Non-Contact Automatic Vital Signs Monitoring of Infants in a Neonatal Intensive Care Unit Based on Neural Networks
Abstract
:1. Introduction
2. Methods and Materials
2.1. Study Design
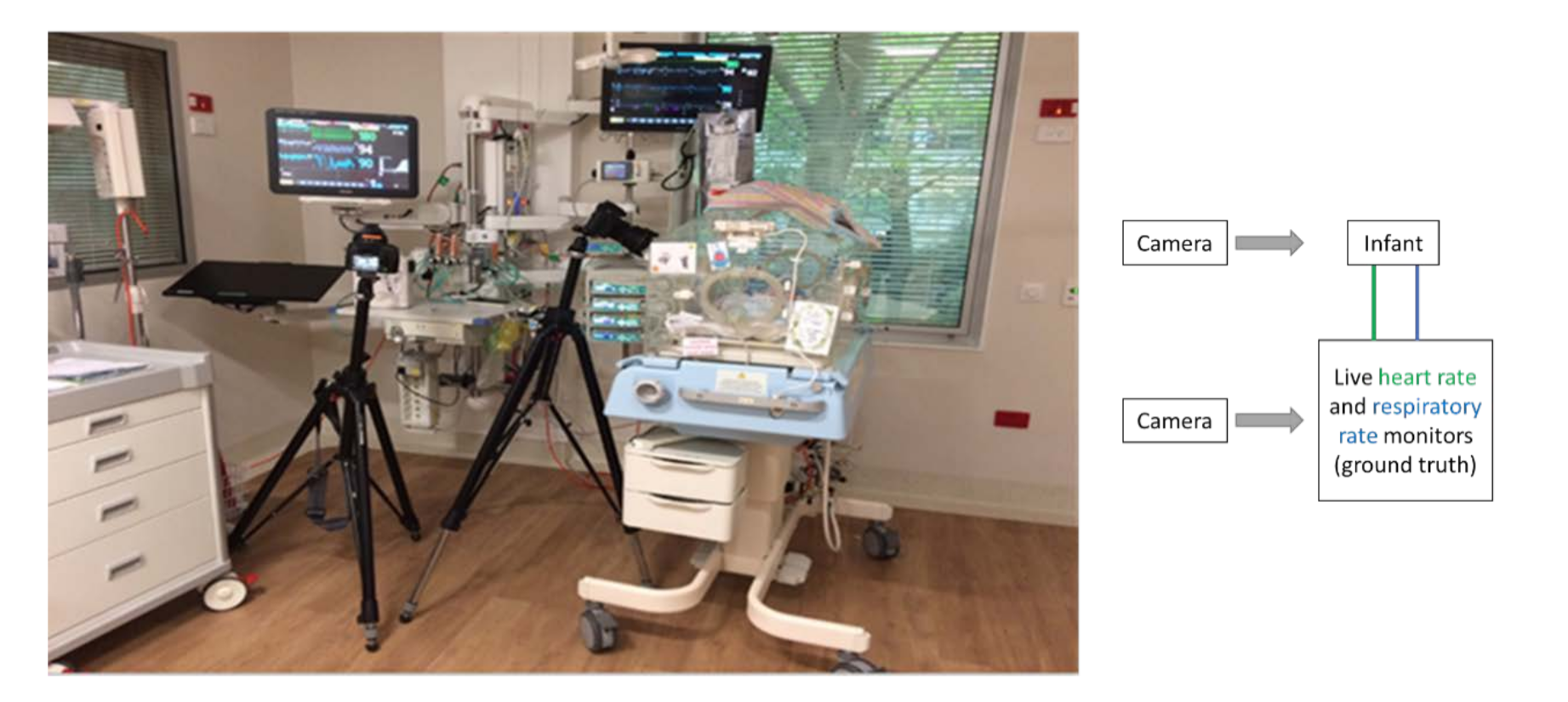
