A System for Real-Time, Online Mixed-Reality Visualization of Cardiac Magnetic Resonance Images
Abstract
:1. Introduction
1.1. Real-Time Magnetic Resonance Imaging and Reconstruction Protocols
1.2. Medical Imaging Visualization Approaches
1.3. Research Aims
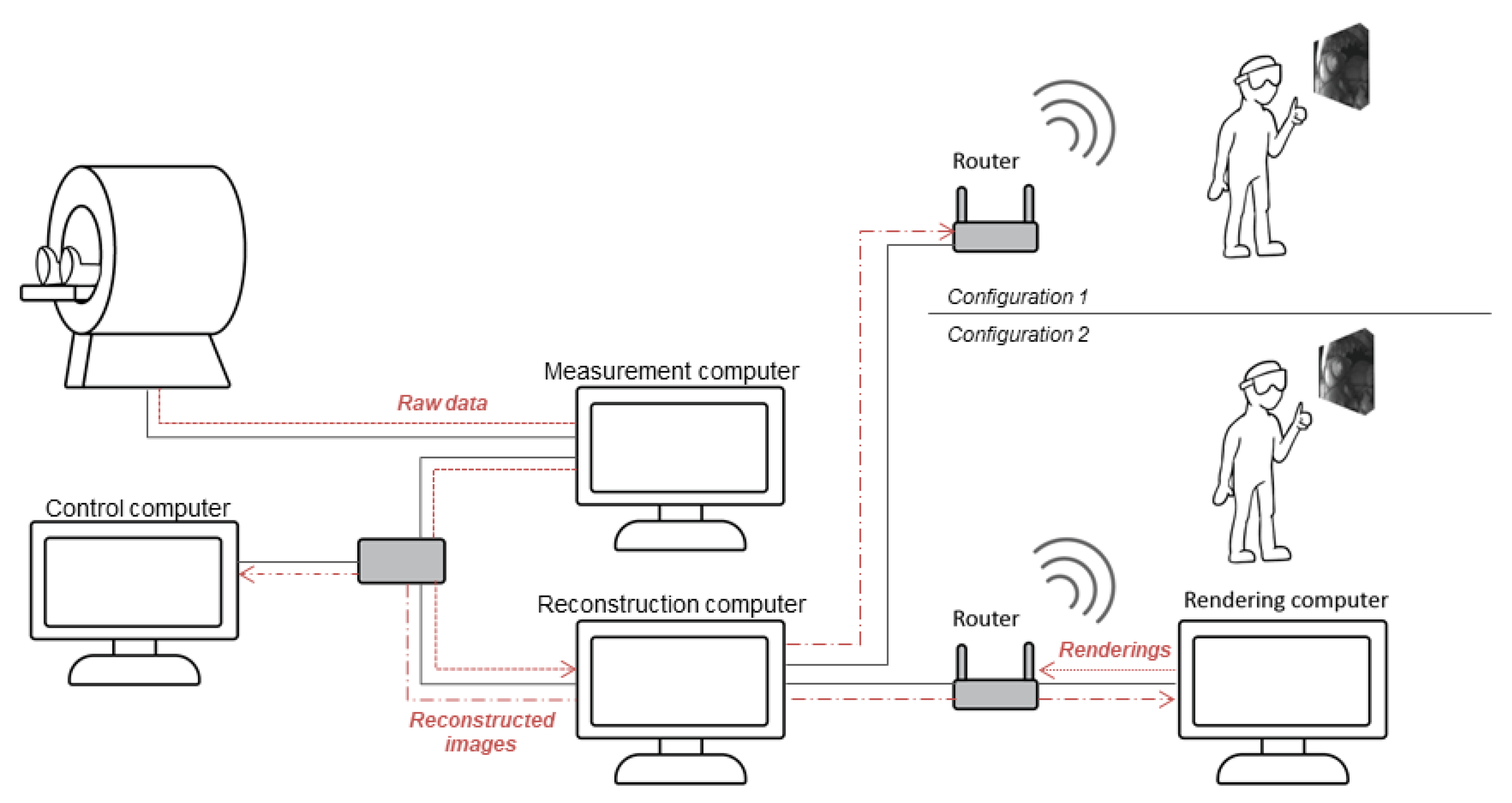
2. Materials and Methods
2.1. Data Acquisition
2.2. Image Reconstruction
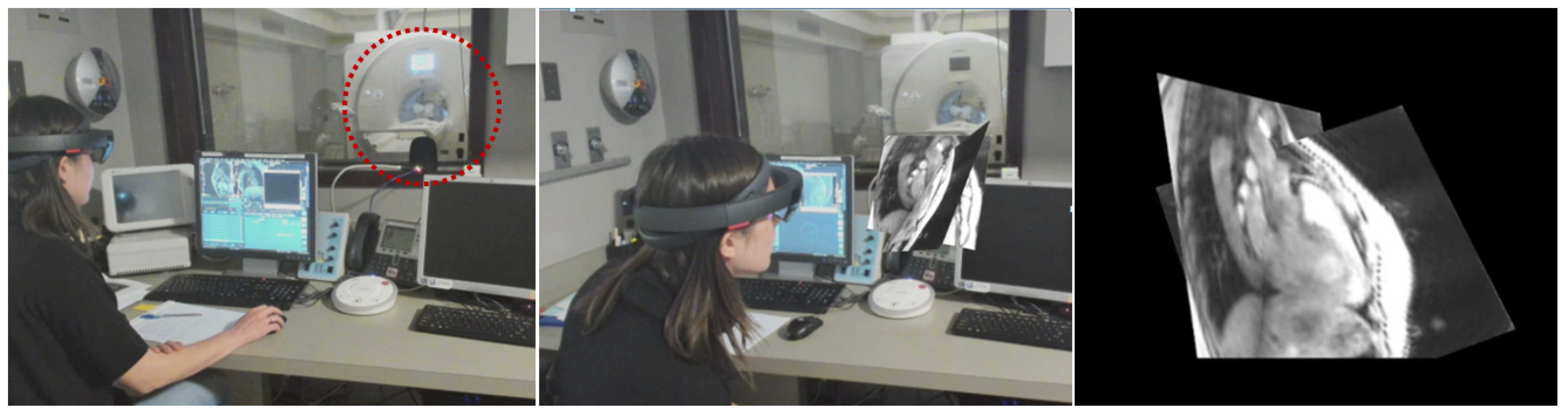
2.3. Rendering
2.4. System Integration
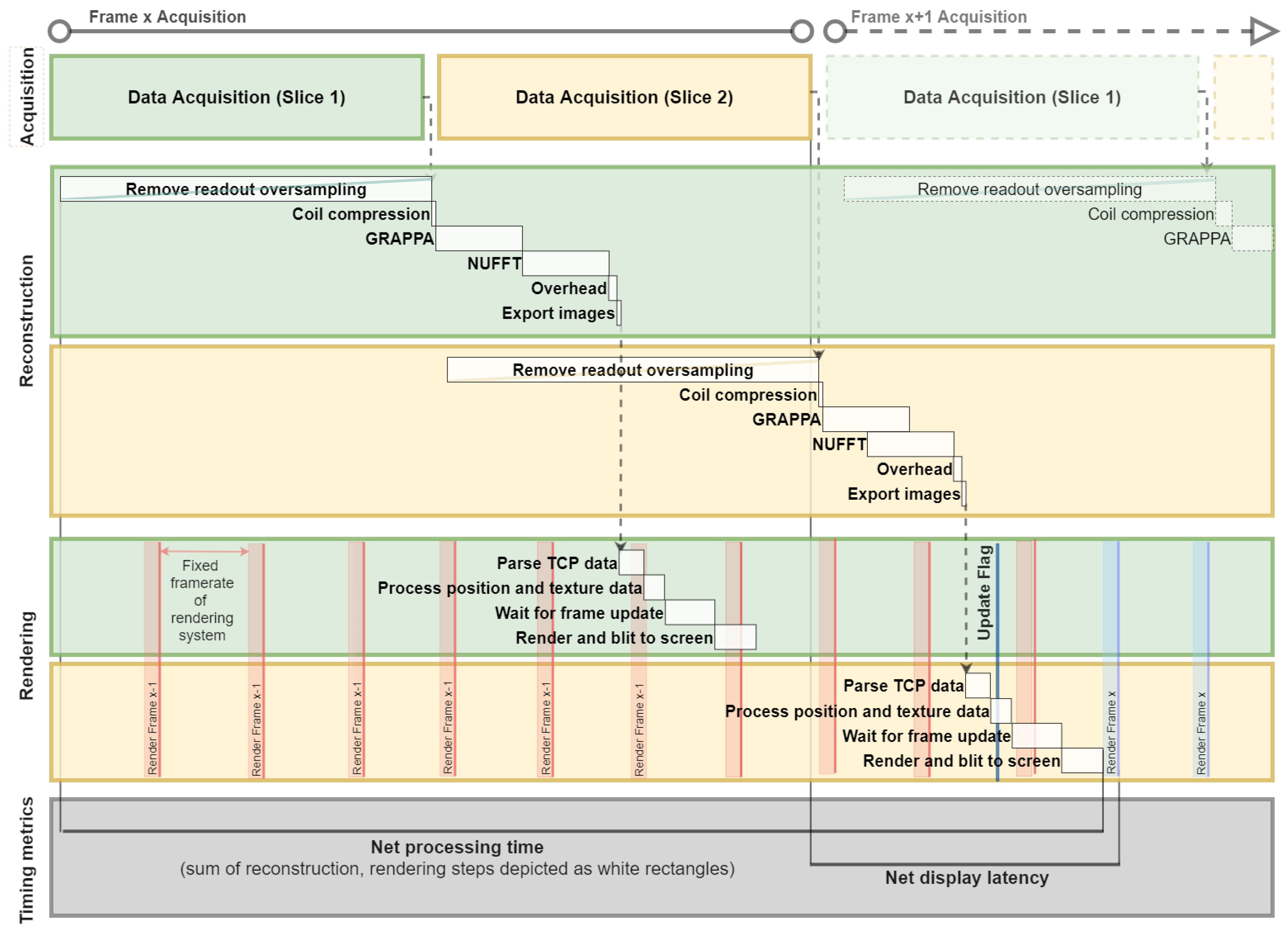
2.5. System Testing and Timing
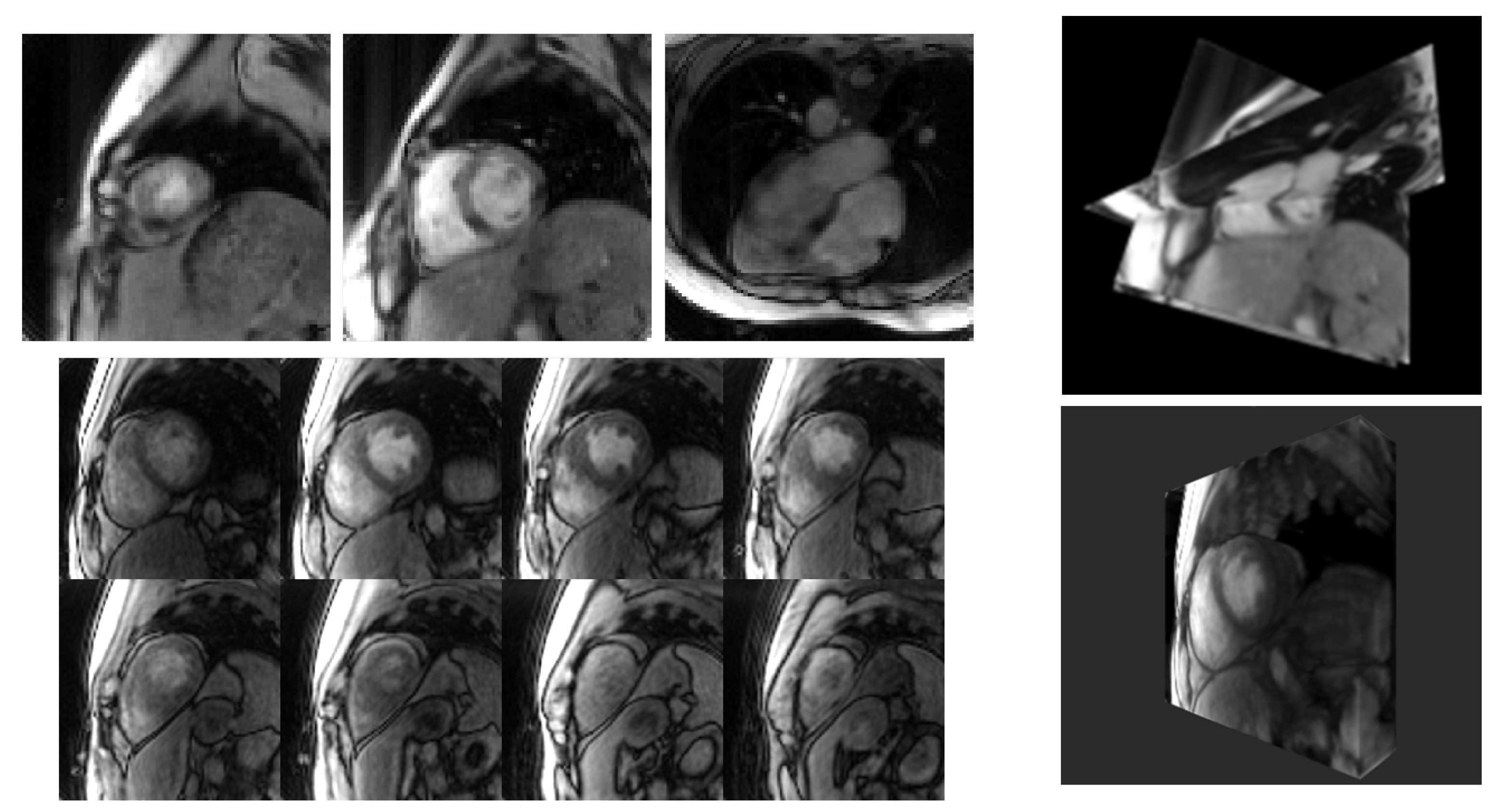
3. Results
4. Discussion
5. Limitations and Future Work
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fetterly, K.A.; Mathew, V.; Lennon, R.; Bell, M.R.; Holmes, D.R.; Rihal, C.S. Radiation dose reduction in the invasive cardiovascular laboratory: Implementing a culture and philosophy of radiation safety. JACC Cardiovasc. Interv. 2012, 5, 866–873. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pyne, C.T.; Gadey, G.; Jeon, C.; Piemonte, T.; Waxman, S.; Resnic, F. Effect of reduction of the pulse rates of fluoroscopy and CINE-acquisition on X-ray dose and angiographic image quality during invasive cardiovascular procedures. Circ. Cardiovasc. Interv. 2014, 7, 441–446. [Google Scholar] [CrossRef] [Green Version]
- Rolf, S.; Hindricks, G.; Sommer, P.; Richter, S.; Arya, A.; Bollmann, A.; Kosiuk, J.; Koutalas, E. Electroanatomical mapping of atrial fibrillation: Review of the current techniques and advances. J. Atr. Fibrillation 2014, 7, 57–68. [Google Scholar] [CrossRef]
