A Survey of Brain Tumor Segmentation and Classification Algorithms
Abstract
:1. Introduction
1.1. Brain Tumor Imaging Modalities
1.1.1. Magnetic Resource Imaging
1.1.2. Computed Tomography Imaging
2. Related Works
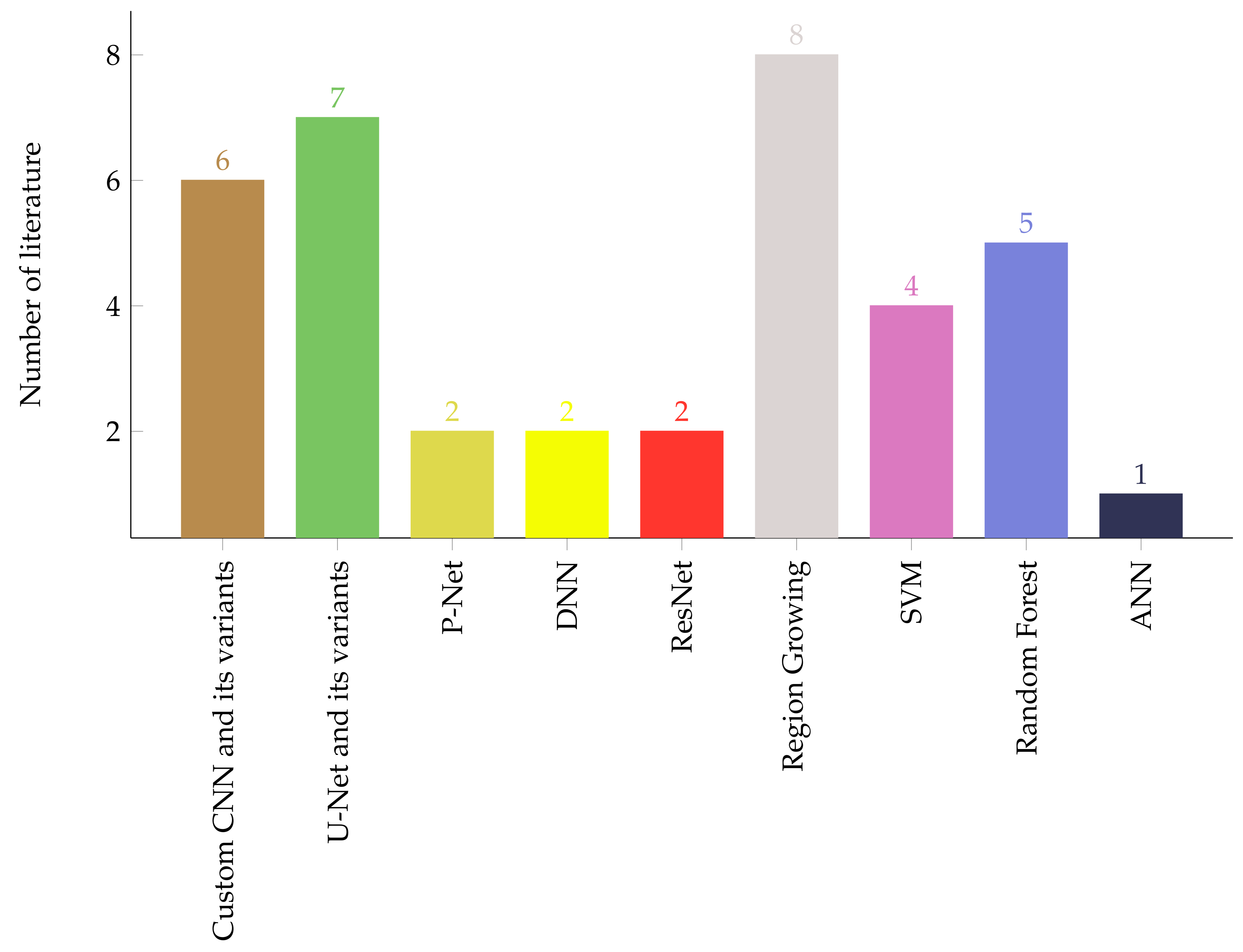
3. Method
| Algorithm 1 Paper search strategy from different search databases. |
|
4. Performance Measuring Metrics
5. Brain Tumor Segmentation Methods
5.1. Region-Based and Shallow Unsupervised Machine Learning Approach
5.2. Supervised Shallow Machine Learning Based Approach
5.3. Deep Learning-Based Approach
6. Brain Tumor Classification Methods
6.1. Conventional Machine Learning Based Approach
6.1.1. Pre-processing
6.1.2. Region of Interest (ROI) Detection
6.1.3. Feature Extraction
6.1.4. Classification
6.2. Deep Learning Approach
7. Discussion
Challenges in Automatic Brain Tumor Segmentation and Classification
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Afework, Y.K.; Debelee, T.G. Detection of Bacterial Wilt on Enset Crop Using Deep Learning Approach. Int. J. Eng. Res. Afr. 2020, 51, 131–146. [Google Scholar] [CrossRef]
- Debelee, T.G.; Schwenker, F.; Ibenthal, A.; Yohannes, D. Survey of deep learning in breast cancer image analysis. Evol. Syst. 2019, 11, 143–163. [Google Scholar] [CrossRef]
- Debelee, T.G.; Kebede, S.R.; Schwenker, F.; Shewarega, Z.M. Deep Learning in Selected Cancers’ Image Analysis—A Survey. J. Imaging 2020, 6, 121. [Google Scholar] [CrossRef]
- Debelee, T.G.; Amirian, M.; Ibenthal, A.; Palm, G.; Schwenker, F. Classification of Mammograms Using Convolutional Neural Network Based Feature Extraction. In Lecture Notes of the Institute for Computer Sciences, Social Informatics and Telecommunications Engineering; Springer International Publishing: Berlin/Heidelberg, Germany, 2018; pp. 89–98. [Google Scholar] [CrossRef]
- Debelee, T.G.; Gebreselasie, A.; Schwenker, F.; Amirian, M.; Yohannes, D. Classification of Mammograms Using Texture and CNN Based Extracted Features. J. Biomimetics Biomater. Biomed. Eng. 2019, 42, 79–97. [Google Scholar] [CrossRef]
- Debelee, T.G.; Schwenker, F.; Rahimeto, S.; Yohannes, D. Evaluation of modified adaptive k-means segmentation algorithm. Comput. Vis. Media 2019, 5, 347–361. [Google Scholar] [CrossRef] [Green Version]
- Kebede, S.R.; Debelee, T.G.; Schwenker, F.; Yohannes, D. Classifier Based Breast Cancer Segmentation. J. Biomimetics Biomater. Biomed. Eng. 2020, 47, 41–61. [Google Scholar] [CrossRef]
- Megersa, Y.; Alemu, G. Brain tumor detection and segmentation using hybrid intelligent algorithms. In Proceedings of the AFRICON 2015, Addis Ababa, Ethiopia, 14–17 September 2015. [Google Scholar] [CrossRef]
- Roberts, T.A.; Hyare, H.; Agliardi, G.; Hipwell, B.; d’Esposito, A.; Ianus, A.; Breen-Norris, J.O.; Ramasawmy, R.; Taylor, V.; Atkinson, D.; et al. Noninvasive diffusion magnetic resonance imaging of brain tumour cell size for the early detection of therapeutic response. Sci. Rep. 2020, 10. [Google Scholar] [CrossRef]
- Villanueva-Meyer, J.E.; Mabray, M.C.; Cha, S. Current Clinical Brain Tumor Imaging. Neurosurgery 2017, 81, 397–415. [Google Scholar] [CrossRef] [Green Version]
- Rosenbloom, M.J.; Pfefferbaum, A. Magnetic resonance imaging of the living brain: Evidence for brain degeneration among alcoholics and recovery with abstinence. Alcohol Res. Health J. Natl. Inst. Alcohol Abus. Alcohol. 2008, 31, 362–376. [Google Scholar]
- Noback, C.R.; Strominger, N.L.; Demarest, R.J.; Ruggiero, A.D. The Human Nervous System: Structure and Function; Humana Press: Totowa, NJ, USA, 2005. [Google Scholar]
- Louis, D.N.; Ohgaki, H.; Wiestler, O.D.; Cavenee, W.K. WHO Classification of Tumors of the Central Nervous System; International Agency for Research on Cancer (IARC): Lyon, France, 2007. [Google Scholar]
- Kayode, A.A.; Shahzadi, A.; Akram, M.; Anwar, H.; Kayode, O.T.; Akinnawo, O.O.; Okoh, S.O. Brain Tumor: An overview of the basic clinical manifestations and treatment. Glob. J. Cancer Ther. 2020, 2020, 38–41. [Google Scholar] [CrossRef]
- Johnson, D.R.; Guerin, J.B.; Giannini, C.; Morris, J.M.; Eckel, L.J.; Kaufmann, T.J. 2016 Updates to the WHO Brain Tumor Classification System: What the Radiologist Needs to Know. RadioGraphics 2017, 37, 2164–2180. [Google Scholar] [CrossRef] [Green Version]
- Roth, P.; Pace, A.; Rhun, E.L.; Weller, M.; Ay, C.; Moyal, E.C.J.; Coomans, M.; Giusti, R.; Jordan, K.; Nishikawa, R.; et al. Neurological and vascular complications of primary and secondary brain tumours: EANO-ESMO Clinical Practice Guidelines for prophylaxis, diagnosis, treatment and follow-up. Ann. Oncol. 2021, 32, 171–182. [Google Scholar] [CrossRef]
- Buckner, J.C.; Brown, P.D.; O’Neill, B.P.; Meyer, F.B.; Wetmore, C.J.; Uhm, J.H. Central Nervous System Tumors. In Mayo Clinic Proceedings; Elsevier: Amsterdam, The Netherlands, 2007; Volume 82, pp. 1271–1286. [Google Scholar]
- Smithuis, R. Neuroradiology: Brain Index. Available online: https://radiologyassistant.nl/neuroradiology/brain (accessed on 3 March 2021).