2.2. Experimental Setup
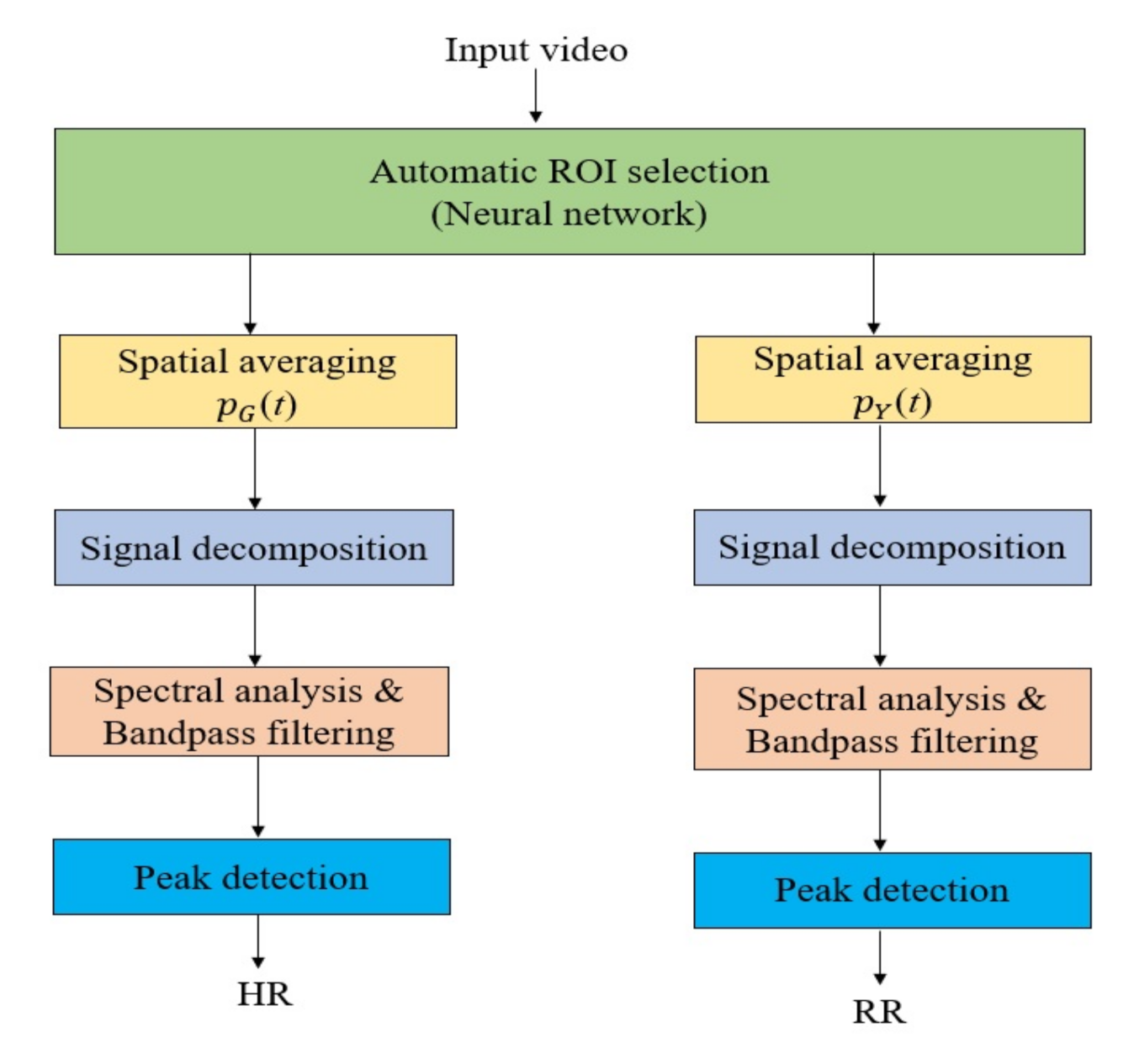
2.3. System Framework
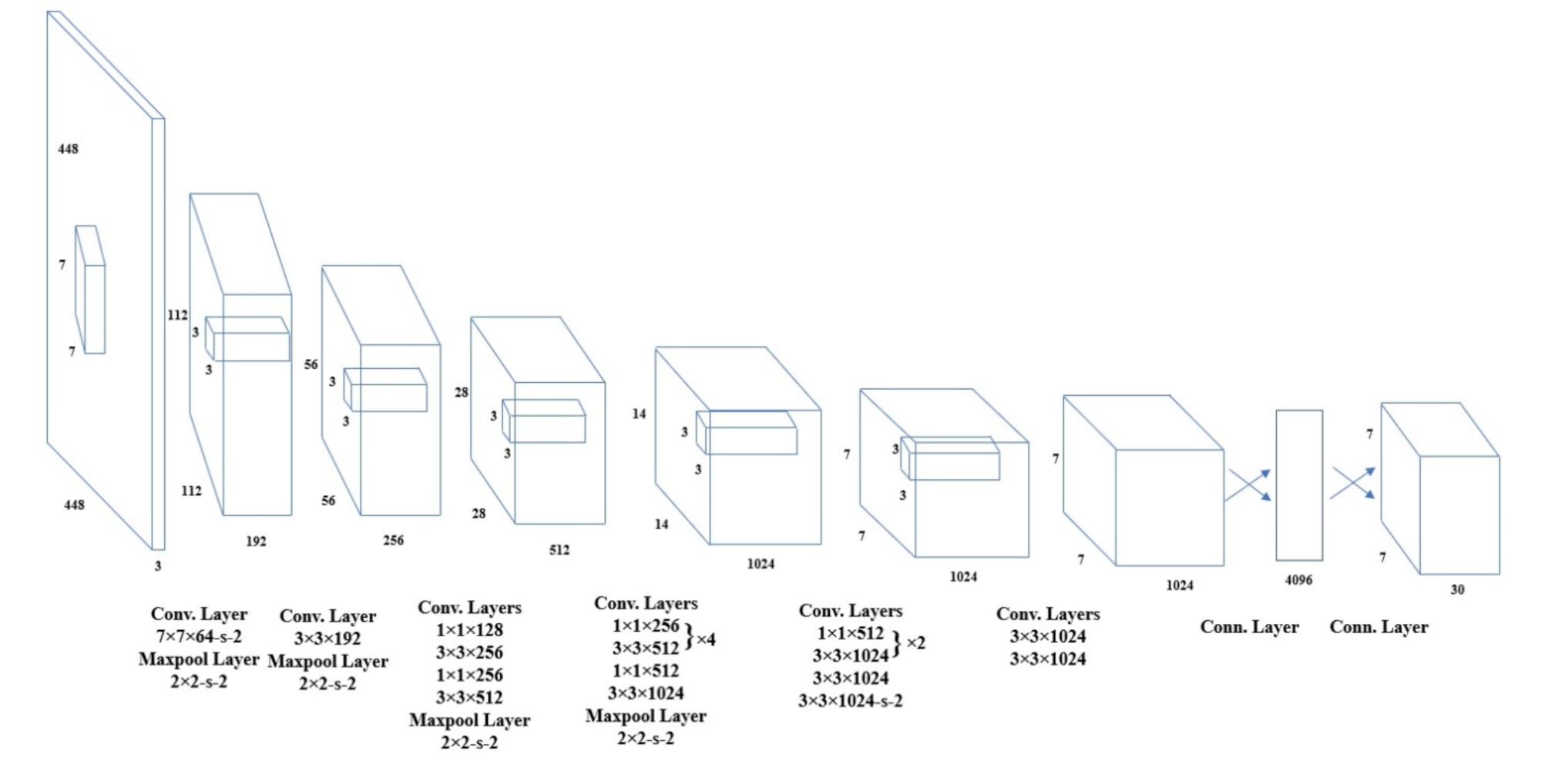
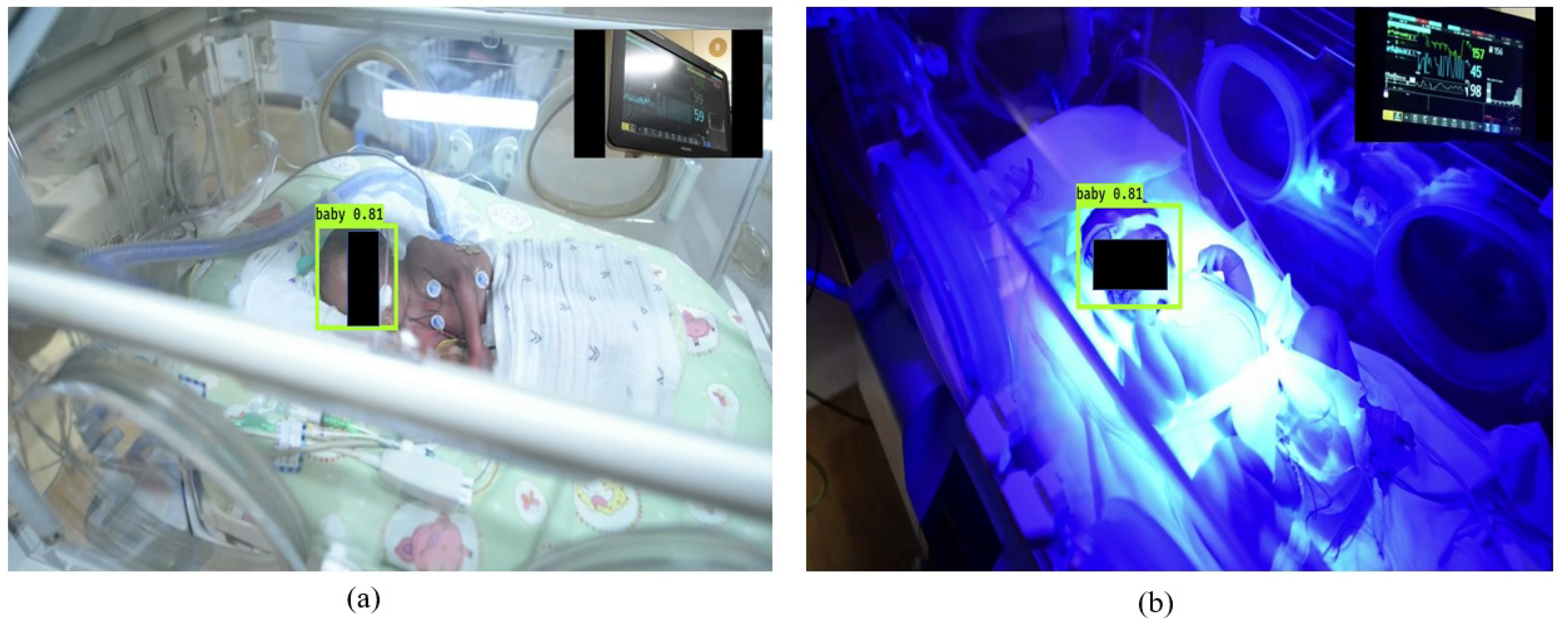
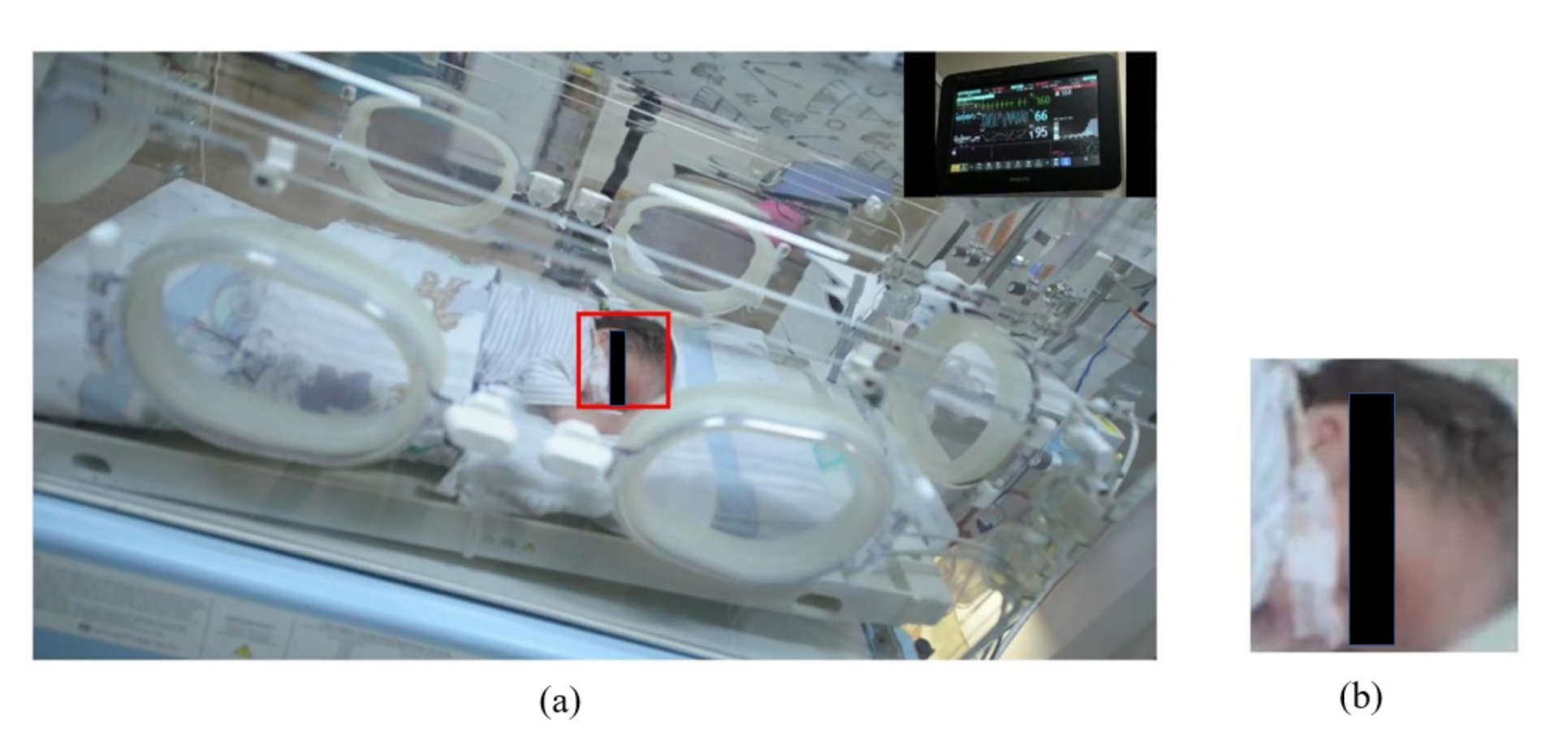