- Weiss, C.R.; Nour, S.G.; Lewin, J.S. MR-guided biopsy: A review of current techniques and applications. J. Magn. Reson. Imaging 2008, 27, 311–325. [Google Scholar] [CrossRef]
- Kaye, E.A.; Granlund, K.L.; Morris, E.A.; Maybody, M.; Solomon, S.B. Closed-bore interventional MRI: Percutaneous biopsies and ablations. Am. J. Roentgenol. 2015, 205, W400–W410. [Google Scholar] [CrossRef]
- Campbell-Washburn, A.E.; Tavallaei, M.A.; Pop, M.; Grant, E.K.; Chubb, H.; Rhode, K.; Wright, G.A. Real-time MRI guidance of cardiac interventions. J. Magn. Reson. Imaging 2017, 46, 935–950. [Google Scholar] [CrossRef]
- Nageotte, S.J.; Lederman, R.J.; Ratnayaka, K. MRI catheterization: Ready for broad adoption. Pediatr. Cardiol. 2020, 41, 503–513. [Google Scholar] [CrossRef] [PubMed]
- Sommer, P.; Grothoff, M.; Eitel, C.; Gaspar, T.; Piorkowski, C.; Gutberlet, M.; Hindricks, G. Feasibility of real-time magnetic resonance imaging-guided electrophysiology studies in humans. Europace 2013, 15, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Chubb, H.; Harrison, J.L.; Weiss, S.; Krueger, S.; Koken, P.; Bloch, L.T.; Kim, W.Y.; Stenzel, G.S.; Wedan, S.R.; Weisz, J.L.; et al. Development, pre-clinical validation, and clinical translation of a cardiac magnetic resonance-electrophysiology system with active catheter tracking for ablation of cardiac arrhythmia. JACC Clin. Electrophysiol. 2017, 3, 89–103. [Google Scholar] [CrossRef]
