] [PubMed]
- Mannes, D.; Lehmann, E.; Masalles, A.; Schmidt-Ott, K.; Schaeppi, K.; Schmid, F.; Peetermans, S.; Hunger, K. The study of cultural heritage relevant objects by means of neutron imaging techniques. Insight—Non Destr. Test. Cond. Monit. 2014, 56, 137. [Google Scholar] [CrossRef]
- Kockelmann, W.; Siano, S.; Bartoli, L.; Visser, D.; Hallebeek, P.; Traum, R.; Linke, R.; Schreiner, M.; Kirfel, A. Applications of TOF neutron diffraction in archaeometry. Appl. Phys. A Mater. Sci. Process 2006, 83, 175–182. [Google Scholar] [CrossRef]
- Kak, A.C.; Slaney, M. Principles of computerized tomographic imaging. Society for Industrial and Applied Mathematics, Philadelphia; Stewart P. Cold neutron imaging for gas turbine inspection. Real Time Radiol. Imag. 2001, 8, 180–198. [Google Scholar]
- Peetermans, S.; Lehmann, E. Simultaneous neutron transmission and diffraction contrast tomography as a non-destructive 3D method for bulk single crystal quality investigations. J. Appl. Phys. 2013, 114, 124905. [Google Scholar] [CrossRef]
- Pfeiffer, F.; Grünzweig, C.; Bunk, O.; Frei, G.; Lehmann, E.; David, C. Neutron phase imaging and tomography. Phys. Rev. Lett. 2006, 96, 215505. [Google Scholar] [CrossRef]
- Festa, G.; Andreani, C.; De Pascale, M.P.; Senesi, R.; Vitali, G.; Porcinai, S.; Giusti, A.M.; Schulze, R.; Canella, L.; Kudejova, P.; et al. A nondestructive stratigraphic and radiographic neutron study of Lorenzo Ghiberti’s reliefs from paradise and north doors of Florence baptistery. J. Appl. Phys. 2009, 106, 074909. [Google Scholar] [CrossRef]
- Festa, G.; Christiansen, T.; Turina, V.; Borla, M.; Kelleher, J.; Arcidiacono, L.; Cartechini, L.; Ponterio, R.C.; Scatigno, C.; Senesi, R.; et al. Egyptian metallic inks on textiles from the 15th century BCE unravelled by non-invasive techniques and chemometric analysis. Sci. Rep. 2019, 9, 7310. [Google Scholar] [CrossRef]
- Andreani, C.; Aliotta, F.; Arcidiacono, L.; Borla, M.; Di Martino, D.; Facchetti, F.; Ferraris, E.; Festa, G.; Gorini, G.; Kockelmann, W.; et al. A neutron study of sealed pottery from the grave-goods of Kha and Merit. J. Anal. At. Spectrom. 2017, 32, 1342–1347. [Google Scholar] [CrossRef]
- Leona, M.; Fukunaga, K.; Liang, H.; Baglioni, P.; Festa, G.; Levchenko, V. From physics to art and back. Nat. Rev. Phys. 2021, 3, 681–684. [Google Scholar] [CrossRef]
- Kardjilov, N.; Manke, I.; Woracek, R.; Hilger, A.; Banhart, J. Advances in neutron imaging. Mater. Today 2018, 21, 652–672. [Google Scholar] [CrossRef]
- Schillinger, B.; Beaudet, A.; Fedrigo, A.; Grazzi, F.; Kullmer, O.; Laaß, M.; Makowska, M.; Werneburg, I.; Zanolli, C. Neutron imaging in cultural heritage research at the FRM II reactor of the Heinz Maier-Leibnitz center. J. Imaging 2018, 4, 22. [Google Scholar] [CrossRef]
- Laaß, M.; Hampe, O.; Schudack, M.; Hoff, C.; Kardjilov, N.; Hilger, A. New insights into the respiration and metabolic physiology of Lystrosaurus. Acta Zool. 2011, 92, 363–371. [Google Scholar] [CrossRef]
- Zanolli, C.; Schillinger, B.; Beaudet, A.; Kullmer, O.; Macchiarelli, R.; Mancini, L.; Schrenk, F.; Tuniz, C.; Vodopivec, V. Exploring hominin and non-hominin primate dental fossil remains with neutron microtomography. Phys. Procedia 2017, 88, 109–115. [Google Scholar] [CrossRef]