2.3.1. Automatic ROI Selection
2.3.2. Spatial Averaging
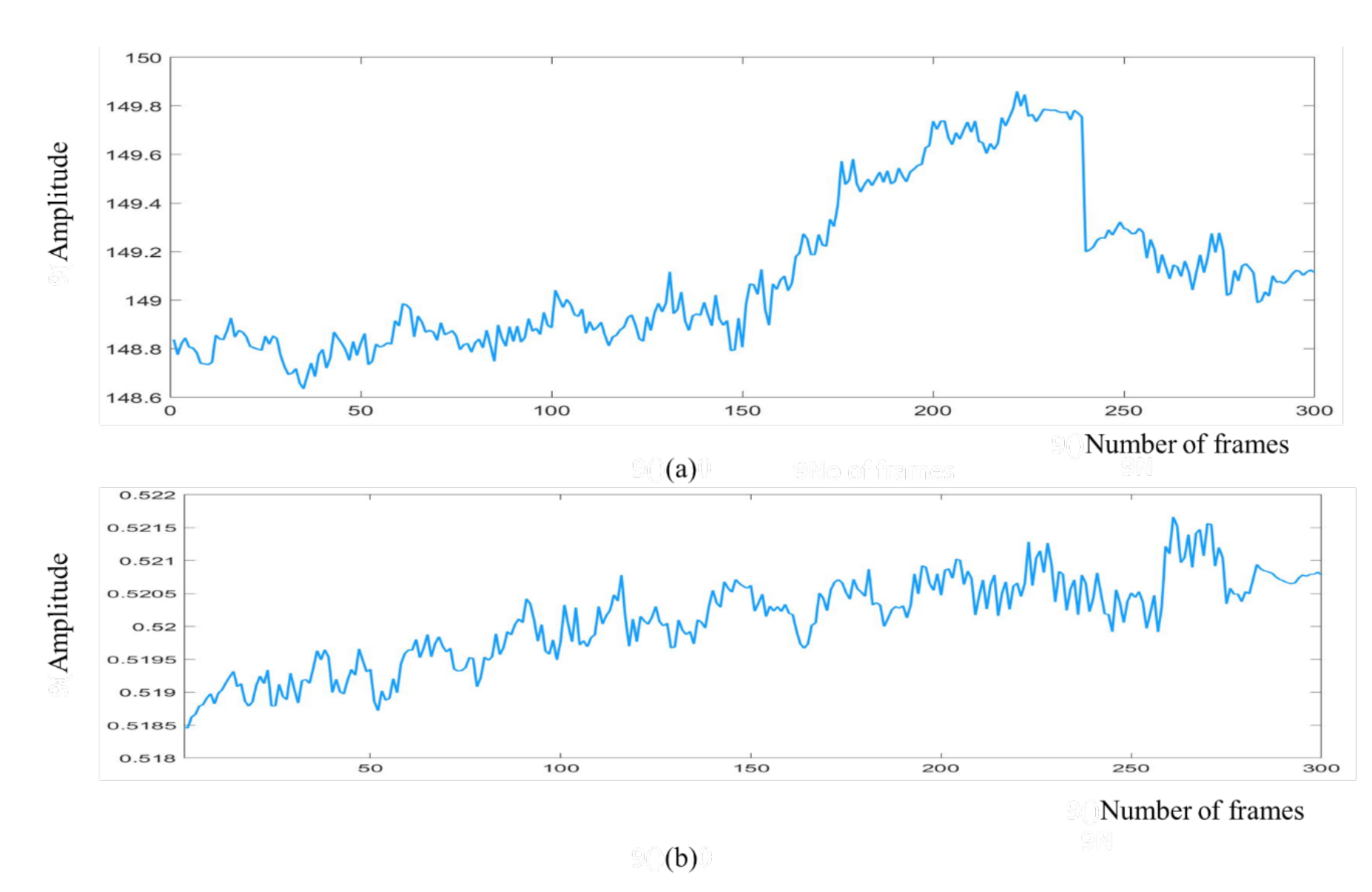
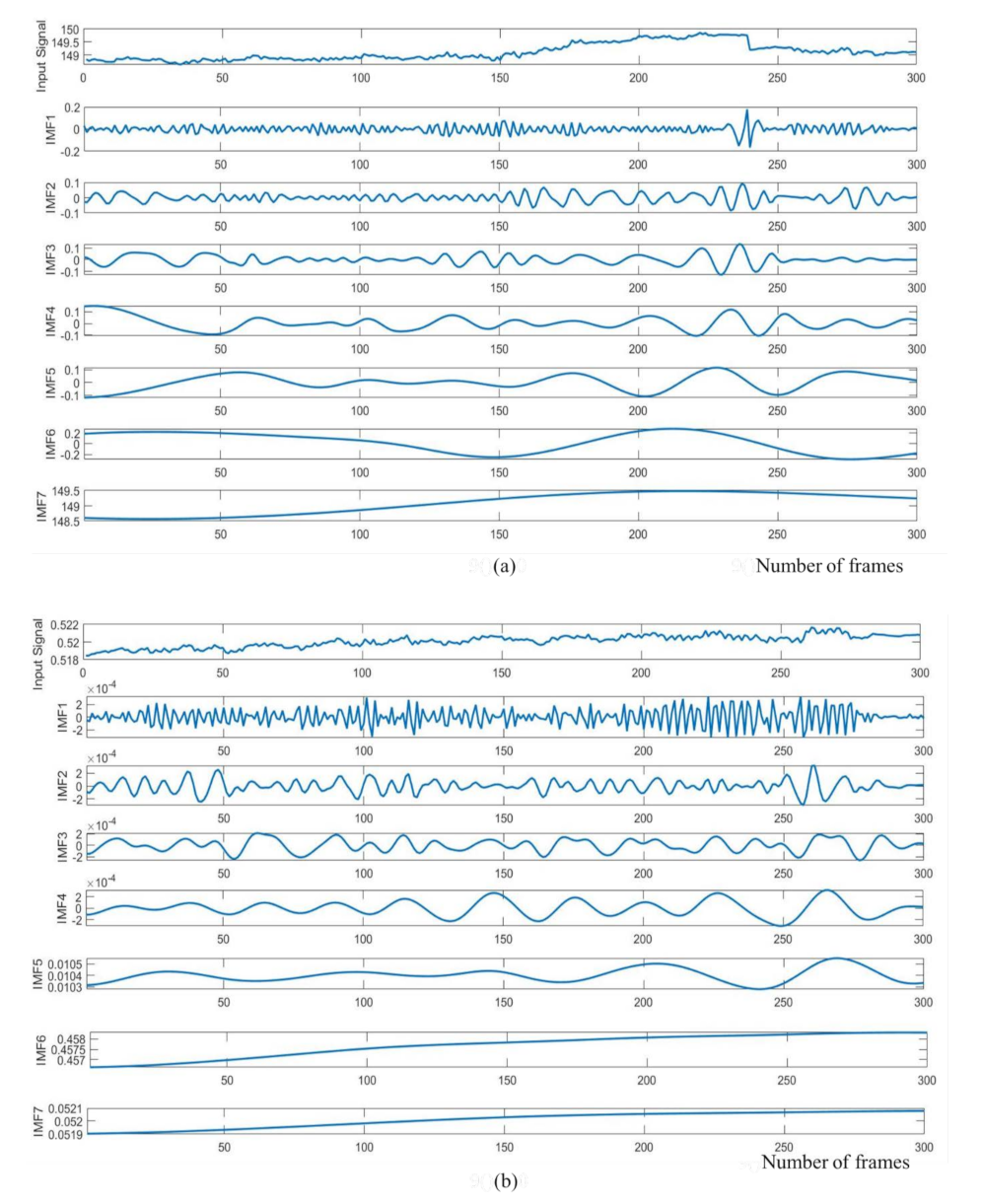
2.3.3. Signal Decomposition
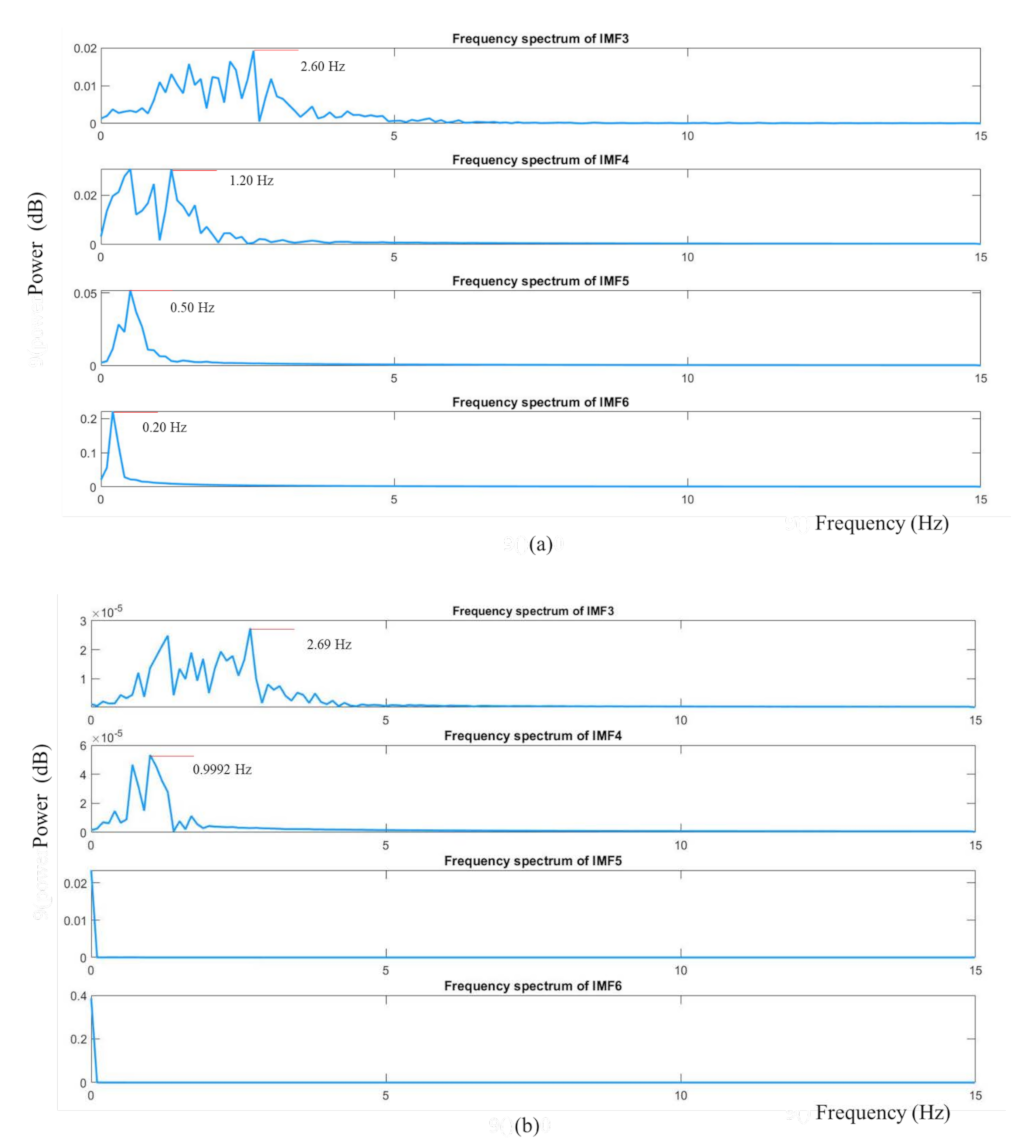