- Alves, A.F.F.; de Arruda Miranda, J.R.; Reis, F.; de Souza, S.A.S.; Alves, L.L.R.; de Moura Feitoza, L.; de Souza de Castro, J.T.; de Pina, D.R. Inflammatory lesions and brain tumors: Is it possible to differentiate them based on texture features in magnetic resonance imaging? J. Venom. Anim. Toxins Incl. Trop. Dis. 2020, 26. [Google Scholar] [CrossRef] [PubMed]
- Kasban, H. A Comparative Study of Medical Imaging Techniques. Int. J. Inf. Sci. Intell. Syst. 2015, 4, 37–58. [Google Scholar]
- Ammari, S.; Pitre-Champagnat, S.; Dercle, L.; Chouzenoux, E.; Moalla, S.; Reuze, S.; Talbot, H.; Mokoyoko, T.; Hadchiti, J.; Diffetocq, S.; et al. Influence of Magnetic Field Strength on Magnetic Resonance Imaging Radiomics Features in Brain Imaging, an In Vitro and In Vivo Study. Front. Oncol. 2021, 10. [Google Scholar] [CrossRef] [PubMed]
- Rajasekaran, K.A.; Gounder, C.C. Advanced Brain Tumour Segmentation from MRI Images. In High-Resolution Neuroimaging—Basic Physical Principles and Clinical Applications; InTech: Vienna, Austria, 2018. [Google Scholar] [CrossRef] [Green Version]
- Foltz, W.D.; Jaffray, D.A. Principles of Magnetic Resonance Imaging. Radiat. Res. 2012, 177, 331–348. [Google Scholar] [CrossRef] [PubMed]
- Hornark, J.P. The Basics of MRI. Available online: http://www.cis.rit.edu/htbooks/mri (accessed on 20 March 2021).
- Mustafa, W.F.; Abbas, M.; Elsorougy, L. Role of diffusion-weighted imaging in differentiation between posterior fossa brain tumors. Egypt. J. Neurol. Psychiatry Neurosurg. 2020, 56. [Google Scholar] [CrossRef]
- Salama, G.R.; Heier, L.A.; Patel, P.; Ramakrishna, R.; Magge, R.; Tsiouris, A.J. Diffusion Weighted/Tensor Imaging, Functional MRI and Perfusion Weighted Imaging in Glioblastoma—Foundations and Future. Front. Neurol. 2018, 8. [Google Scholar] [CrossRef] [Green Version]
- Fink, J.R.; Muzi, M.; Peck, M.; Krohn, K.A. Multimodality Brain Tumor Imaging: MR Imaging, PET, and PET/MR Imaging. J. Nucl. Med. 2015, 56, 1554–1561. [Google Scholar] [CrossRef] [Green Version]
- Luo, Q.; Li, Y.; Luo, L.; Diao, W. Comparisons of the accuracy of radiation diagnostic modalities in brain tumor. Medicine 2018, 97, e11256. [Google Scholar] [CrossRef]
- Sharma, P.; Shukla, A.P. A Review on Brain Tumor Segmentation and Classification for MRI Images. In Proceedings of the 2021 International Conference on Advance Computing and Innovative Technologies in Engineering (ICACITE), Greater Noida, India, 30–31 December 2021. [Google Scholar] [CrossRef]
- Rao, C.S.; Karunakara, K. A comprehensive review on brain tumor segmentation and classification of MRI images. Multimed. Tools Appl. 2021, 80, 17611–17643. [Google Scholar] [CrossRef]
- Magadza, T.; Viriri, S. Deep Learning for Brain Tumor Segmentation: A Survey of State-of-the-Art. J. Imaging 2021, 7, 19. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, A.; Srivastava, S.; Pant, M. Brain tumor segmentation and classification from magnetic resonance images: Review of selected methods from 2014 to 2019. Pattern Recognit. Lett. 2020, 131, 244–260. [Google Scholar] [CrossRef]
- Kumari, N.; Saxena, S. Review of Brain Tumor Segmentation and Classification. In Proceedings of the 2018 International Conference on Current Trends towards Converging Technologies (ICCTCT), Coimbatore, India, 1–3 March 2018. [Google Scholar] [CrossRef]
- Meier, R.; Knecht, U.; Loosli, T.; Bauer, S.; Slotboom, J.; Wiest, R.; Reyes, M. Clinical Evaluation of a Fully-automatic Segmentation Method for Longitudinal Brain Tumor Volumetry. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef] [Green Version]
- Pohle, R.; Toennies, K.D. Segmentation of medical images using adaptive region growing. In Medical Imaging 2001: Image Processing; Sonka, M., Hanson, K.M., Eds.; SPIE: Bellingham, WA, USA, 2001. [Google Scholar] [CrossRef] [Green Version]
- Dey, N.; Ashour, A.S. Computing in Medical Image Analysis. In Soft Computing Based Medical Image Analysis; Elsevier: Amsterdam, The Netherlands, 2018; pp. 3–11. [Google Scholar] [CrossRef]
- Dhanachandra, N.; Manglem, K.; Chanu, Y.J. Image Segmentation Using K -means Clustering Algorithm and Subtractive Clustering Algorithm. Procedia Comput. Sci. 2015, 54, 764–771. [Google Scholar] [CrossRef] [Green Version]
- Hooda, H.; Verma, O.P.; Singhal, T. Brain tumor segmentation: A performance analysis using K-Means, Fuzzy C-Means and Region growing algorithm. In Proceedings of the 2014 IEEE International Conference on Advanced Communications, Control and Computing Technologies, Ramanathapuram, India, 8–10 May 2014. [Google Scholar] [CrossRef]
- Bal, A.; Banerjee, M.; Sharma, P.; Maitra, M. Brain Tumor Segmentation on MR Image Using K-Means and Fuzzy-Possibilistic Clustering. In Proceedings of the 2018 2nd International Conference on Electronics, Materials Engineering & Nano-Technology (IEMENTech), Kolkata, India, 4–5 April 2018. [Google Scholar] [CrossRef]
- Kumar, D.V.; Krishniah, V.J.R. Segmentation of Brain Tumor Using K-Means Clustering Algorithm. J. Eng. Appl. Sci. 2018, 13, 3942–3945. [Google Scholar]
- Selvakumar, J.; Lakshmi, A.; Arivoli, T. Brain tumor segmentation and its area calculation in brain MR images using K-mean clustering and Fuzzy C-mean algorithm. In Proceedings of the IEEE-International Conference on Advances in Engineering, Science And Management (ICAESM-2012), Nagapattinam, India, 30–31 March 2012; pp. 186–190. [Google Scholar]
- Shanker, R.; Singh, R.; Bhattacharya, M. Segmentation of tumor and edema based on K-mean clustering and hierarchical centroid shape descriptor. In Proceedings of the 2017 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Kansas City, MO, USA, 13–16 November 2017; pp. 1105–1109. [Google Scholar] [CrossRef]
- Kaur, N.; Sharma, M. Brain tumor detection using self-adaptive K-means clustering. In Proceedings of the 2017 International Conference on Energy, Communication, Data Analytics and Soft Computing (ICECDS), Chennai, India, 1–2 August 2017; pp. 1861–1865. [Google Scholar] [CrossRef]
- Mahmud, M.R.; Mamun, M.A.; Hossain, M.A.; Uddin, M.P. Comparative Analysis of K-Means and Bisecting K-Means Algorithms for Brain Tumor Detection. In Proceedings of the 2018 International Conference on Computer, Communication, Chemical, Material and Electronic Engineering (IC4ME2), Rajshahi, Bangladesh, 8–9 February 2018; pp. 1–4. [Google Scholar] [CrossRef]
- Shasidhar, M.; Raja, V.S.; Kumar, B.V. MRI Brain Image Segmentation Using Modified Fuzzy C-Means Clustering Algorithm. In Proceedings of the 2011 International Conference on Communication Systems and Network Technologies, Katra, India, 3–5 June 2011; pp. 473–478. [Google Scholar] [CrossRef]
- Agrawal, R.; Sharma, M.; Singh, B.K. Segmentation of Brain Tumour Based on Clustering Technique: Performance Analysis. J. Intell. Syst. 2019, 28, 291–306. [Google Scholar] [CrossRef]
- Pitchai, R.; Supraja, P.; Victoria, A.H.; Madhavi, M. Brain Tumor Segmentation Using Deep Learning and Fuzzy K-Means Clustering for Magnetic Resonance Images. Neural Process. Lett. 2020. [Google Scholar] [CrossRef]
- Almahfud, M.A.; Setyawan, R.; Sari, C.A.; Setiadi, D.R.I.M.; Rachmawanto, E.H. An Effective MRI Brain Image Segmentation using Joint Clustering (K-Means and Fuzzy C-Means). In Proceedings of the 2018 International Seminar on Research of Information Technology and Intelligent Systems (ISRITI), Yogyakarta, Indonesia, 21–22 November 2018; pp. 11–16. [Google Scholar] [CrossRef]