- Laaß, M.; Schillinger, B.; Kaestner, A. What did the “Unossified zone” of the non-mammalian therapsid braincase house? J. Morphol. 2017, 278, 1020–1032. [Google Scholar] [CrossRef]
- Trtik, P.; Hovind, J.; Grünzweig, C.; Bollhalder, A.; Thominet, V.; David, C.; Kaestner, A.; Lehmann, E.H. Improving the spatial resolution of neutron imaging at paul scherrer institut–the neutron microscope project. Phys. Procedia 2015, 69, 169–176. [Google Scholar] [CrossRef]
- Wu, H.; Khaykovich, B.; Wang, X.; Hussey, D.S. Wolter mirrors for neutron imaging. Phys. Procedia 2017, 88, 184–189. [Google Scholar] [CrossRef]
- Hussey, D.S.; Wen, H.; Wu, H.; Gentile, T.R.; Chen, W.; Jacobson, D.L.; LaManna, J.M.; Khaykovich, B. Demonstration of focusing Wolter mirrors for neutron phase and magnetic imaging. J. Imaging 2018, 4, 50. [Google Scholar] [CrossRef]
- Lehmann, E.H.; Hartmann, S.; Speidel, M.O. Investigation of the content of ancient Tibetan metallic Buddha statues by means of neutron imaging methods. Archaeometry 2010, 52, 416–428. [Google Scholar] [CrossRef]
- Qiao, S.; Li, J.; Zhao, C.; Zhang, T. No-reference quality assessment for neutron radiographic image based on a deep bilinear convolutional neural network. Nucl. Instrum. Methods Phys. Res. Sect. A Accel. Spectrometers Detect. Assoc. Equip. 2021, 1005, 165406. [Google Scholar] [CrossRef]
- Festa, G.; Andreani, C.; Arcidiacono, L.; Burca, G.; Kockelmann, W.; Minniti, T.; Senesi, R. Characterization of γ-ray background at IMAT beamline of ISIS Spallation Neutron Source. J. Instrum. 2017, 12, 08005. [Google Scholar] [CrossRef][Green Version]
- Miceli, A.; Festa, G.; Senesi, R.; Cippo, E.P.; Giacomelli, L.; Tardocchi, M.; Scherillo, A.; Schooneveld, E.; Frost, C.; Gorini, G.; et al. Measurements of gamma-ray background spectra at spallation neutron source beamlines. J. Anal. At. Spectrom. 2014, 29, 1897–1903. [Google Scholar] [CrossRef]
- Lee, S.; Oh, O.; Kim, Y.; Kim, D.; Hussey, D.S.; Wang, G.; Lee, S.W. Deep learning for high-resolution and high-sensitivity interferometric phase contrast imaging. Sci. Rep. 2020, 10, 9891. [Google Scholar] [CrossRef]
- Llamas, J.; Lerones, P.M.; Zalama, E.; Gómez-García-Bermejo, J. Applying Deep Learning Techniques to Cultural Heritage Images within the INCEPTION Project; Springer: Cham, Switzerland, 2016; pp. 25–32. [Google Scholar]
- Pierdicca, R.; Paolanti, M.; Matrone, F.; Martini, M.; Morbidoni, C.; Malinverni, E.S.; Frontoni, E.; Lingua, A.M. Point Cloud Semantic Segmentation Using a Deep Learning Framework for Cultural Heritage. Remote Sens. 2020, 12, 1005. [Google Scholar] [CrossRef]
- Garcia-Garcia, A.; Orts-Escolano, S.; Oprea, S.; Villena-Martinez, V.; Garcia-Rodriguez, J. A review on deep learning techniques applied to semantic segmentation. arXiv 2017, arXiv:1704.06857. [Google Scholar]
- Scatigno, C.; Festa, G. FTIR coupled with machine learning to unveil spectroscopic benchmarks in the Italian EVOO. Int. J. Food Sci. Technol. 2022, 57, 4156–4162. [Google Scholar] [CrossRef]