2.3.4. Spectral Analysis and Band-Pass Filtering
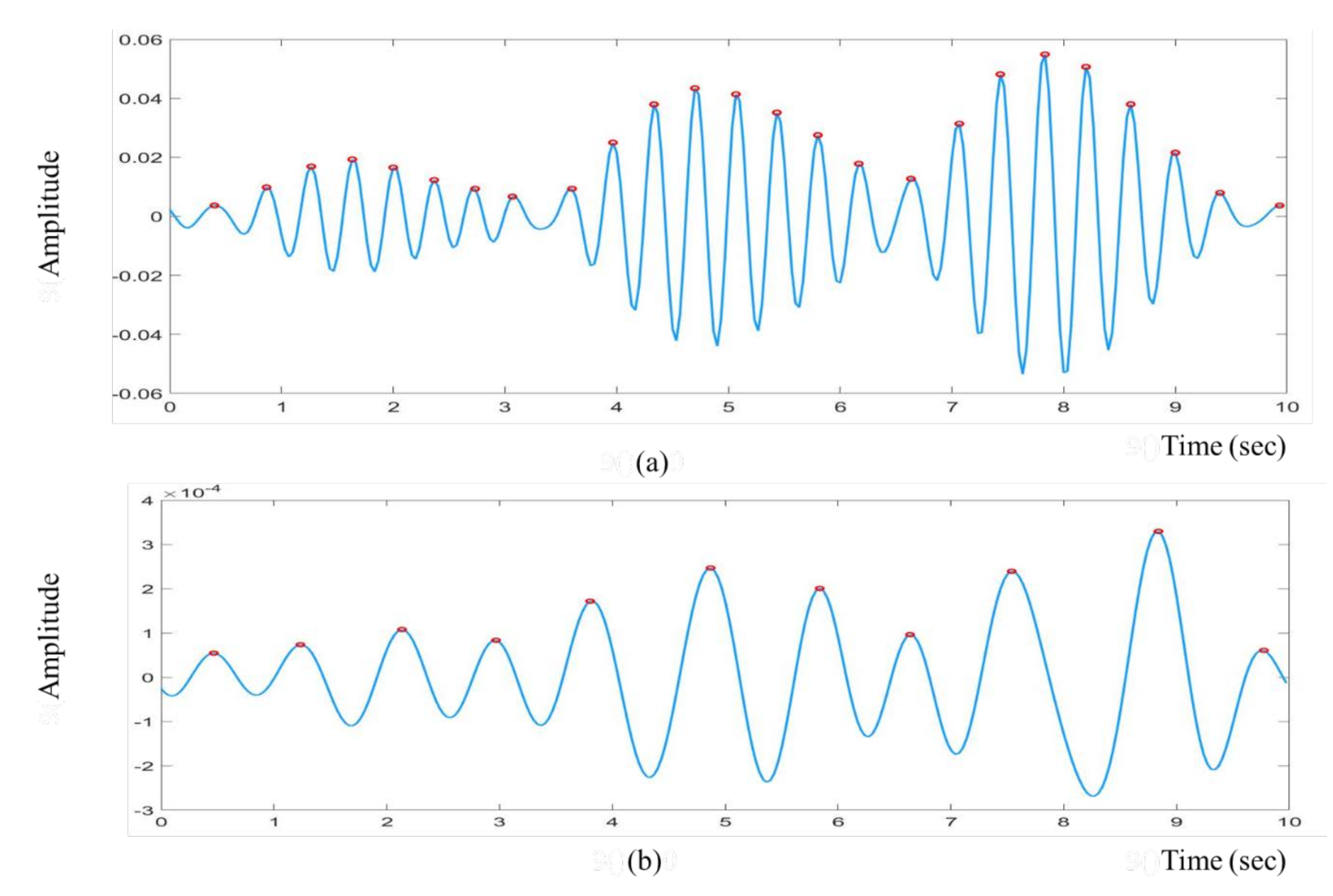
2.3.5. Peak Detection
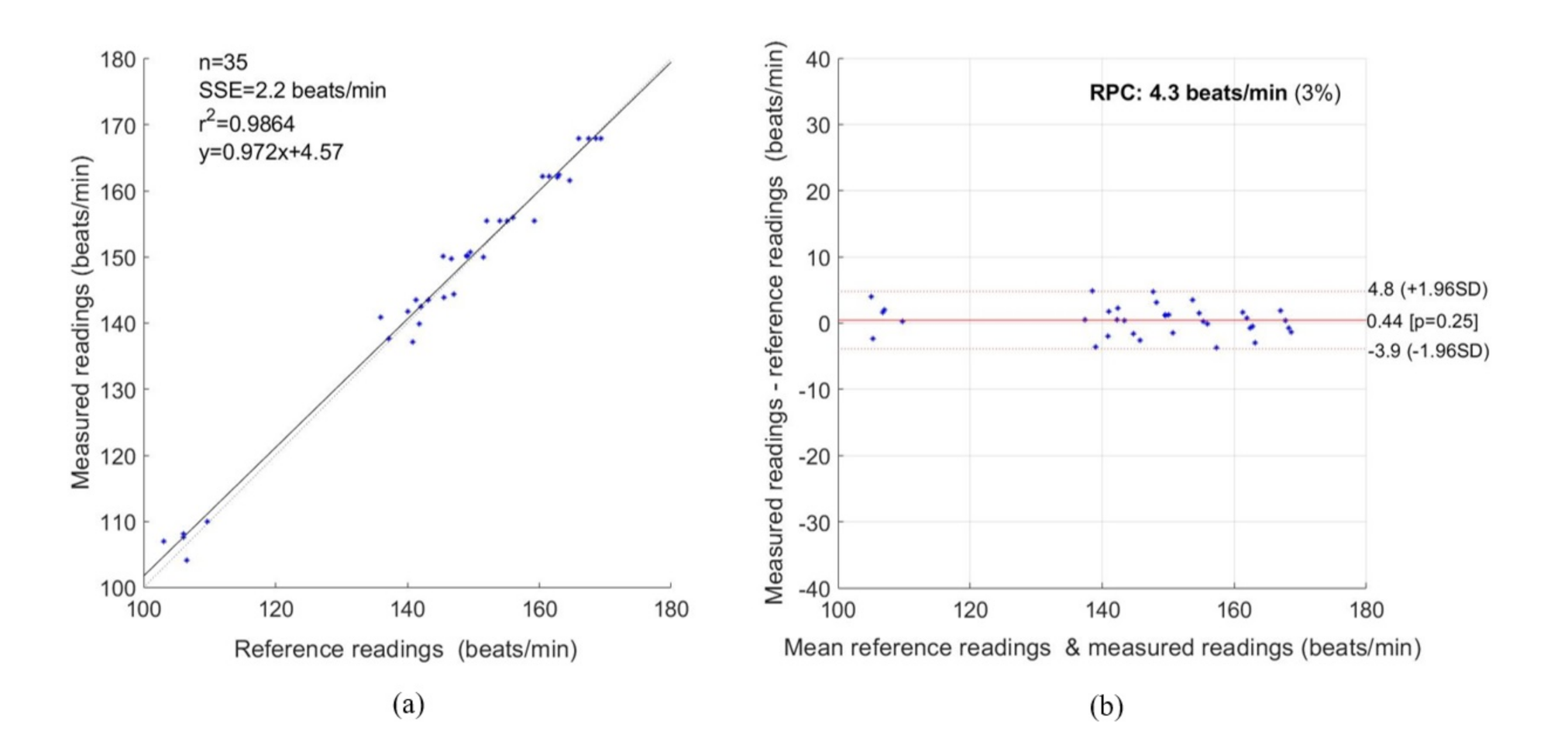
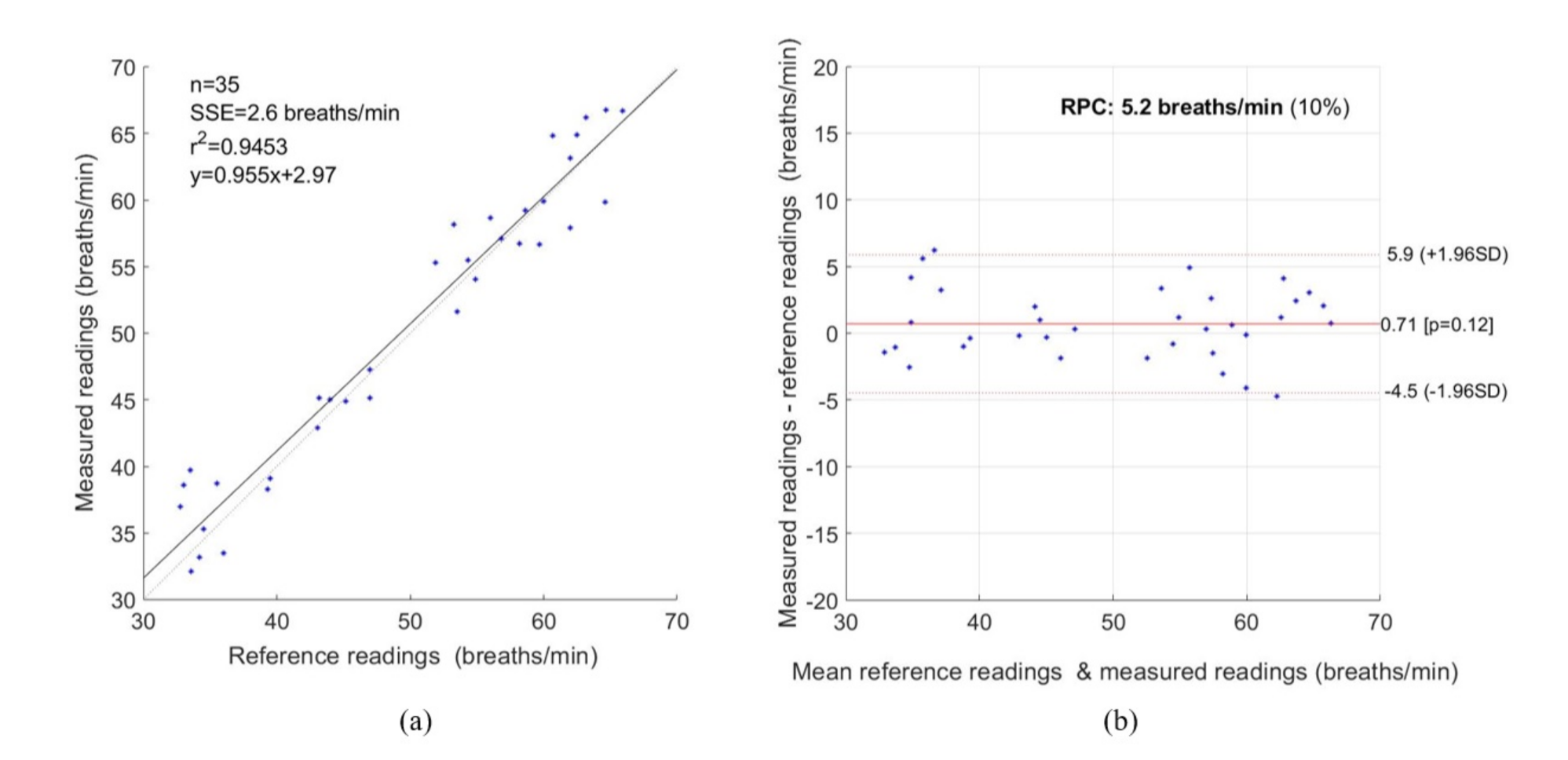
3. Experimental Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO. International Statistical Classification of Diseases and Related Health Problems. Available online: https://www.who.int/standards/classifications/classification-of-diseases (accessed on 1 March 2021).