- Abdel-Maksoud, E.; Elmogy, M.; Al-Awadi, R. Brain tumor segmentation based on a hybrid clustering technique. Egypt. Informatics J. 2015, 16, 71–81. [Google Scholar] [CrossRef]
- Mannor, S.; Jin, X.; Han, J.; Jin, X.; Han, J.; Jin, X.; Han, J.; Zhang, X. K-Medoids Clustering. In Encyclopedia of Machine Learning; Springer: New York, NY, USA, 2011; pp. 564–565. [Google Scholar] [CrossRef]
- Bezdek, J.C.; Hall, L.O.; Clarke, L.P. Review of MR image segmentation techniques using pattern recognition. Med. Phys. 1993, 20, 1033–1048. [Google Scholar] [CrossRef]
- Blessy, S.A.P.S.; Sulochana, C.H. Performance analysis of unsupervised optimal fuzzy clustering algorithm for MRI brain tumor segmentation. Technol. Health Care 2014, 23, 23–35. [Google Scholar] [CrossRef] [PubMed]
- Arakeri, M.P.; Reddy, G.R.M. Efficient Fuzzy Clustering Based Approach to Brain Tumor Segmentation on MR Images. In Communications in Computer and Information Science; Springer: Berlin/Heidelberg, Germany, 2011; pp. 790–795. [Google Scholar] [CrossRef]
- Dubey, Y.K.; Mushrif, M.M. FCM Clustering Algorithms for Segmentation of Brain MR Images. Adv. Fuzzy Syst. 2016, 2016, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Badmera, M.S.; Nilawar, A.P.; Karwankar, A.R. Modified FCM approach for MR brain image segmentation. In Proceedings of the 2013 International Conference on Circuits, Power and Computing Technologies (ICCPCT), Nagercoil, India, 20–21 March 2013; pp. 891–896. [Google Scholar] [CrossRef]
- Sheela, C.J.J.; Suganthi, G. Automatic Brain Tumor Segmentation from MRI using Greedy Snake Model and Fuzzy C-Means Optimization. J. King Saud Univ. Comput. Inf. Sci. 2019. [Google Scholar] [CrossRef]
- Wang, Y. Tutorial: Image Segmentation; Graduate Institute of Communication Engineering National Taiwan University: Taipei, Taiwan, 2010. [Google Scholar]
- Rajinikanth, V.; Fernandes, S.L.; Bhushan, B.; Harisha; Sunder, N.R. Segmentation and Analysis of Brain Tumor Using Tsallis Entropy and Regularised Level Set. Proceedings of 2nd International Conference on Micro-Electronics, Electromagnetics and Telecommunications, Singapore, 7 September 2017; Springer: Singapore; pp. 313–321. [Google Scholar] [CrossRef]
- Cabria, I.; Gondra, I. Automated Localization of Brain Tumors in MRI Using Potential-K-Means Clustering Algorithm. In Proceedings of the 2015 12th Conference on Computer and Robot Vision, Halifax, NS, Canada, 3–5 June 2015; pp. 125–132. [Google Scholar] [CrossRef]
- Suraj, N.S.S.K.; Muppalla, V.; Sanghani, P.; Ren, H. Comparative Study of Unsupervised Segmentation Algorithms for Delineating Glioblastoma Multiforme Tumour. In Proceedings of the 2018 3rd International Conference on Advanced Robotics and Mechatronics (ICARM), Singapore, 18–20 July 2018; pp. 468–473. [Google Scholar] [CrossRef]
- Mehidi, I.; Belkhiat, D.E.C.; Jabri, D. An Improved Clustering Method Based on K-Means Algorithm for MRI Brain Tumor Segmentation. In Proceedings of the 2019 6th International Conference on Image and Signal Processing and their Applications (ISPA), Mostaganem, Algeria, 24–25 November 2019; pp. 1–4. [Google Scholar] [CrossRef]
- Rundo, L.; Militello, C.; Tangherloni, A.; Russo, G.; Vitabile, S.; Gilardi, M.C.; Mauri, G. NeXt for neuro-radiosurgery: A fully automatic approach for necrosis extraction in brain tumor MRI using an unsupervised machine learning technique. Int. J. Imaging Syst. Technol. 2017, 28, 21–37. [Google Scholar] [CrossRef]
- Chandra, G.R.; Rao, K.R.H. Tumor Detection In Brain Using Genetic Algorithm. Procedia Comput. Sci. 2016, 79, 449–457. [Google Scholar] [CrossRef] [Green Version]
- Rundo, L.; Militello, C.; Russo, G.; Vitabile, S.; Gilardi, M.C.; Mauri, G. GTVcut for neuro-radiosurgery treatment planning: An MRI brain cancer seeded image segmentation method based on a cellular automata model. Nat. Comput. 2017, 17, 521–536. [Google Scholar] [CrossRef]
- Ayachi, R.; Ben Amor, N. Brain Tumor Segmentation Using Support Vector Machines. In Symbolic and Quantitative Approaches to Reasoning with Uncertainty; Sossai, C., Chemello, G., Eds.; Springer: Berlin/Heidelberg, Germany, 2009; pp. 736–747. [Google Scholar]
- Cui, B.; Xie, M.; Wang, C. A Deep Convolutional Neural Network Learning Transfer to SVM-Based Segmentation Method for Brain Tumor. In Proceedings of the 2019 IEEE 11th International Conference on Advanced Infocomm Technology (ICAIT), Jinan, China, 18–20 October 2019; pp. 1–5. [Google Scholar] [CrossRef]
- Zhang, N.; Ruan, S.; Lebonvallet, S.; Liao, Q.; Zhu, Y. Multi-kernel SVM based classification for brain tumor segmentation of MRI multi-sequence. In Proceedings of the 2009 16th IEEE International Conference on Image Processing (ICIP), Cairo, Egypt, 7–10 November 2009; pp. 3373–3376. [Google Scholar] [CrossRef]
- Chen, W.; Qiao, X.; Liu, B.; Qi, X.; Wang, R.; Wang, X. Automatic brain tumor segmentation based on features of separated local square. In Proceedings of the 2017 Chinese Automation Congress (CAC), Jinan, China., 20–22 October 2017. [Google Scholar] [CrossRef]
- Chithambaram, T.; Perumal, K. Brain tumor segmentation using genetic algorithm and ANN techniques. In Proceedings of the 2017 IEEE International Conference on Power, Control, Signals and Instrumentation Engineering (ICPCSI), Chennai, India, 21–22 September 2017. [Google Scholar] [CrossRef]
- Bougacha, A.; Boughariou, J.; Slima, M.B.; Hamida, A.B.; Mahfoudh, K.B.; Kammoun, O.; Mhiri, C. Comparative study of supervised and unsupervised classification methods: Application to automatic MRI glioma brain tumors segmentation. In Proceedings of the 2018 4th International Conference on Advanced Technologies for Signal and Image Processing (ATSIP), Tunisia, 21–24 March 2018. [Google Scholar] [CrossRef]
- Ma, C.; Luo, G.; Wang, K. Concatenated and Connected Random Forests With Multiscale Patch Driven Active Contour Model for Automated Brain Tumor Segmentation of MR Images. IEEE Trans. Med. Imaging 2018, 37, 1943–1954. [Google Scholar] [CrossRef]
- Tang, H.; Lu, H.; Liu, W.; Tao, X. Tumor segmentation from single contrast MR images of human brain. In Proceedings of the 2015 IEEE 12th International Symposium on Biomedical Imaging (ISBI), New York, NY, USA, 16–19 April 2015. [Google Scholar] [CrossRef]
- Csaholczi, S.; Kovacs, L.; Szilagyi, L. Automatic Segmentation of Brain Tumor Parts from MRI Data Using a Random Forest Classifier. In Proceedings of the 2021 IEEE 19th World Symposium on Applied Machine Intelligence and Informatics (SAMI), Herl’any, Slovakia, 21–23 January 2021. [Google Scholar] [CrossRef]
- Pinto, A.; Pereira, S.; Dinis, H.; Silva, C.A.; Rasteiro, D.M.L.D. Random decision forests for automatic brain tumor segmentation on multi-modal MRI images. In Proceedings of the 2015 IEEE 4th Portuguese Meeting on Bioengineering (ENBENG), Porto, Portugal, 26–28 February 2015. [Google Scholar] [CrossRef]
- Hatami, T.; Hamghalam, M.; Reyhani-Galangashi, O.; Mirzakuchaki, S. A Machine Learning Approach to Brain Tumors Segmentation Using Adaptive Random Forest Algorithm. In Proceedings of the 2019 5th Conference on Knowledge Based Engineering and Innovation (KBEI), Tehran, Iran, 28 February–1 March 2019. [Google Scholar] [CrossRef]
- Fulop, T.; Gyorfi, A.; Csaholczi, S.; Kovacs, L.; Szilagyi, L. Brain Tumor Segmentation from Multi-Spectral MRI Data Using Cascaded Ensemble Learning. In Proceedings of the 2020 IEEE 15th International Conference of System of Systems Engineering (SoSE), Hungary, 2–4 June 2020 Budapest. [Google Scholar] [CrossRef]