- Rogers, T.; Ratnayaka, K.; Khan, J.M.; Stine, A.; Schenke, W.H.; Grant, L.P.; Mazal, J.R.; Grant, E.K.; Campbell-Washburn, A.; Hansen, M.S.; et al. CMR fluoroscopy right heart catheterization for cardiac output and pulmonary vascular resistance: R in 102 patients. J. Cardiovasc. Magn. Reson. 2017, 19, 54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ratnayaka, K.; Kanter, J.P.; Faranesh, A.Z.; Grant, E.K.; Olivieri, L.J.; Cross, R.R.; Cronin, I.F.; Hamann, K.S.; Campbell-Washburn, A.E.; Brien, K.J.O.; et al. Radiation-free CMR diagnostic heart catheterization in children. J. Cardiovasc. Comput. Tomogr. 2017, 19, 65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Veeram Reddy, S.R.; Arar, Y.; Zahr, R.A.; Gooty, V.; Hernandez, J.; Potersnak, A.; Douglas, P.; Blair, Z.; Greer, J.S.; Roujol, S.; et al. Invasive cardiovascular magnetic resonance (iCMR) for diagnostic right and left heart catheterization using an MR-conditional guidewire and passive visualization in congenital heart disease. J. Cardiovasc. Magn. Reson. 2020, 22, 20. [Google Scholar] [CrossRef] [PubMed]
- Velasco Forte, M.N.; Roujol, S.; Ruijsink, B.; Valverde, I.; Duong, P.; Byrne, N.; Krueger, S.; Weiss, S.; Arar, Y.; Reddy, S.R.; et al. MRI for guided right and left heart cardiac catheterization: A prospective study in congenital heart disease. J. Magn. Reson. Imaging 2020, 53, 1446–1457. [Google Scholar] [CrossRef]
- Pruessmann, K.P.; Weiger, M.; Scheidegger, M.B.; Boesiger, P. SENSE: Sensitivity Encoding for Fast MRI. Magn. Reson. Med. 1999, 42, 952–962. [Google Scholar] [CrossRef]