- Nunes, J.D.; Carvalho, M.; Carneiro, D.; Cardoso, J.S. Spiking Neural Networks: A Survey. IEEE Access 2022, 10, 60738–60764. [Google Scholar] [CrossRef]
- Cao, Y.; Chen, Y.; Khosla, D. Spiking deep convolutional neural networks for energy-efficient object recognition. Int. J. Comput. Vis. 2015, 113, 54–66. [Google Scholar] [CrossRef]
- Yuan, X.; Han, S. Single-pixel neutron imaging with artificial intelligence: Breaking the barrier in multi-parameter imaging, sensitivity, and spatial resolution. Innovation 2021, 2, 100100. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Saniie, J.; Heifetz, A. Detection of defects in additively manufactured stainless steel 316L with a compact infrared camera and machine learning algorithms. JOM 2020, 72, 4244–4253. [Google Scholar] [CrossRef]
- Butler, K.T.; Davies, D.W.; Cartwright, H.; Isayev, O.; Walsh, A. Machine learning for molecular and materials science. Nature 2018, 559, 547–555. [Google Scholar] [CrossRef]
- Tembely, M.; AlSumaiti, A.M.; Alameri, W.S. Machine and deep learning for estimating the permeability of complex carbonate rock from X-ray micro-computed tomography. Energy Rep. 2021, 7, 1460–1472. [Google Scholar] [CrossRef]
- Jia, Y.; Yu, G.; Du, J.; Gao, X.; Song, Y.; Wang, F. Adopting traditional image algorithms and deep learning to build the finite model of a 2.5 D composite based on X-Ray computed tomography. Compos. Struct. 2021, 275, 114440. [Google Scholar] [CrossRef]
- Aoki, H.; Liu, Y.; Yamashita, T. Deep learning approach for an interface structure analysis with a large statistical noise in neutron reflectometry. Sci. Rep. 2021, 11, 22711. [Google Scholar] [CrossRef]
- Turkson, R.E.; Qu, H.; Mawuli, C.B.; Eghan, M.J. Classification of Alzheimer’s disease using deep convolutional spiking neural network. Neural Process. Lett. 2021, 53, 2649–2663. [Google Scholar] [CrossRef]
- Ahmadi, M.; Sharifi, A.; Hassantabar, S.; Enayati, S. QAIS-DSNN: Tumor area segmentation of MRI image with optimized quantum matched-filter technique and deep spiking neural network. BioMed Res. Int. 2021, 2021, 6653879. [Google Scholar] [CrossRef]
- Garain, A.; Basu, A.; Giampaolo, F.; Velasquez, J.D.; Sarkar, R. Detection of COVID-19 from CT scan images: A spiking neural network-based approach. Neural Comput. Appl. 2021, 33, 12591–12604. [Google Scholar] [CrossRef]
- Dansana, D.; Kumar, R.; Bhattacharjee, A.; Hemanth, D.J.; Gupta, D.; Khanna, A.; Castillo, O. Early diagnosis of COVID-19-affected patients based on X-ray and computed tomography images using deep learning algorithm. Soft Comput. 2020, 26, 1–9. [Google Scholar] [CrossRef]
- Tajbakhsh, N.; Jeyaseelan, L.; Li, Q.; Chiang, J.N.; Wu, Z.; Ding, X. Embracing imperfect datasets: A review of deep learning solutions for medical image segmentation. Med. Image Anal. 2020, 63, 101693. [Google Scholar] [CrossRef] [PubMed]
- Amyar, A.; Modzelewski, R.; Li, H.; Ruan, S. Multi-task deep learning based CT imaging analysis for covid-19 pneumonia: Classification and segmentation. Comput. Biol. Med. 2020, 126, 104037. [Google Scholar] [CrossRef]