- Bakas, S.; Akbari, H.; Sotiras, A.; Bilello, M.; Rozycki, M.; Kirby, J.S.; Freymann, J.B.; Farahani, K.; Davatzikos, C. Advancing The Cancer Genome Atlas glioma MRI collections with expert segmentation labels and radiomic features. Sci. Data 2017, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bakas, S.; Akbari, H.; Sotiras, A.; Bilello, M.; Rozycki, M.; Kirby, J.; Freymann, J.; Farahani, K.; Davatzikos, C. Segmentation Labels for the Pre-operative Scans of the TCGA-GBM collection. Cancer Imaging Arch. 2017. [Google Scholar] [CrossRef]
- Tobon-Gomez, C.; Geers, A.J.; Peters, J.; Weese, J.; Pinto, K.; Karim, R.; Ammar, M.; Daoudi, A.; Margeta, J.; Sandoval, Z.; et al. Benchmark for Algorithms Segmenting the Left Atrium From 3D CT and MRI Datasets. IEEE Trans. Med Imaging 2015, 34, 1460–1473. [Google Scholar] [CrossRef]
- Menze, B.H.; Jakab, A.; Bauer, S.; Kalpathy-Cramer, J.; Farahani, K.; Kirby, J.; Burren, Y.; Porz, N.; Slotboom, J.; Wiest, R.; et al. The Multimodal Brain Tumor Image Segmentation Benchmark (BRATS). IEEE Trans. Med Imaging 2015, 34, 1993–2024. [Google Scholar] [CrossRef] [PubMed]
- Pereira, S.; Pinto, A.; Alves, V.; Silva, C.A. Brain Tumor Segmentation Using Convolutional Neural Networks in MRI Images. IEEE Trans. Med. Imaging 2016, 35, 1240–1251. [Google Scholar] [CrossRef] [PubMed]
- Deng, W.; Shi, Q.; Wang, M.; Zheng, B.; Ning, N. Deep Learning-Based HCNN and CRF-RRNN Model for Brain Tumor Segmentation. IEEE Access 2020, 8, 26665–26675. [Google Scholar] [CrossRef]
- Ding, Y.; Li, C.; Yang, Q.; Qin, Z.; Qin, Z. How to Improve the Deep Residual Network to Segment Multi-Modal Brain Tumor Images. IEEE Access 2019, 7, 152821–152831. [Google Scholar] [CrossRef]
- Ding, Y.; Chen, F.; Zhao, Y.; Wu, Z.; Zhang, C.; Wu, D. A Stacked Multi-Connection Simple Reducing Net for Brain Tumor Segmentation. IEEE Access 2019, 7, 104011–104024. [Google Scholar] [CrossRef]
- Ali, M.; Gilani, S.O.; Waris, A.; Zafar, K.; Jamil, M. Brain Tumour Image Segmentation Using Deep Networks. IEEE Access 2020, 8, 153589–153598. [Google Scholar] [CrossRef]
- Razzak, M.I.; Imran, M.; Xu, G. Efficient Brain Tumor Segmentation With Multiscale Two-Pathway-Group Conventional Neural Networks. IEEE J. Biomed. Health Inform. 2019, 23, 1911–1919. [Google Scholar] [CrossRef]
- Aboelenein, N.M.; Songhao, P.; Koubaa, A.; Noor, A.; Afifi, A. HTTU-Net: Hybrid Two Track U-Net for Automatic Brain Tumor Segmentation. IEEE Access 2020, 8, 101406–101415. [Google Scholar] [CrossRef]
- Wang, G.; Li, W.; Zuluaga, M.A.; Pratt, R.; Patel, P.A.; Aertsen, M.; Doel, T.; David, A.L.; Deprest, J.; Ourselin, S.; et al. Interactive Medical Image Segmentation Using Deep Learning With Image-Specific Fine Tuning. IEEE Trans. Med. Imaging 2018, 37, 1562–1573. [Google Scholar] [CrossRef]
- Hao, J.; Li, X.; Hou, Y. Magnetic Resonance Image Segmentation Based on Multi-Scale Convolutional Neural Network. IEEE Access 2020, 8, 65758–65768. [Google Scholar] [CrossRef]
- Zhou, T.; Canu, S.; Ruan, S. Fusion based on attention mechanism and context constraint for multi-modal brain tumor segmentation. Comput. Med Imaging Graph. 2020, 86, 101811. [Google Scholar] [CrossRef] [PubMed]
- Ye, F.; Zheng, Y.; Ye, H.; Han, X.; Li, Y.; Wang, J.; Pu, J. Parallel pathway dense neural network with weighted fusion structure for brain tumor segmentation. Neurocomputing 2021, 425, 1–11. [Google Scholar] [CrossRef]
- Sun, J.; Peng, Y.; Guo, Y.; Li, D. Segmentation of the multimodal brain tumor image used the multi-pathway architecture method based on 3D FCN. Neurocomputing 2021, 423, 34–45. [Google Scholar] [CrossRef]
- Ben naceur, M.; Akil, M.; Saouli, R.; Kachouri, R. Fully automatic brain tumor segmentation with deep learning-based selective attention using overlapping patches and multi-class weighted cross-entropy. Med. Image Anal. 2020, 63, 101692. [Google Scholar] [CrossRef] [PubMed]
- Naser, M.A.; Deen, M.J. Brain tumor segmentation and grading of lower-grade glioma using deep learning in MRI images. Comput. Biol. Med. 2020, 121, 103758. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; He, Z.; Jia, Y. AFPNet: A 3D fully convolutional neural network with atrous-convolution feature pyramid for brain tumor segmentation via MRI images. Neurocomputing 2020, 402, 235–244. [Google Scholar] [CrossRef]
- Li, H.; Li, A.; Wang, M. A novel end-to-end brain tumor segmentation method using improved fully convolutional networks. Comput. Biol. Med. 2019, 108, 150–160. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zeng, J.; Qin, P.; Zhao, L. Brain tumor segmentation of multi-modality MR images via triple intersecting U-Nets. Neurocomputing 2021, 421, 195–209. [Google Scholar] [CrossRef]
- Xu, F.; Ma, H.; Sun, J.; Wu, R.; Liu, X.; Kong, Y. LSTM Multi-modal UNet for Brain Tumor Segmentation. In Proceedings of the 2019 IEEE 4th International Conference on Image, Vision and Computing (ICIVC), Xiamen, China, 5–7 July 2019; pp. 236–240. [Google Scholar] [CrossRef]
- Kleihues, P.; Louis, D.N.; Scheithauer, B.W.; Rorke, L.B.; Reifenberger, G.; Burger, P.C.; Cavenee, W.K. The WHO Classification of Tumors of the Nervous System. J. Neuropathol. Exp. Neurol. 2002, 61, 215–225. [Google Scholar] [CrossRef]
- Badža, M.M.; Barjaktarović, M.Č. Classification of Brain Tumors from MRI Images Using a Convolutional Neural Network. Appl. Sci. 2020, 10, 1999. [Google Scholar] [CrossRef] [Green Version]
- Tandel, G.S.; Biswas, M.; Kakde, O.G.; Tiwari, A.; Suri, H.S.; Turk, M.; Laird, J.; Asare, C.; Ankrah, A.A.; Khanna, N.N.; et al. A Review on a Deep Learning Perspective in Brain Cancer Classification. Cancers 2019, 11, 111. [Google Scholar] [CrossRef] [Green Version]
- Quon, J.; Bala, W.; Chen, L.; Wright, J.; Kim, L.; Han, M.; Shpanskaya, K.; Lee, E.; Tong, E.; Iv, M.; et al. Deep Learning for Pediatric Posterior Fossa Tumor Detection and Classification: A Multi-Institutional Study. Am. J. Neuroradiol. 2020. [Google Scholar] [CrossRef]
- Díaz-Pernas, F.J.; Martínez-Zarzuela, M.; Antón-Rodríguez, M.; González-Ortega, D. A Deep Learning Approach for Brain Tumor Classification and Segmentation Using a Multiscale Convolutional Neural Network. Healthcare 2021, 9, 153. [Google Scholar] [CrossRef] [PubMed]
- Deepak, S.; Ameer, P. Brain tumor classification using deep CNN features via transfer learning. Comput. Biol. Med. 2019, 111, 103345. [Google Scholar] [CrossRef] [PubMed]
- Paul, J.S.; Plassard, A.J.; Landman, B.A.; Fabbri, D. Deep learning for brain tumor classification. In Medical Imaging 2017: Biomedical Applications in Molecular, Structural, and Functional Imaging; Krol, A., Gimi, B., Eds.; SPIE: Bellingham, WA, USA, 2017. [Google Scholar] [CrossRef] [Green Version]
- Khan, H.A.; Jue, W.; Mushtaq, M.; Mushtaq, M.U. Brain tumor classification in MRI image using convolutional neural network. Math. Biosci. Eng. 2020, 17, 6203–6216. [Google Scholar] [CrossRef]
- Dangeti, P. Statistics for Machine Learning; Packt Publishing: Birmingham, UK, 2017. [Google Scholar]
- Ahmmed, R.; Swakshar, A.S.; Hossain, M.F.; Rafiq, M.A. Classification of tumors and it stages in brain MRI using support vector machine and artificial neural network. In Proceedings of the 2017 International Conference on Electrical, Computer and Communication Engineering (ECCE), Cox’s Bazar, Bangladesh, 16–18 Feb. 2017; pp. 229–234. [Google Scholar] [CrossRef]
- Ismael, M.R.; Abdel-Qader, I. Brain Tumor Classification via Statistical Features and Back-Propagation Neural Network. In Proceedings of the 2018 IEEE International Conference on Electro/Information Technology (EIT), Rochester, MI, USA, 3–5 May 2018; pp. 0252–0257. [Google Scholar] [CrossRef]