- Griswold, M.A.; Jakob, P.M.; Heidemann, R.M.; Nittka, M.; Jellus, V.; Wang, J.; Kiefer, B.; Haase, A. Generalized Autocalibrating Partially Parallel Acquisitions (GRAPPA). Magn. Reson. Med. 2002, 47, 1202–1210. [Google Scholar] [CrossRef] [Green Version]
- Sørensen, T.S.; Atkinson, D.; Schaeffter, T.; Hansen, M.S. Real-time reconstruction of sensitivity encoded radial magnetic resonance imaging using a graphics processing unit. IEEE Trans. Med. Imaging 2009, 28, 1974–1985. [Google Scholar] [CrossRef]
- Uecker, M.; Zhang, S.; Voit, D.; Karaus, A.; Merboldt, K.D.; Frahm, J. Real-time MRI at a resolution of 20 ms. NMR Biomed. 2010, 23, 986–994. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nam, S.; Akçakaya, M.; Basha, T.; Stehning, C.; Manning, W.J.; Tarokh, V.; Nezafat, R. Compressed sensing reconstruction for whole-heart imaging with 3D radial trajectories: A graphics processing unit implementation. Magn. Reson. Med. 2013, 69, 91–102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shin, T.; Nayak, K.S.; Santos, J.M.; Nishimura, D.G.; Hu, B.S.; McConnell, M.V. Three-dimensional first-pass myocardial perfusion MRI using a stack-of-spirals acquisition. Magn. Reson. Med. 2013, 69, 839–844. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bilgic, B.; Gagoski, B.A.; Cauley, S.F.; Fan, A.P.; Polimeni, J.R.; Grant, P.E.; Wald, L.L.; Setsompop, K. Wave-CAIPI for highly accelerated 3D imaging. Magn. Reson. Med. 2015, 73, 2152–2162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baron, C.A.; Dwork, N.; Pauly, J.M.; Nishimura, D.G. Rapid compressed sensing reconstruction of 3D non-Cartesian MRI. Magn. Reson. Med. 2018, 79, 2685–2692. [Google Scholar] [CrossRef] [PubMed]