- Kobayashi, K.; Hwang, S.-W.; Okochi, T.; Lee, W.-H.; Sugiyama, J. Non-destructive method for wood identification using conventional X-ray computed tomography data. J. Cult. Herit. 2019, 38, 88–93. [Google Scholar] [CrossRef]
- Bouibed, M.L.; Nemmour, H.; Chibani, Y. SVM-based writer retrieval system in handwritten document images. Multimed. Tools Appl. 2021, 81, 22629–22651. [Google Scholar] [CrossRef]
- Massimiliano, P.; Alfio, V.S.; Costantino, D.; Scaringi, D. Data for 3D reconstruction and point cloud classification using machine learning in cultural heritage environment. Data Brief 2022, 42, 108250. [Google Scholar]
- Kambau, R.A.; Hasibuan, Z.A.; Pratama, M.O. Classification for multiformat object of cultural heritage using deep learning. In Proceedings of the 2018 Third International Conference on Informatics and Computing (ICIC), Palembang, Indonesia, 17–18 October 2018; pp. 1–7. [Google Scholar]
- Cintas, C.; Lucena, M.; Fuertes, J.M.; Delrieux, C.; Navarro, P.; González-José, R.; Molinos, M. Automatic feature extraction and classification of Iberian ceramics based on deep convolutional networks. J. Cult. Herit. 2020, 41, 106–112. [Google Scholar] [CrossRef]
- Lu, P.; Peng, X.; Yu, J.; Peng, X. Gated CNN for visual quality assessment based on color perception. Signal Process. Image Commun. 2019, 72, 105–112. [Google Scholar] [CrossRef]
- Rachmadi, M.F.; Valdés-Hernández, M.d.C.; Agan, M.L.F.; Komura, T. Deep learning vs. conventional machine learning: Pilot study of wmh segmentation in brain MRI with absence or mild vascular pathology. J. Imaging 2017, 3, 66. [Google Scholar] [CrossRef]
- Klöppel, S.; Abdulkadir, A.; Hadjidemetriou, S.; Issleib, S.; Frings, L.; Thanh, T.N.; Mader, I.; Teipel, S.J.; Hüll, M.; Ronneberger, O. A comparison of different automated methods for the detection of white matter lesions in MRI data. NeuroImage 2011, 57, 416–422. [Google Scholar] [CrossRef]
- Leite, M.; Rittner, L.; Appenzeller, S.; Ruocco, H.H.; Lotufo, R. Etiology-based classification of brain white matter hyperintensity on magnetic resonance imaging. J. Med. Imaging 2015, 2, 014002. [Google Scholar] [CrossRef]
- Ithapu, V.; Singh, V.; Lindner, C.; Austin, B.P.; Hinrichs, C.; Carlsson, C.M.; Bendlin, B.B.; Johnson, S.C. Extracting and summarizing white matter hyperintensities using supervised segmentation methods in Alzheimer’s disease risk and ageing studies. Hum. Brain Mapp. 2014, 35, 4219–4235. [Google Scholar] [CrossRef] [PubMed]
- Kamnitsas, K.; Ledig, C.; Newcombe, V.F.; Simpson, J.P.; Kane, A.D.; Menon, D.K.; Rueckert, D.; Glocker, B. Efficient multi-scale 3D {CNN} with fully connected {CRF} for accurate brain lesion segmentation. Med. Image Anal. 2017, 36, 61–78. [Google Scholar] [CrossRef] [PubMed]
- Brosch, T.; Tang, L.Y.; Yoo, Y.; Li, D.K.; Traboulsee, A.; Tam, R. Deep 3D convolutional encoder networks with shortcuts for multiscale feature integration applied to multiple sclerosis lesion segmentation. IEEE Trans. Med. Imaging 2016, 35, 1229–1239. [Google Scholar] [CrossRef] [PubMed]
- Brosch, T.; Yoo, Y.; Tang, L.Y.; Li, D.K.; Traboulsee, A.; Tam, R. Deep convolutional encoder networks for multiple sclerosis lesion segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; Springer: Cham, Switzerland, 2015; pp. 3–11. [Google Scholar]