- Sathi, K.A.; Islam, M.S. Hybrid Feature Extraction Based Brain Tumor Classification using an Artificial Neural Network. In Proceedings of the 2020 IEEE 5th International Conference on Computing Communication and Automation (ICCCA), Greater Noida, India, 30–31 October 2020; pp. 155–160. [Google Scholar] [CrossRef]
- Shree, N.V.; Kumar, T.N.R. Identification and classification of brain tumor MRI images with feature extraction using DWT and probabilistic neural network. Brain Inform. 2018, 5, 23–30. [Google Scholar] [CrossRef] [Green Version]
- Ramdlon, R.H.; Kusumaningtyas, E.M.; Karlita, T. Brain Tumor Classification Using MRI Images with K-Nearest Neighbor Method. In Proceedings of the 2019 International Electronics Symposium (IES), Surabaya, Indonesia, 27–28 September 2019; pp. 660–667. [Google Scholar] [CrossRef]
- Garg, G.; Garg, R. Brain Tumor Detection and Classification based on Hybrid Ensemble Classifier. arXiv 2021, arXiv:2101.00216. [Google Scholar]
- Engy, N.; Salam, N.M.; Al-Atabany, W. Evaluating the Efficiency of different Feature Sets on Brain Tumor Classification in MR Images. Int. J. Comput. Appl. 2018, 180, 1–7. [Google Scholar] [CrossRef]
- Gurbina, M.; Lascu, M.; Lascu, D. Tumor Detection and Classification of MRI Brain Image using Different Wavelet Transforms and Support Vector Machines. In Proceedings of the 2019 42nd International Conference on Telecommunications and Signal Processing (TSP), Budapest, Hungary, 1–3 July 2019; pp. 505–508. [Google Scholar] [CrossRef]
- Ali, H.M. MRI Medical Image Denoising by Fundamental Filters. In High-Resolution Neuroimaging—Basic Physical Principles and Clinical Applications; InTech: Vienna, Austria, 2018. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Yang, H.; Fan, J.; Liu, R.W.; Duan, Y. Rician noise and intensity nonuniformity correction (NNC) model for MRI data. Biomed. Signal Process. Control 2019, 49, 506–519. [Google Scholar] [CrossRef]
- Ramesh, S.; Sasikala, S.; Paramanandham, N. Segmentation and classification of brain tumors using modified median noise filter and deep learning approaches. Multimed. Tools Appl. 2021, 80, 11789–11813. [Google Scholar] [CrossRef]
- Ravikumar Gurusamy, D.V.S. A Machine Learning Approach for MRI Brain Tumor Classification. Comput. Mater. Contin. 2017, 53, 91–108. [Google Scholar] [CrossRef]
- Li, M.; Wang, H.; Shang, Z.; Yang, Z.; Zhang, Y.; Wan, H. Ependymoma and pilocytic astrocytoma: Differentiation using radiomics approach based on machine learning. J. Clin. Neurosci. 2020, 78, 175–180. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, K.; Kaya, Y.; Kuncan, M.; Ertunç, H.M. Brain tumor classification using modified local binary patterns (LBP) feature extraction methods. Med. Hypotheses 2020, 139, 109696. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.; Ullah, Z.; Gwak, J. MRI-Based Brain Tumor Classification Using Ensemble of Deep Features and Machine Learning Classifiers. Sensors 2021, 21, 2222. [Google Scholar] [CrossRef]
- Amin, J.; Sharif, M.; Raza, M.; Saba, T.; Rehman, A. Brain Tumor Classification: Feature Fusion. In Proceedings of the 2019 International Conference on Computer and Information Sciences (ICCIS), Sakaka, Saudi Arabia, 3–4 April 2019; pp. 1–6. [Google Scholar] [CrossRef]
- Baranwal, S.K.; Jaiswal, K.; Vaibhav, K.; Kumar, A.; Srikantaswamy, R. Performance analysis of Brain Tumour Image Classification using CNN and SVM. In Proceedings of the 2020 Second International Conference on Inventive Research in Computing Applications (ICIRCA), Coimbatore, India, 15– 17 July 2020; pp. 537–542. [Google Scholar] [CrossRef]
- Gumaei, A.; Hassan, M.M.; Hassan, M.R.; Alelaiwi, A.; Fortino, G. A Hybrid Feature Extraction Method With Regularized Extreme Learning Machine for Brain Tumor Classification. IEEE Access 2019, 7, 36266–36273. [Google Scholar] [CrossRef]
- Minz, A.; Mahobiya, C. MR Image Classification Using Adaboost for Brain Tumor Type. In Proceedings of the 2017 IEEE 7th International Advance Computing Conference (IACC), Hyderabad, India, 5–7 January 2017; pp. 701–705. [Google Scholar] [CrossRef]
- Gayathri, S.; Wise, D.J.W.; Janani, V.; Eleaswari, M.; Hema, S. Analyzing, Detecting and Automatic Classification of Different Stages of Brain Tumor Using Region Segmentation and Support Vector Machine. In Proceedings of the 2020 International Conference on Electronics and Sustainable Communication Systems (ICESC), Coimbatore, India, 2–4 July 2020; pp. 404–408. [Google Scholar] [CrossRef]
- Sarkar, A.; Maniruzzaman, M.; Ahsan, M.S.; Ahmad, M.; Kadir, M.I.; Islam, S.M.T. Identification and Classification of Brain Tumor from MRI with Feature Extraction by Support Vector Machine. In Proceedings of the 2020 International Conference for Emerging Technology (INCET), Belgaum, India, 5–7 June 2020; pp. 1–4. [Google Scholar] [CrossRef]
- Mathew, A.R.; Anto, P.B. Tumor detection and classification of MRI brain image using wavelet transform and SVM. In Proceedings of the 2017 International Conference on Signal Processing and Communication (ICSPC), Coimbatore, India, 28–29 July 2017; pp. 75–78. [Google Scholar] [CrossRef]
- Cinarer, G.; Emiroglu, B.G. Classificatin of Brain Tumors by Machine Learning Algorithms. In Proceedings of the 2019 3rd International Symposium on Multidisciplinary Studies and Innovative Technologies (ISMSIT), Ankara, Turkey, 11–13 October 2019; pp. 1–4. [Google Scholar] [CrossRef]
- Lavanyadevi, R.; Machakowsalya, M.; Nivethitha, J.; Kumar, A.N. Brain tumor classification and segmentation in MRI images using PNN. In Proceedings of the 2017 IEEE International Conference on Electrical, Instrumentation and Communication Engineering (ICEICE), Karur, India, 27–28 April 2017; pp. 1–6. [Google Scholar] [CrossRef]
- Amin, J.; Sharif, M.; Yasmin, M.; Fernandes, S.L. A distinctive approach in brain tumor detection and classification using MRI. Pattern Recognit. Lett. 2020, 139, 118–127. [Google Scholar] [CrossRef]
- Prabha, S.; Raghav, R.; Moulya, C.; Preethi, K.G.; Sankaran, K. Fusion based Brain Tumor Classification using Multiscale Transform Methods. In Proceedings of the 2020 International Conference on Communication and Signal Processing (ICCSP), Chennai, India, 28–30 July 2020; pp. 1390–1393. [Google Scholar] [CrossRef]
- Wasule, V.; Sonar, P. Classification of brain MRI using SVM and KNN classifier. In Proceedings of the 2017 Third International Conference on Sensing, Signal Processing and Security (ICSSS), Chennai, India, 4–5 May 2017; pp. 218–223. [Google Scholar] [CrossRef]
- Sachdeva, J.; Kumar, V.; Gupta, I.; Khandelwal, N.; Ahuja, C.K. A package-SFERCB-“Segmentation, feature extraction, reduction and classification analysis by both SVM and ANN for brain tumors”. Appl. Soft Comput. 2016, 47, 151–167. [Google Scholar] [CrossRef]
- Keerthana, K.; Xavier, S. An Intelligent System for Early Assessment and Classification of Brain Tumor. In Proceedings of the 2018 Second International Conference on Inventive Communication and Computational Technologies (ICICCT), Coimbatore, India, 20–21 April 2018; pp. 1265–1268. [Google Scholar] [CrossRef]
- Yin, B.; Wang, C.; Abza, F. New brain tumor classification method based on an improved version of whale optimization algorithm. Biomed. Signal Process. Control 2020, 56, 101728. [Google Scholar] [CrossRef]
- Cheng, J. Brain Tumor Dataset. 2017. Available online: https://figshare.com/articles/dataset/brain_tumor_dataset/1512427 (accessed on 2 June 2021).