- Blencowe, H.; Cousens, S.; Oestergaard, M.Z.; Chou, D.; Moller, A.-B.; Narwal, R.; Adler, A.; Garcia, C.V.; Rohde, S.; Say, L. National, regional, and worldwide estimates of preterm birth rates in the year 2010 with time trends since 1990 for selected countries: A systematic analysis and implications. Lancet 2012, 379, 2162–2172. [Google Scholar] [CrossRef] [Green Version]
- Kenner, C.; Lott, J.W. Comprehensive Neonatal Care: An Interdisciplinary Approach; Elsevier Health Sciences: London, UK, 2007. [Google Scholar]
- Behrman, R.E.; Butler, A.S. Mortality and acute complications in preterm infants. In Preterm Birth: Causes, Consequences, and Prevention; National Academies Press: Washington, DC, USA, 2007. [Google Scholar]
- Khanam, F.-T.-Z.; Al-Naji, A.; Chahl, J. Remote Monitoring of Vital Signs in Diverse Non-Clinical and Clinical Scenarios Using Computer Vision Systems: A Review. Appl. Sci. 2019, 9, 4474. [Google Scholar] [CrossRef] [Green Version]
- Khanam, F.-T.-Z.; Chahl, L.A.; Chahl, J.S.; Al-Naji, A.; Perera, A.G.; Wang, D.; Lee, Y.; Ogunwa, T.T.; Teague, S.; Nguyen, T.X.B. Noncontact Sensing of Contagion. J. Imaging 2021, 7, 28. [Google Scholar] [CrossRef]
- Cretikos, M.A.; Bellomo, R.; Hillman, K.; Chen, J.; Finfer, S.; Flabouris, A. Respiratory rate: The neglected vital sign. Med. J. Aust. 2008, 188, 657–659. [Google Scholar] [CrossRef]
- Zhao, F.; Li, M.; Qian, Y.; Tsien, J.Z. Remote measurements of heart and respiration rates for telemedicine. PLoS ONE 2013, 8, e71384. [Google Scholar] [CrossRef] [Green Version]
- Baharestani, M.M. An overview of neonatal and pediatric wound care knowledge and considerations. Ostomy/Wound Manag. 2007, 53, 34–36, 38, 40. [Google Scholar]
- Lloyd, R.; Goulding, R.; Filan, P.; Boylan, G. Overcoming the practical challenges of electroencephalography for very preterm infants in the neonatal intensive care unit. Acta Paediatr. 2015, 104, 152–157. [Google Scholar] [CrossRef]
- Steffen, M.; Aleksandrowicz, A.; Leonhardt, S. Mobile noncontact monitoring of heart and lung activity. IEEE Trans. Biomed. Circuits Syst. 2007, 1, 250–257. [Google Scholar] [CrossRef] [PubMed]
- Lv, W.; He, W.; Lin, X.; Miao, J. Non-Contact Monitoring of Human Vital Signs Using FMCW Millimeter Wave Radar in the 120 GHz Band. Sensors 2021, 21, 2732. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Yang, C.; Mao, S. On CSI-based vital sign monitoring using commodity WiFi. ACM Trans. Comput. Healthc. 2020, 1, 1–27. [Google Scholar] [CrossRef]
- Wang, X.; Yang, C.; Mao, S. PhaseBeat: Exploiting CSI phase data for vital sign monitoring with commodity WiFi devices. In Proceedings of the 2017 IEEE 37th International Conference on Distributed Computing Systems (ICDCS), Atlanta, GA, USA, 5–8 June 2017; pp. 1230–1239. [Google Scholar]
- Springer, D.B.; Brennan, T.; Hitzeroth, J.; Mayosi, B.M.; Tarassenko, L.; Clifford, G.D. Robust heart rate estimation from noisy phonocardiograms. In Proceedings of the Computing in Cardiology, Cambridge, MA, USA, 7–10 September 2014; pp. 613–616. [Google Scholar]
- Abbas, A.K.; Heimann, K.; Jergus, K.; Orlikowsky, T.; Leonhardt, S. Neonatal non-contact respiratory monitoring based on real-time infrared thermography. Biomed. Eng. Online 2011, 10, 1–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lorato, I.; Stuijk, S.; Meftah, M.; Kommers, D.; Andriessen, P.; van Pul, C.; de Haan, G. Multi-camera infrared thermography for infant respiration monitoring. Biomed. Opt. Express 2020, 11, 4848–4861. [Google Scholar] [CrossRef] [PubMed]
- Kevat, A.C.; Bullen, D.V.; Davis, P.G.; Kamlin, C.O.F. A systematic review of novel technology for monitoring infant and newborn heart rate. Acta Paediatr. 2017, 106, 710–720. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.; Li, M.; Tsien, J.Z. Technology platforms for remote monitoring of vital signs in the new era of telemedicine. Expert Rev. Med. Devices 2015, 12, 411–429. [Google Scholar] [CrossRef]
- Tarjan, P.P.; McFee, R. Electrodeless measurements of the effective resistivity of the human torso and head by magnetic induction. IEEE Trans. Biomed. Eng. 1968, 266–278. [Google Scholar] [CrossRef] [PubMed]
- Guardo, R.; Trudelle, S.; Adler, A.; Boulay, C.; Savard, P. Contactless recording of cardiac related thoracic conductivity changes. In Proceedings of the 17th International Conference of the Engineering in Medicine and Biology Society, Montreal, QC, Canada, 20–23 September 1995; pp. 1581–1582. [Google Scholar]
- Vetter, P.; Leicht, L.; Leonhardt, S.; Teichmann, D. Integration of an electromagnetic coupled sensor into a driver seat for vital sign monitoring: Initial insight. In Proceedings of the 2017 IEEE International Conference on Vehicular Electronics and Safety (ICVES), Vienna, Austria, 27–28 June 2017; pp. 185–190. [Google Scholar]
- Liu, H.; Allen, J.; Zheng, D.; Chen, F. Recent development of respiratory rate measurement technologies. Physiol. Meas. 2019, 40, 07TR01. [Google Scholar] [CrossRef] [Green Version]
- Abdelnasser, H.; Harras, K.A.; Youssef, M. UbiBreathe: A ubiquitous non-invasive WiFi-based breathing estimator. In Proceedings of the 16th ACM International Symposium on Mobile Ad Hoc Networking and Computing, New York, NY, USA, 22–25 June 2015; pp. 277–286. [Google Scholar]
- Dalal, H.; Basu, A.; Abegaonkar, M.P. Remote sensing of vital sign of human body with radio frequency. CSI Trans. ICT 2017, 5, 161–166. [Google Scholar] [CrossRef]
- Rabbani, M.S.; Ghafouri-Shiraz, H. Ultra-wide patch antenna array design at 60 GHz band for remote vital sign monitoring with Doppler radar principle. J. Infrared Millim. Terahertz Waves 2017, 38, 548–566. [Google Scholar] [CrossRef] [Green Version]
- Yang, M.; Liu, Q.; Turner, T.; Wu, Y. Vital sign estimation from passive thermal video. In Proceedings of the 2008 IEEE Conference on Computer Vision and Pattern Recognition, Anchorage, AK, USA, 23–28 June 2008; pp. 1–8. [Google Scholar]
- Garbey, M.; Sun, N.; Merla, A.; Pavlidis, I. Contact-free measurement of cardiac pulse based on the analysis of thermal imagery. IEEE Trans. Biomed. Eng. 2007, 54, 1418–1426. [Google Scholar] [CrossRef]
- Al-Naji, A.; Gibson, K.; Lee, S.-H.; Chahl, J. Monitoring of cardiorespiratory signal: Principles of remote measurements and review of methods. IEEE Access 2017, 5, 15776–15790. [Google Scholar] [CrossRef]