- Zhang, Y.; Jiang, H.; Ye, T.; Juhas, M. Deep learning for imaging and detection of microorganisms. Trends Microbiol. 2021, 29, 569–572. [Google Scholar] [CrossRef] [PubMed]
- Lugagne, B.; Lin, H.; Dunlop, M.J. DeLTA: Automated cell segmentation, tracking, and lineage reconstruction using deep learning. PLoS Comput. Biol. 2020, 16, e1007673. [Google Scholar] [CrossRef]
- Micieli, D.; Minniti, T.; Evans, L.M.; Gorini, G. Accelerating Neutron tomography experiments through Artificial Neural Network based reconstruction. Sci. Rep. 2019, 9, 2450. [Google Scholar] [CrossRef]
- Venkatakrishnan, S.; Ziabari, A.; Hinkle, J.; Needham, A.W.; Warren, J.M.; Bilheux, H.Z. Convolutional neural network based non-iterative reconstruction for accelerating neutron tomography. Mach. Learn. Sci. Technol. 2021, 2, 025031. [Google Scholar] [CrossRef]
- Lehmann, E.H.; Peetermans, S.; Josic, L.; Leber, H.; van Swygenhoven, H. Energy-selective neutron imaging with high spatial resolution and its impact on the study of crystalline-structured materials. Nucl. Instrum. Methods Phys. Res. Sect. A Accel. Spectrometers Detect. Assoc. Equip. 2014, 735, 102–109. [Google Scholar] [CrossRef]
- Kamiyama, T.; Hirano, K.; Sato, H.; Ono, K.; Suzuki, Y.; Ito, D.; Saito, Y. Application of Machine Learning Methods to Neutron Transmission Spectroscopic Imaging for Solid–Liquid Phase Fraction Analysis. Appl. Sci. 2021, 11, 5988. [Google Scholar] [CrossRef]
- Venkatakrishnan, S.; Zhang, Y.; Dessieux, L.; Hoffmann, C.; Bingham, P.; Bilheux, H. Improved Acquisition and Reconstruction for Wavelength-Resolved Neutron Tomography. J. Imaging 2021, 7, 10. [Google Scholar] [CrossRef]
- Mannes, D.; Lehmann, E.H. Neutron Imaging of Cultural Heritage Objects. In Handbook of Cultural Heritage Analysis; Springer International Publishing: Cham, Switzerland, 2022; pp. 211–237. [Google Scholar]
- Xu, K.; Tremsin, A.S.; Li, J.; Ushizima, D.M.; Davy, C.A.; Bouterf, A.; Su, Y.T.; Marroccoli, M.; Mauro, A.M.; Osanna, M.; et al. Microstructure and water absorption of ancient concrete from Pompeii: An integrated synchrotron microtomography and neutron radiography characterization. Cem. Concr. Res. 2021, 139, 106282. [Google Scholar] [CrossRef]
- Rymarczyk, T.; Kłosowski, G.; Hoła, A.; Sikora, J.; Wołowiec, T.; Tchórzewski, P.; Skowron, S. Comparison of machine learning methods in electrical tomography for detecting moisture in building walls. Energies 2021, 14, 2777. [Google Scholar] [CrossRef]
- Du, G.; Cao, X.; Liang, J.; Chen, X.; Zhan, Y. Medical image segmentation based on u-net: A review. J. Imaging Sci. Technol. 2020, 64, 1–12. [Google Scholar] [CrossRef]
- Zhang, M.; Li, W.; Du, Q.; Gao, L.; Zhang, B. Feature extraction for classification of hyperspectral and LiDAR data using patch-to-patch CNN. IEEE Trans. Cybern. 2018, 50, 100–111. [Google Scholar] [CrossRef] [PubMed]
| Method | Operation | Aim | Improvement | Limitation |
|---|---|---|---|---|
| CNN | Feature extraction, modeling, and fine-tuning | Early detect disease | Noise reduction | Preprocessing phase |