- Seiberlich, N.; Ehses, P.; Duerk, J.; Gilkeson, R.; Griswold, M. Improved radial GRAPPA calibration for real-time free-breathing cardiac imaging. Magn. Reson. Med. 2011, 65, 492–505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barkauskas, K.J.; Hamilton, J.I.; Spottiswoode, B.; Zuehlsdorff, S.; Griswold, M.; Seiberlich, N. First-pass contrast-enhanced cardiac perfusion with 3D coverage per heartbeat with 3D through-time radial GRAPPA. In Proceedings of the ISMRM, Salt Lake City, UT, USA, 20–26 April 2013; p. 94557. [Google Scholar]
- Todd, N.; Prakash, J.; Od, H.; Bever, J.D.; Payne, A.; Yalavarthy, P.; Parker, D.L. Toward real-time availability of 3D temperature maps created with temporally constrained reconstruction. Magn. Reson. Med. 2014, 71, 1394–1404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ozenne, V.; Toupin, S.; Bour, P.; de Senneville, B.D.; Lepetit-Coiffe, M.; Boissenin, M.; Benois-Pineau, J.; Hansen, M.S.; Inati, S.J.; Govari, A.; et al. Improved cardiac magnetic resonance thermometry and dosimetry for monitoring lesion formation during catheter ablation. Magn. Reson. Med. 2017, 77, 673–683. [Google Scholar] [CrossRef] [PubMed]
- Unterberg-Buchwald, C.; Ritter, C.O.; Reupke, V.; Wilke, R.N.; Stadelmann, C.; Steinmetz, M.; Schuster, A.; Hasenfuß, G.; Lotz, J.; Uecker, M. Targeted endomyocardial biopsy guided by real-time cardiovascular magnetic resonance. J. Cardiovasc. Magn. Reson. 2017, 19, 45. [Google Scholar] [CrossRef] [PubMed]