- Gaikwad, S.B.; Joshi, M.S. Brain Tumor Classification using Principal Component Analysis and Probabilistic Neural Network. Int. J. Comput. Appl. 2015, 120, 5–9. [Google Scholar] [CrossRef]
- Kumar, A.; Ashok, A.; Ansari, M.A. Brain Tumor Classification Using Hybrid Model Of PSO And SVM Classifier. In Proceedings of the 2018 International Conference on Advances in Computing, Communication Control and Networking (ICACCCN), Greater Noida, India, 12–13 October 2018; pp. 1022–1026. [Google Scholar] [CrossRef]
- Ge, C.; Gu, I.Y.H.; Jakola, A.S.; Yang, J. Enlarged Training Dataset by Pairwise GANs for Molecular-Based Brain Tumor Classification. IEEE Access 2020, 8, 22560–22570. [Google Scholar] [CrossRef]
- Bakas, S.; Akbari, H.; Sotiras, A.; Bilello, M.; Rozycki, M.; Kirby, J.; Freymann, J.; Farahani, K.; Davatzikos, C. Segmentation Labels for the Pre-operative Scans of the TCGA-LGG collection. Cancer Imaging Arch. 2017. [Google Scholar] [CrossRef]
- Sultan, H.H.; Salem, N.M.; Al-Atabany, W. Multi-Classification of Brain Tumor Images Using Deep Neural Network. IEEE Access 2019, 7, 69215–69225. [Google Scholar] [CrossRef]
- Scarpace, L.; Flanders, A.E.; Jain, R.; Mikkelsen, T.; Andrews, D.W. Data from Rembrandt. 2019. Available online: https://wiki.cancerimagingarchive.net/display/Public/REMBRANDT (accessed on 3 May 2021).
- Huang, Z.; Du, X.; Chen, L.; Li, Y.; Liu, M.; Chou, Y.; Jin, L. Convolutional Neural Network Based on Complex Networks for Brain Tumor Image Classification With a Modified Activation Function. IEEE Access 2020, 8, 89281–89290. [Google Scholar] [CrossRef]
- Afshar, P.; Mohammadi, A.; Plataniotis, K.N. BayesCap: A Bayesian Approach to Brain Tumor Classification Using Capsule Networks. IEEE Signal Process. Lett. 2020, 27, 2024–2028. [Google Scholar] [CrossRef]
- Ucuzal, H.; YASAR, S.; Colak, C. Classification of brain tumor types by deep learning with convolutional neural network on magnetic resonance images using a developed web-based interface. In Proceedings of the 2019 3rd International Symposium on Multidisciplinary Studies and Innovative Technologies (ISMSIT), Ankara, Turkey, 11–13 October 2019; pp. 1–5. [Google Scholar] [CrossRef]
- Noreen, N.; Palaniappan, S.; Qayyum, A.; Ahmad, I.; Imran, M.; Shoaib, M. A Deep Learning Model Based on Concatenation Approach for the Diagnosis of Brain Tumor. IEEE Access 2020, 8, 55135–55144. [Google Scholar] [CrossRef]
- Rehman, A.; Naz, S.; Razzak, M.I.; Akram, F.; Imran, M. A Deep Learning-Based Framework for Automatic Brain Tumors Classification Using Transfer Learning. Circuits Syst. Signal Process. 2019, 39, 757–775. [Google Scholar] [CrossRef]
- Cheng, Y.; Qin, G.; Zhao, R.; Liang, Y.; Sun, M. ConvCaps: Multi-input Capsule Network for Brain Tumor Classification. In Neural Information Processing; Springer International Publishing: Berlin/Heidelberg, Germany, 2019; pp. 524–534. [Google Scholar] [CrossRef]
- Kurup, R.V.; Sowmya, V.; Soman, K.P. Effect of Data Pre-processing on Brain Tumor Classification Using Capsulenet. In ICICCT 2019 – System Reliability, Quality Control, Safety, Maintenance and Management; Springer: Singapore, 2019; pp. 110–119. [Google Scholar] [CrossRef]
- Liu, D.; Liu, Y.; Dong, L. G-ResNet: Improved ResNet for Brain Tumor Classification. In Neural Information Processing; Springer International Publishing: Berlin/Heidelberg, Germany, 2019; pp. 535–545. [Google Scholar] [CrossRef]
- Kokkalla, S.; Kakarla, J.; Venkateswarlu, I.B.; Singh, M. Three-class brain tumor classification using deep dense inception residual network. Soft Comput. 2021. [Google Scholar] [CrossRef]
- Çinarer, G.; Emiroğlu, B.G.; Yurttakal, A.H. Prediction of Glioma Grades Using Deep Learning with Wavelet Radiomic Features. Appl. Sci. 2020, 10, 6296. [Google Scholar] [CrossRef]
- Erickson, B.; Akkus, Z.; Sedlar, J.; Korfiatis, P. Data from LGG-1p19qDeletion. 2017. Available online: https://wiki.cancerimagingarchive.net/display/Public/LGG-1p19qDeletion (accessed on 3 May 2021). [CrossRef]
- Abiwinanda, N.; Hanif, M.; Hesaputra, S.T.; Handayani, A.; Mengko, T.R. Brain Tumor Classification Using Convolutional Neural Network. In IFMBE Proceedings; Springer: Singapore, 2018; pp. 183–189. [Google Scholar] [CrossRef]
- Sharif, M.I.; Khan, M.A.; Alhussein, M.; Aurangzeb, K.; Raza, M. A decision support system for multimodal brain tumor classification using deep learning. Complex Intell. Syst. 2021. [Google Scholar] [CrossRef]
- Irmak, E. Multi-Classification of Brain Tumor MRI Images Using Deep Convolutional Neural Network with Fully Optimized Framework. Iran. J. Sci. Technol. Trans. Electr. Eng. 2021. [Google Scholar] [CrossRef]
- Clark, K.; Vendt, B.; Smith, K.; Freymann, J.; Kirby, J.; Koppel, P.; Moore, S.; Phillips, S.; Maffitt, D.; Pringle, M.; et al. The Cancer Imaging Archive (TCIA): Maintaining and Operating a Public Information Repository. J. Digit. Imaging 2013, 26, 1045–1057. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pei, L.; Vidyaratne, L.; Rahman, M.M.; Iftekharuddin, K.M. Context aware deep learning for brain tumor segmentation, subtype classification, and survival prediction using radiology images. Sci. Rep. 2020, 10. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.N.; Fred, A.L.; Varghese, P.S. An Overview of Segmentation Algorithms for the Analysis of Anomalies on Medical Images. J. Intell. Syst. 2018, 29, 612–625. [Google Scholar] [CrossRef]
- Biratu, E.S.; Schwenker, F.; Debelee, T.G.; Kebede, S.R.; Negera, W.G.; Molla, H.T. Enhanced Region Growing for Brain Tumor MR Image Segmentation. J. Imaging 2021, 7, 22. [Google Scholar] [CrossRef] [PubMed]
- Miotto, R.; Wang, F.; Wang, S.; Jiang, X.; Dudley, J.T. Deep learning for healthcare: Review, opportunities and challenges. Briefings Bioinform. 2017, 19, 1236–1246. [Google Scholar] [CrossRef] [PubMed]
- Zhu, G.; Jiang, B.; Tong, L.; Xie, Y.; Zaharchuk, G.; Wintermark, M. Applications of Deep Learning to Neuro-Imaging Techniques. Front. Neurol. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Sert, E.; Özyurt, F.; Doğantekin, A. A new approach for brain tumor diagnosis system: Single image super resolution based maximum fuzzy entropy segmentation and convolutional neural network. Med. Hypotheses 2019, 133, 109413. [Google Scholar] [CrossRef]
- Natekar, P.; Kori, A.; Krishnamurthi, G. Demystifying Brain Tumor Segmentation Networks: Interpretability and Uncertainty Analysis. Front. Comput. Neurosci. 2020, 14. [Google Scholar] [CrossRef] [Green Version]
- Saleem, H.; Shahid, A.R.; Raza, B. Visual interpretability in 3D brain tumor segmentation network. Comput. Biol. Med. 2021, 133, 104410. [Google Scholar] [CrossRef]
- Zeng, Y.; Zhang, B.; Zhao, W.; Xiao, S.; Zhang, G.; Ren, H.; Zhao, W.; Peng, Y.; Xiao, Y.; Lu, Y.; et al. Magnetic Resonance Image Denoising Algorithm Based on Cartoon, Texture, and Residual Parts. Comput. Math. Methods Med. 2020, 2020, 1–10. [Google Scholar] [CrossRef]
- Heo, Y.C.; Kim, K.; Lee, Y. Image Denoising Using Non-Local Means (NLM) Approach in Magnetic Resonance (MR) Imaging: A Systematic Review. Appl. Sci. 2020, 10, 7028. [Google Scholar] [CrossRef]
- López, M.M.; Frederick, J.M.; Ventura, J. Evaluation of MRI Denoising Methods Using Unsupervised Learning. Front. Artif. Intell. 2021, 4. [Google Scholar] [CrossRef]