| SVR *, LR *, RF *, IGB *, CNN | Feature extraction | Predict petrophysical properties | Computation time reduction | A large calibration dataset is required |
| GLCM *, LBPs *, k-NN * | Feature extraction and classification | Material identification | Classification accuracies | Pretreatment data |
| Method | Dataset | Aim | Limitation | CH in NI |
|---|---|---|---|---|
| SVM | Digitized documents | Handwriting recognition | No replicability for other datasets |  |
| RF | Photogrammetric images | Features classification and segmentation for 3D reconstruction | Several steps |  |
| CNN | Binary profile images | Pottery classification | Resizing images |  |
| Method | Aim | Improvement | Challenge |
|---|---|---|---|
| CNNs | Automatic detection of microorganisms | Accuracy (quantification) and speed of diagnosis | Transforming lower resolution into super-resolution images and constructing 3D images such as with fluorescence microscopy |
| Virus classification (structure, size, and morphology) | Fast and cost-efficient classification | ||
| U-Net | Segmentation of images | Detection and counting of colonies |
| Method | NI Technique | Aim | Applicability to CH-NI | Improvement |
|---|---|---|---|---|
| CNN-GMSD * | Radiography | Extracting features |  | Classification of materials |
| NN-FBP * | Tomography | Reducing data acquisition time and data storage |  | Especially for active materials |
| CNN-GAN * | Interferometric phase-contrast imaging | Improving resolution and the sensitivity |  | Shapes complex |
| pCNN, DnCNN, CAE * | Reflectometry | Extracting hidden profiles in large statistical noise |  | Multimaterial, different shapes |
| Material Investigated | Aim | Challenge | DL Applicable | Improvement |
|---|---|---|---|---|
| Metallic statue | Origin, purpose, manufacturing process, provenance | Quantitative purpose (filled voxels are hardly considered) | CNNs/U-Net | Automatic counting of the voxels |
| restoration and conservation purpose (distinguishing different corrosion zones) | Classification of different corrosion products | CNNs/Patch-CNN | Detection and quantification of the damaged products by automatic segmentation | |
| Ancient concrete | Studying the microstructure, cracks, and water adsorption | Dynamics of water penetration | U-Net/SBL * | High-resolution estimation |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Scatigno, C.; Festa, G. Neutron Imaging and Learning Algorithms: New Perspectives in Cultural Heritage Applications. J. Imaging 2022, 8, 284. https://doi.org/10.3390/jimaging8100284
Scatigno C, Festa G. Neutron Imaging and Learning Algorithms: New Perspectives in Cultural Heritage Applications. Journal of Imaging. 2022; 8(10):284. https://doi.org/10.3390/jimaging8100284
Chicago/Turabian StyleScatigno, Claudia, and Giulia Festa. 2022. "Neutron Imaging and Learning Algorithms: New Perspectives in Cultural Heritage Applications" Journal of Imaging 8, no. 10: 284. https://doi.org/10.3390/jimaging8100284
APA StyleScatigno, C., & Festa, G. (2022). Neutron Imaging and Learning Algorithms: New Perspectives in Cultural Heritage Applications. Journal of Imaging, 8(10), 284. https://doi.org/10.3390/jimaging8100284