- Fielden, S.W.; Feng, X.; Zhao, L.; Miller, G.W.; Geeslin, M.; Dallapiazza, R.F.; Elias, W.J.; Wintermark, M.; Butts Pauly, K.; Meyer, C.H. A spiral-based volumetric acquisition for MR temperature imaging. Magn. Reson. Med. 2018, 79, 3122–3127. [Google Scholar] [CrossRef] [PubMed]
- Saybasili, H.; Herzka, D.A.; Seiberlich, N.; Griswold, M.A. Real-time imaging with radial GRAPPA: Implementation on a heterogeneous architecture for low-latency reconstructions. Magn. Reson. Imaging 2014, 32, 747–758. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hansen, M.S.; Sørensen, T.S. Gadgetron: An open source framework for medical image reconstruction. Magn. Reson. Med. 2013, 69, 1768–1776. [Google Scholar] [CrossRef] [Green Version]
- Uecker, M.; Ong, F.; Tamir, J.; Bahri, D.; Virtue, P.; Cheng, J.; Zhang, T.; Lustig, M. Berkeley Advanced Reconstruction Toolbox. In Proceedings of the ISMRM, Toronto, ON, Canada, 30 May–5 June 2015; p. 2486. [Google Scholar]
- Center for Advanced Imaging Innovation and Research, New York, NY, USA, Yarra Framework—Open-Source Toolkit for Clinical-Translational MRI Research. Available online: https://yarra-framework.org/ (accessed on 8 December 2021).
- Sørensen, T.S.; Therkildsen, S.V.; Makowski, P.; Knudsen, J.L.; Pedersen, E.M. A new virtual reality approach for planning of cardiac interventions. Artif. Intell. Med. 2001, 22, 193–214. [Google Scholar] [CrossRef]
- Novotny, J.; Miller, W.R.; Luks, F.I.; Merck, D.; Collins, S.; Laidlaw, D.H. Towards placental surface vasculature exploration in virtual reality. IEEE Comput. Graph. Appl. 2018, 40, 28–39. [Google Scholar] [CrossRef]