- Kidoh, M.; Shinoda, K.; Kitajima, M.; Isogawa, K.; Nambu, M.; Uetani, H.; Morita, K.; Nakaura, T.; Tateishi, M.; Yamashita, Y. Deep Learning Based Noise Reduction for Brain MR Imaging: Tests on Phantoms and Healthy Volunteers. Magn. Reson. Med Sci. 2020, 19, 195–206. [Google Scholar] [CrossRef] [Green Version]
- Higaki, T.; Nakamura, Y.; Tatsugami, F.; Nakaura, T.; Awai, K. Improvement of image quality at CT and MRI using deep learning. Jpn. J. Radiol. 2018, 37, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.H.; Do, W.J.; Park, S.H. Improving resolution of MR images with an adversarial network incorporating images with different contrast. Med. Phys. 2018, 45, 3120–3131. [Google Scholar] [CrossRef] [PubMed]

| Author and Publication Year | Strength | Limitation |
|---|---|---|
| Sharma and Shukla [29] 2021 | Thresholding, conventional supervised and unsupervised based segmentation techniques are briefly described. |
|
| Rao and Karunakara [30] 2021 |
|
|
| Magadza and Viriri [31] 2021 |
|
|
| Tiwari et al. [32] 2020 |
|
|
| Kumari and Saxena [33] 2018 |
|
|
| IC | EC |
|---|---|
| IC1: Paper must be peer reviewed. | EC1: Duplicate studies in different databases. |
| IC2: Journals on which papers published must be either scopus or web of science indexed | EC2: Study that uses imaging techniques other than MRI. |
| IC3: The paper should use only MRI brain images | EC3: Study which is less cited by other peer reviewed papers. |
| EC4: MSc and PhD papers. | |
| EC5: Case study papers. |
| Paper | Dataset | Segmentation Technique | Objective Function | Performance |
|---|---|---|---|---|
| [58] | BRATS 2015 BRATS-MICCAI | Multi-level thresholding with level-set segmentation | Euclidean distance | JI 81.94%, DSC 89.91% |
| [48] | https://radiopaedia.org/ (accessed on 3 May 2021) | K-means and FCM | Euclidean distance | ACC 56.4 % |
| [43] | BRATS | K-means with histogram peaks centroid initialization | Euclidean distance | - |
| [39] | BRATS | Patch based k-means with FCM | Euclidean distance | SI 91% |
| [42] | BRATS 2012 | Random | Sum of Squared Error | DSC 91% |
| [44] | MRI images collected by authors | Bi-secting (No initialization) | Sum of Squared Error | ACC 83.05% |
| [59] | BRATS | Force Clustering | Distance (in pixels) | - |
| [60] | BRATS 2017 | Random | Euclidean distance | DSC 62.5% |
| [61] | MRI images collected by authors | DPSO 1 | Euclidean distance | ACC 99.98%, SEN 95.02%, SPE 99.92% DSC 93.09% |
| [62] | MRI images collected by authors | FCM preceded by gross tumor volume segmentation with random centroid intialization | Inter-cluster variance | DSC 95.93 ± 4.23%, JI 92.81 ± 6.56%, SPE 95.31 ± 6.56%, SEN 98.09 ± 1.75% |
| [63] | MRI images collected by authors | DWT 2 based genetic algorithm (GA) | fitness function variance | ACC 97% |
| [64] | MRI images collected by authors | semi-automatic cellular automata seeded segmentation with morphological post-processing | pixel similarity function | DSC 90.88 ± 4.19%, JI 84.11 ± 6.74%, SPE 99.99 ± 0.01%, SEN 91.20 ± 7.00% |
| Paper | Dataset | Preprocessing | Features | Model | Post- Processing | Performance |
|---|---|---|---|---|---|---|
| [66] | Clinically collected MRI | N4ITK | deep features from CNN | SVM | - | DSC 88%, SEN 89%, PR 83% |
| [67] | Clinically collected MRI | Registration | Intensity texture | Multi- kernel SVM | Region growing | TP 98.9%, FP 4.5%, FN 3.1% |
| [68] | BRATS 2013 | N4ITK, histogram matching, SLIC 1 | Gray statistical, GLCM | SVM | - | DSC 86.12%, SEN 79.69%, SPE 99.48% |
| [70] | BRATS 2015 | - | Intensity, texture | ANN, SVM | - | SVM: DSC 88.7%, IOU 79.7%, ANN: DSC 90.79%, IOU 83.1% |
| [71] | BRATS 2015, [77,78,79] | - | Dual pathway tree based features | ccRF 2 | mpAC 3 | DSC 89%, SPE 90%, SEN 85% |
| [72] | BRATS 2012 | registration, normalization | intensity, similarity, blobness | RF | Independent connected component analysis | DSC 96.5% |
| [74] | [80] | N4ITK, normalization, histogram matching | intensity, gradient, context | RDF 4 | morphological filtering | DSC 86.41%, SEN 82%, PR 92.92% |
| [75] | BRATS 2015 | noise removal, enhancement | first higher order features, texture | RF | morphological other filtering | DSC 98.4%, SEN 97.9%, SPE 80.7%, ACC 97.7% |
| [76] | BRATS 2015 | histogram enhancement | Gabor wavelet, intensity | RF | morphological other filtering | DSC 85.5%, SEN 77.1%, SPE 99.3% |
| Paper | Dataset | Preprocessing | Model Architecture | Performance |
|---|---|---|---|---|
| [81] | BRATS 2013 & 2015 | bias field correction, intensity and patch normalization, augmentation | Custom CNN | DSC 88%, SEN 89%, PR 87% |
| [82] | BRATS 2013 | intensity normalization, augmentation | HCNN + CRF-RRNN 1 | SEN 95%, SPE 95.5%, PR 96.5%, RE 97.8%, ACC 98.6% |
| [83] | BRATS 2015 | Z-score normalization on the image, | Residual Network+ Dilated convolution RDM-Net 2 | DSC 86% |
| [84] | BRATS 2015 | Z-score normalization | Stack Multi-connection Simple Reducing_Net (SMCSRNet) | DSC 83.42%, PR 78.96%, SEN 90.24% |
| [85] | BRATS 2019 | - | Ensemble of a 3D-CNN and U-net | DSC 90.6% |
| [86] | BRATS 2015 | Bias correction, intensity normalization | Two-PathGroup-CNN (2PG-CNN) | DSC 89.2%, PR 88.22%, SEN 88.32% |
| [87] | BRATS 2018 | - | Hybrid two track U-Net (HTTU-Net) | DSC 86.5%, SEN 88.3%, SPE 99.9% |
| [88] | BRATS 2015 | - | P-Net with bounding box and image specific fine tunning (BIFSeg) | DSC 86.29% |
| [89] | ADNI | denoising, Skull stripping, sub-sampling | Multi-scale CNN (MSCNN) | ACC 90.1% |
| [90] | BRATS 2017 | Intensity normalization, resizing, Bias field correction | Cascaded 3D U-nets | DSC 89.4% |
| [91] | BRATS 2015 & 2017 | Down sampling | 3D Center-crop Dense Block | BRATS 2015: DSC 88.4%, SEN 83.8% BRATS 2017: DSC 88.7%, SEN 84.3% |
| [92] | BRATS 2018 & 2019 | Z-score normalization, cropping | 3D FCN 3 | BRATS 2018: DSC 90%, SEN 90.3, SPE 99.48%; BRATS 2019: DSC 89%, SEN 88.3%, SPE 99.51% |
| [93] | BRATS 2018 | intensity normalization, removing 1% of highest & lowest intensity | DCNN (Dense-MultiOCM 4) | BRATS 2018: DSC 86.2%, SEN 84.8 %, SPE 99.5% |
| [94] | TCIA | Image cropping, padding, resizing, intensity normalization | U-Net | DSC 84%, SEN 92%, SPE 92%, ACC 92% |
| [95] | BRATS 2013, 2015, 2018 | - | AFPNet 5 + 3D CRF | BRATS 2013 DSC 86%, BRATS 2015 DSC 82%, BRATS 2018 86.58% |
| [96] | BRATS 2015, 2017 | z-score normalization | Inception-based U-Net + up skip connection + cascaded training strategy | DSC 89%, PR 78.5%, SEN 89.5% |
| [97] | BRATS 2015, BrainWeb | cropping, z-score normalization, min-max normalization (BrainWeb) | Tripple intersecting UNets (TIU-Net) | BRATS 2015: DSC 85%, BrainWeb DSC 99.5% |
| [98] | BRATS 2015 | - | LSTM multi-modal UNet | DSC 73.09%, SEN 63.76%, PR 89.79% |
| Paper | Dataset | Preprocessing | ROI Detection | Feature Extraction | Classifier | Tumor Types | Performance |
|---|---|---|---|---|---|---|---|
| [108] | Local dataset | Median and weiner filter | k-means modified FCM | shape features, statistical features | ANN | Benign malignant stage (I-IV) | SPE 100%, SEN 98%, ACC 97.73%, BER 0.0294 |