- Kranjec, J.; Beguš, S.; Geršak, G.; Drnovšek, J. Non-contact heart rate and heart rate variability measurements: A review. Biomed. Signal Process. Control. 2014, 13, 102–112. [Google Scholar] [CrossRef]
- Al-Naji, A.; Chahl, J. Detection of cardiopulmonary activity and related abnormal events using Microsoft Kinect sensor. Sensors 2018, 18, 920. [Google Scholar] [CrossRef] [Green Version]
- Al-Naji, A.; Chahl, J. Remote Optical Cardiopulmonary Signal Extraction with Noise Artifact Removal, Multiple Subject Detection & Long-Distance. IEEE Access 2018, 6, 11573–11595. [Google Scholar]
- Al-Naji, A.; Gibson, K.; Lee, S.-H.; Chahl, J. Real time apnoea monitoring of children using the Microsoft Kinect sensor: A pilot study. Sensors 2017, 17, 286. [Google Scholar] [CrossRef] [Green Version]
- Al-Naji, A.; Perera, A.G.; Chahl, J. Remote monitoring of cardiorespiratory signals from a hovering unmanned aerial vehicle. Biomed. Eng. Online 2017, 16, 101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verkruysse, W.; Svaasand, L.O.; Nelson, J.S. Remote plethysmographic imaging using ambient light. Opt. Express 2008, 16, 21434–21445. [Google Scholar] [CrossRef] [Green Version]
- De Haan, G.; Jeanne, V. Robust pulse rate from chrominance-based rPPG. IEEE Trans. Biomed. Eng. 2013, 60, 2878–2886. [Google Scholar] [CrossRef]
- De Haan, G.; Van Leest, A. Improved motion robustness of remote-PPG by using the blood volume pulse signature. Physiol. Meas. 2014, 35, 1913. [Google Scholar] [CrossRef]
- Al-Naji, A.; Chahl, J. Remote respiratory monitoring system based on developing motion magnification technique. Biomed. Signal Process. Control. 2016, 29, 1–10. [Google Scholar] [CrossRef]
- Bal, U. Non-contact estimation of heart rate and oxygen saturation using ambient light. Biomed. Opt. Express 2015, 6, 86–97. [Google Scholar] [CrossRef] [Green Version]
- Wieringa, F.P.; Mastik, F.; van der Steen, A.F. Contactless multiple wavelength photoplethysmographic imaging: A first step toward “SpO 2 camera” technology. Ann. Biomed. Eng. 2005, 33, 1034–1041. [Google Scholar] [CrossRef] [PubMed]
- Scalise, L.; Bernacchia, N.; Ercoli, I.; Marchionni, P. Heart rate measurement in neonatal patients using a webcamera. In Proceedings of the 2012 IEEE International Symposium on Medical Measurements and Applications Proceedings, Budapest, Hungary, 18–19 May 2012; pp. 1–4. [Google Scholar]
- Poh, M.-Z.; McDuff, D.J.; Picard, R.W. Advancements in noncontact, multiparameter physiological measurements using a webcam. IEEE Trans. Biomed. Eng. 2010, 58, 7–11. [Google Scholar] [CrossRef] [Green Version]
- Aarts, L.A.; Jeanne, V.; Cleary, J.P.; Lieber, C.; Nelson, J.S.; Oetomo, S.B.; Verkruysse, W. Non-contact heart rate monitoring utilizing camera photoplethysmography in the neonatal intensive care unit—A pilot study. Early Hum. Dev. 2013, 89, 943–948. [Google Scholar] [CrossRef] [PubMed]
- Klaessens, J.H.; van den Born, M.; van der Veen, A.; Sikkens-van de Kraats, J.; van den Dungen, F.A.; Verdaasdonk, R.M. Development of a baby friendly non-contact method for measuring vital signs: First results of clinical measurements in an open incubator at a neonatal intensive care unit. In Proceedings of the Advanced Biomedical and Clinical Diagnostic Systems XII, San Francisco, CA, USA, 27 February 2014; International Society for Optics and Photonics: Bellingham, WA, USA, 2014; p. 89351P-1-7. [Google Scholar]
- Gibson, K.; Al-Naji, A.; Fleet, J.; Steen, M.; Esterman, A.; Chahl, J.; Huynh, J.; Morris, S. Non-contact heart and respiratory rate monitoring of preterm infants based on a computer vision system: A method comparison study. Pediatric Res. 2019, 86, 738–741. [Google Scholar] [CrossRef] [PubMed]
- Lorato, I.; Stuijk, S.; Meftah, M.; Kommers, D.; Andriessen, P.; van Pul, C.; de Haan, G. Towards Continuous Camera-Based Respiration Monitoring in Infants. Sensors 2021, 21, 2268. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, J.; McDuff, D.; Quigley, K.; Maes, P.; Picard, R.W. Wearable motion-based heart rate at rest: A workplace evaluation. IEEE J. Biomed. Health Inform. 2018, 23, 1920–1927. [Google Scholar] [CrossRef]
- Hernandez, J.; McDuff, D.; Picard, R.W. Biowatch: Estimation of heart and breathing rates from wrist motions. In Proceedings of the 2015 9th International Conference on Pervasive Computing Technologies for Healthcare (PervasiveHealth), Istanbul, Turkey, 20–23 May 2015; pp. 169–176. [Google Scholar]
- Cobos-Torres, J.-C.; Abderrahim, M.; Martínez-Orgado, J. Non-Contact, Simple Neonatal Monitoring by Photoplethysmography. Sensors 2018, 18, 4362. [Google Scholar] [CrossRef] [Green Version]
- Paul, M.; Karthik, S.; Joseph, J.; Sivaprakasam, M.; Kumutha, J.; Leonhardt, S.; Antink, C.H. Non-contact sensing of neonatal pulse rate using camera-based imaging: A clinical feasibility study. Physiol. Meas. 2020, 41, 024001. [Google Scholar] [CrossRef]
- Mestha, L.K.; Kyal, S.; Xu, B.; Lewis, L.E.; Kumar, V. Towards continuous monitoring of pulse rate in neonatal intensive care unit with a webcam. In Proceedings of the 2014 36th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Chicago, IL, USA, 26–30 August 2014; pp. 3817–3820. [Google Scholar]
- Villarroel, M.; Guazzi, A.; Jorge, J.; Davis, S.; Watkinson, P.; Green, G.; Shenvi, A.; McCormick, K.; Tarassenko, L. Continuous non-contact vital sign monitoring in neonatal intensive care unit. Healthc. Technol. Lett. 2014, 1, 87–91. [Google Scholar] [CrossRef] [Green Version]
- Jorge, J.; Villarroel, M.; Chaichulee, S.; Guazzi, A.; Davis, S.; Green, G.; McCormick, K.; Tarassenko, L. Non-contact monitoring of respiration in the neonatal intensive care unit. In Proceedings of the 2017 12th IEEE International Conference on Automatic Face & Gesture Recognition (FG 2017), Washington, DC, USA, 30 May–3 June 2017; pp. 286–293. [Google Scholar]
- LeCun, Y.; Bengio, Y. Convolutional networks for images, speech, and time series. Handb. Brain Theory Neural Netw. 1995, 3361, 1–14. [Google Scholar]
- Chaichulee, S.; Villarroel, M.; Jorge, J.; Arteta, C.; Green, G.; McCormick, K.; Zisserman, A.; Tarassenko, L. Multi-task convolutional neural network for patient detection and skin segmentation in continuous non-contact vital sign monitoring. In Proceedings of the 2017 12th IEEE International Conference on Automatic Face & Gesture Recognition (FG 2017), Washington, DC, USA, 30 May–3 June 2017; pp. 266–272. [Google Scholar]