| [109] | [138] | Median and weiner filter | manually | 2-D DWT 2-D Gabor feature | ANN | Glioma (GL), Meningioma (MG) Pituitary tumor (PT) | overall ACC 91.9%, SPE (GL) 96.29%, SPE (MG) 96%, SPE (PT) 96.2%, SEN (GL) 95.1%, SEN(MG) 86.97%, SEN(PT) 91.24% |
| [110] | Local dataset | resizing skull removing | Canny | Gabor filter, GLCM DWT | ANN | Benign and malignant stage (I-IV) | SPE 98.5%, SEN 99.1%, ACC 98.9% |
| [139] | Local dataset | resizing | - | PCA 1 | PNN | Benign malignant stage | SPE 100%, SEN 92.3%, ACC 97.4% |
| [112] | TCIA | resizing, cropping, median filtering | morphological, watersheed | shape features | KNN | Astrocytoma Glioblastoma Oligodendroglioma | ACC 89.5% |
| [115] | Local dataset | wavelets | thresholding | DWT coeficients statistical features | SVM | Benign malignant | ACC (linear) 92%, ACC (kernel) 99% |
| [134] | BRATS and Local dataset | enhancement median filter | Morphological | GLCM features | SVM | Benign malignant | BRATS: SVM (linear):SPE 100%, SEN 72%, ACC 82.5% SVM (Quadratic):SPE 73.3%, SEN 88%, ACC 82.5% SVM (RBF): SPE 100%, SEN 76%, ACC 85% Clinical: SVM (linear):SPE 60%, SEN 76%, ACC 68% SVM (Quadratic):SPE 88%, SEN 100%, ACC 94% SVM (RBF): SPE 100%, SEN 92%, ACC 96% |
| [120] | Local dataset | Gabor transform texture wavelet | SVM | Ependymoma Pilocytic Astrocytoma | SPE 80%, SEN 93%, ACC 88%, AUC 0.86 | ||
| [140] | BRATS-2015 | wavelet filters, inhomogeneity correction | edge detection, morphological operations | shape, texture, intensity | PSO 2-SVM | Benign, malignant | SPE 94.8%, SEN 100% |
| [136] | - | median filtering skull removing | thresholding | GLCM | GA-SVM | Benign, malignant | - |
| [130] | REMBRANDT | - | - | texture features | SVM | Multifocal, Multicentric, Gliomatosis | PR 90%, SEN 90%, ACC 90%, F1-Score 90% |
| [133] | Local dataset | Image fusion with contourlet transform | Otsu’s thresholding | curvlet transform GLCM features | SVM | Benign, Malignant | ACC 93% |
| [125] | [138] | min-max normalization, | - | NGIST features | RELM 3 | Meningioma, Glioma, Pituitary | ACC 94.23% |
| [126] | Local dataset | median filter | thresholding | GLCM texture features | Adaboost | Benign, Malignant | SPE 62.5%, SEN 88.25%, ACC 89.90% |
| [127] | Local dataset | resizing enhancement | morphological, thresholding | GLCM statistical texture features | SVM | Benign, Malignant | SPE 62.5%, SEN 88.25%, ACC 89.90% |
| [128] | Local dataset | noise removal, enhancement | Expectation maximization, levelset | GA, statistical features | SVM | Benign, Malignant | SPE 100%, SEN 98%, ACC 98.30% |
| [124] | [138] | down sampling Gabor filter | - | statistical features | SVM | Meningioma, Glioma, Pituitary | Meningioma: SVM (linear):RE 0.63, PR 0.66, ACC 82.38% SVM (poly):RE 0.62,Pr. 0.73, ACC 84.33% Glioma: SVM (linear):RE 0.82, PR 0.82, ACC 83.01% SVM (poly):RE 0.88, PR 0.79, ACC 84.01% Pituitary: SVM (linear):RE 0.94, PR 0.90, ACC 95.27% SVM (poly):RE 0.91,PR 0.94, ACC 95.43% |
| [122] | Kaggle Brain Tumor Detection 2020 | cropping, resizing using bicubic interpolation | - | Deep features from pretrained CNN | SVM | Meningioma, Glioma, Pituitary | ACC 90.19% |
| Paper | Dataset | Preprocessing | Classifier Model | Tumor Types | Performance |
|---|---|---|---|---|---|
| [100] | [138] | normalization, resizing, augmentation | Custom CNN model | Meningioma, Glioma, Pituitary | ACC 91.9%, precision 94.81%, RE 95.07%, F1-score 94.94%, SPE(GL) 96.2%, SPE(MG) 92%, SPE(PT) 97.7%, SEN(GL) 96.2%, SEN(MG) 89.8%, SEN(PT) 98.4% |
| [141] | [78,142] | Augmentation using GAN | Multi-stream 2D-CNN model | Glioma subtypes: Isocitrate dehydrogenase 1 mutation (IDH1), & IDH1 wild-type | mean ACC 88.82% mean SEN 81.81% mean SPE 92.17% |
| [143] | [138,144] | resizing augmentation | Custom CNN model | Meningioma, Glioma & Pituitary and Glioma (grade:II-IV) | MG: PR 95.8%, SEN 95.5%, SPE 98.7%, ACC 97.54%, GL: PR 97.2%, SEN 94.4%, SPE 95.1%, ACC 95.81%, PT: PR 95.2%, SEN 93.4%, SPE 97%, ACC 96.89% Grade II: PR 100%, SEN 100%, SPE 100%, ACC 100%, III: PR 100%, SEN 95%, SPE 100%, ACC 95%, IV:PR 96.3%, PR 100%, SEN 95%, SPE 100%, ACC 95%SEN 100%, SPE 98%, ACC 100% |
| [145] | [138] | - | CNNBCN 1 | Meningioma, Glioma& Pituitary | ACC 95.49% |
| [146] | [138] | - | BayesCap: captures prediction uncertainity | Meningioma, Glioma& Pituitary | mean ACC 73.9% CI 2:(73.4%, 74.4%) |
| [147] | [138] | Image rotation, resizing | AutoML 3 | Meningioma, Glioma & Pituitary | MG: PR 94.51%, SEN 87.76%, SPE 98.7%, ACC 96.29%, F1-Score 91.01%, MCC 4 88.77%, G-Mean 96.09% GL: PR 96.97%, SEN 95.32%, SPE 96.88%, ACC 96.08%, F1-Score 96.14%, MCC 92.17%, G-Mean 96.09% PT: PR 91.61%, SEN 99.24%, SPE 96.27%, ACC 97.14%, F1-Score 95.27%, MCC 93.38%, G-Mean 97.75% |
| [148] | [138] | - | Iception-V3 DensNet201 | Meningioma, Glioma& Pituitary | Iception-V3: ACC 99.34% DensNet201: ACC 99.51% |
| [149] | [138] | augmentation, contrast- stretching | AlexNet, GoogleNet & VGG16 5 | Meningioma, Glioma& Pituitary | AlexNet: ACC 95.46% GoogleNet: ACC 98.04% VGG16 98.69% |
| [150] | [138] | - | ConvCaps | Meningioma, Glioma& Pituitary | ACC 93.5% |
| [151] | [138] | flipping, patching | CapsulNet | Meningioma, Glioma& Pituitary | MG: PR 85%, RE 94%, F1-Score 94, %GL: PR 85%, RE 94%, F1-Score 94%, PT: PR 85%, RE 94%, F1-Score 94% |
| [152] | [138] | - | G-ResNet | Meningioma, Glioma& Pituitary | ACC 95% |
| [153] | [138] | - | DDIRNet 6 | Meningioma, Glioma& Pituitary | ACC 99.69%, PR 99.6%, RE 99.4%, F1-score 99.4% |
| [103] | [138] | - | Multiscale CNN | Meningioma, Glioma& Pituitary | ACC 97.3% |
| [154] | [155] | DWT | DNN | Meningioma, Glioma& Pituitary | ACC 96.15%, PR 94.12%, AUC 98.75%,F1-score 96.97%, RE 100% |
| [156] | [138] | - | Custom CNN model | Meningioma, Glioma& Pituitary | ACC 84.19% |
| [157] | BraTS 2018 & 2019 | - | Pre-trained DenseNet201 | HGG 7 & LGG 8 | HGG: ACC 99.8%, LGG: ACC 99.3% |
| [158] | [138], [144,159] | - | Custom CNN model | Class 1: Normal, Metastatic, Meningioma, Glioma& Pitiutary Class 2: Grade II, III & IV | Class 1: ACC 92.66% Class 2: ACC 98.14% |
| [160] | BraTS 2019 | - | Custom CNN model | Astrocytoma, Glioblastoma, Oligodendrogloma, | Class 1: ACC 92.66% Class 2: ACC 98.14% |
| [94] | TCIA | cropping, padding, resizing, normalization | VGG16 | Grade II & III | ACC 89%, SEN 87%, SPE 92% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Biratu, E.S.; Schwenker, F.; Ayano, Y.M.; Debelee, T.G. A Survey of Brain Tumor Segmentation and Classification Algorithms. J. Imaging 2021, 7, 179. https://doi.org/10.3390/jimaging7090179
Biratu ES, Schwenker F, Ayano YM, Debelee TG. A Survey of Brain Tumor Segmentation and Classification Algorithms. Journal of Imaging. 2021; 7(9):179. https://doi.org/10.3390/jimaging7090179
Chicago/Turabian StyleBiratu, Erena Siyoum, Friedhelm Schwenker, Yehualashet Megersa Ayano, and Taye Girma Debelee. 2021. "A Survey of Brain Tumor Segmentation and Classification Algorithms" Journal of Imaging 7, no. 9: 179. https://doi.org/10.3390/jimaging7090179
APA StyleBiratu, E. S., Schwenker, F., Ayano, Y. M., & Debelee, T. G. (2021). A Survey of Brain Tumor Segmentation and Classification Algorithms. Journal of Imaging, 7(9), 179. https://doi.org/10.3390/jimaging7090179