- Jorge, J.; Villarroel, M.; Chaichulee, S.; McCormick, K.; Tarassenko, L. Data fusion for improved camera-based detection of respiration in neonates. In Proceedings of the Optical Diagnostics and Sensing XVIII: Toward Point-of-Care Diagnostics, San Francisco, CA, USA, 20 February 2018; International Society for Optics and Photonics: Bellingham, WA, USA, 2018; p. 1050112. [Google Scholar]
- Villarroel, M.; Chaichulee, S.; Jorge, J.; Davis, S.; Green, G.; Arteta, C.; Zisserman, A.; McCormick, K.; Watkinson, P.; Tarassenko, L. Non-contact physiological monitoring of preterm infants in the neonatal intensive care unit. NPJ Digit. Med. 2019, 2, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Chaichulee, S.; Villarroel, M.; Jorge, J.; Arteta, C.; Green, G.; McCormick, K.; Zisserman, A.; Tarassenko, L. Localised photoplethysmography imaging for heart rate estimation of pre-term infants in the clinic. In Proceedings of the Optical Diagnostics and Sensing XVIII: Toward Point-of-Care Diagnostics, San Francisco, CA, USA, 20 February 2018; International Society for Optics and Photonics: Bellingham, WA, USA, 2018; p. 105010R. [Google Scholar]
- Redmon, J.; Farhadi, A. Yolov3: An incremental improvement. arXiv 2018, arXiv:1804.02767. [Google Scholar]
- Lin, T.-Y.; Maire, M.; Belongie, S.; Hays, J.; Perona, P.; Ramanan, D.; Dollár, P.; Zitnick, C.L. Microsoft coco: Common objects in context. In European Conference on Computer Vision; Springer: Berlin/Heidelberg, Germany, 2014; pp. 740–755. [Google Scholar]
- Girshick, R.; Donahue, J.; Darrell, T.; Malik, J. Rich feature hierarchies for accurate object detection and semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Columbus, OH, USA, 23–28 June 2014; pp. 580–587. [Google Scholar]
- Girshick, R. Fast r-cnn. In Proceedings of the IEEE International Conference on Computer Vision, Santiago, Chile, 7–13 December 2015; pp. 1440–1448. [Google Scholar]
- Eichenwald, E.C. Apnea of prematurity. Pediatrics 2016, 137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, Z.; Hidalgo, G.; Simon, T.; Wei, S.-E.; Sheikh, Y. OpenPose: Realtime multi-person 2D pose estimation using Part Affinity Fields. IEEE Trans. Pattern Anal. Mach. Intell. 2019, 43, 172–186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Redmon, J.; Divvala, S.; Girshick, R.; Farhadi, A. You only look once: Unified, real-time object detection. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 779–788. [Google Scholar]
- Chen, X.; Cheng, J.; Song, R.; Liu, Y.; Ward, R.; Wang, Z.J. Video-Based Heart Rate Measurement: Recent Advances and Future Prospects. IEEE Trans. Instrum. Meas. 2018. [Google Scholar] [CrossRef]
- Chen, D.-Y.; Wang, J.-J.; Lin, K.-Y.; Chang, H.-H.; Wu, H.-K.; Chen, Y.-S.; Lee, S.-Y. Image sensor-based heart rate evaluation from face reflectance using Hilbert–Huang transform. IEEE Sens. J. 2014, 15, 618–627. [Google Scholar] [CrossRef]
- Poh, M.-Z.; McDuff, D.J.; Picard, R.W. Non-contact, automated cardiac pulse measurements using video imaging and blind source separation. Opt. Express 2010, 18, 10762–10774. [Google Scholar] [CrossRef]
- Huang, N.E.; Shen, Z.; Long, S.R.; Wu, M.C.; Shih, H.H.; Zheng, Q.; Yen, N.-C.; Tung, C.C.; Liu, H.H. The empirical mode decomposition and the Hilbert spectrum for nonlinear and non-stationary time series analysis. Proc. R. Soc. Lond. Ser. A Math. Phys. Eng. Sci. 1998, 454, 903–995. [Google Scholar] [CrossRef]
- Blanco-Velasco, M.; Weng, B.; Barner, K.E. ECG signal denoising and baseline wander correction based on the empirical mode decomposition. Comput. Biol. Med. 2008, 38, 1–13. [Google Scholar] [CrossRef]
- Zhang, X.; Zhou, P. Filtering of surface EMG using ensemble empirical mode decomposition. Med. Eng. Phys. 2013, 35, 537–542. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Liu, A.; Chiang, J.; Wang, Z.J.; McKeown, M.J.; Ward, R.K. Removing muscle artifacts from EEG data: Multichannel or single-channel techniques? IEEE Sens. J. 2015, 16, 1986–1997. [Google Scholar] [CrossRef]
- Liu, S.; He, Q.; Gao, R.X.; Freedson, P. Empirical mode decomposition applied to tissue artifact removal from respiratory signal. In Proceedings of the 2008 30th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Vancouver, BC, Canada, 20–25 August 2008; pp. 3624–3627. [Google Scholar]
- Song, R.; Li, J.; Wang, M.; Cheng, J.; Li, C.; Chen, X. Remote Photoplethysmography with an EEMD-MCCA Method Robust Against Spatially Uneven Illuminations. IEEE Sens. J. 2021. [Google Scholar] [CrossRef]
- Wu, Z.; Huang, N.E. Ensemble empirical mode decomposition: A noise-assisted data analysis method. Adv. Adapt. Data Anal. 2009, 1, 1–41. [Google Scholar] [CrossRef]
- Zaunseder, S.; Trumpp, A.; Wedekind, D.; Malberg, H. Cardiovascular assessment by imaging photoplethysmography—A review. Biomed. Eng. Biomed. Tech. 2018, 63, 617–634. [Google Scholar] [CrossRef] [PubMed]
- Akar, S.A.; Kara, S.; Latifoğlu, F.; Bilgic, V. Spectral analysis of photoplethysmographic signals: The importance of preprocessing. Biomed. Signal Process. Control. 2013, 8, 16–22. [Google Scholar] [CrossRef]
- Liu, H.; Allen, J.; Khalid, S.G.; Chen, F.; Zheng, D. Filtering-induced time shifts in photoplethysmography pulse features measured at different body sites: The importance of filter definition and standardization. Physiol. Meas. 2021, accepted. [Google Scholar]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khanam, F.-T.-Z.; Perera, A.G.; Al-Naji, A.; Gibson, K.; Chahl, J. Non-Contact Automatic Vital Signs Monitoring of Infants in a Neonatal Intensive Care Unit Based on Neural Networks. J. Imaging 2021, 7, 122. https://doi.org/10.3390/jimaging7080122
Khanam F-T-Z, Perera AG, Al-Naji A, Gibson K, Chahl J. Non-Contact Automatic Vital Signs Monitoring of Infants in a Neonatal Intensive Care Unit Based on Neural Networks. Journal of Imaging. 2021; 7(8):122. https://doi.org/10.3390/jimaging7080122
Chicago/Turabian StyleKhanam, Fatema-Tuz-Zohra, Asanka G. Perera, Ali Al-Naji, Kim Gibson, and Javaan Chahl. 2021. "Non-Contact Automatic Vital Signs Monitoring of Infants in a Neonatal Intensive Care Unit Based on Neural Networks" Journal of Imaging 7, no. 8: 122. https://doi.org/10.3390/jimaging7080122
APA StyleKhanam, F.-T.-Z., Perera, A. G., Al-Naji, A., Gibson, K., & Chahl, J. (2021). Non-Contact Automatic Vital Signs Monitoring of Infants in a Neonatal Intensive Care Unit Based on Neural Networks. Journal of Imaging, 7(8), 122. https://doi.org/10.3390/jimaging7080122