- Shattuck, D.W. Multiuser virtual reality environment for visualising neuroimaging data. Healthc. Technol. Lett. 2018, 5, 183–188. [Google Scholar] [CrossRef]
- Jang, J.; Tschabrunn, C.M.; Barkagan, M.; Anter, E.; Menze, B.; Nezafat, R. Three-dimensional holographic visualization of high-resolution myocardial scar on HoloLens. PLoS ONE 2018, 13, e0205188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morales Mojica, C.M.; Velazco Garcia, J.D.; Navkar, N.V.; Balakrishnan, S.; Abinahed, J.; El Ansari, W.; Al-Rumaihi, K.; Darweesh, A.; Al-Ansari, A.; Gharib, M.; et al. A prototype holographic augmented reality interface for image-guided prostate cancer interventions. In Proceedings of the VCBM 2018—The Annual Eurographics Workshop on Visual Computing for Biology and Medicine, Granada, Spain, 20–21 September 2018; pp. 17–21. [Google Scholar] [CrossRef]
- Petersen, M.V.; Mlakar, J.; Haber, S.N.; Strick, P.L.; Griswold, M.A.; Mcintyre, C.C.; Petersen, M.V.; Mlakar, J.; Haber, S.N.; Parent, M.; et al. Holographic reconstruction of axonal pathways in the human brain. Neuron 2019, 104, 1056–1064. [Google Scholar] [CrossRef]
- Shirk, J.D.; Thiel, D.D.; Wallen, E.M.; Linehan, J.M.; White, W.M.; Badani, K.K.; Porter, J.R. Effect of 3-dimensional virtual reality models for surgical planning of robotic-assisted partial nephrectomy on surgical outcomes: A randomized clinical trial. JAMA Netw. Open 2019, 2, e1911598. [Google Scholar] [CrossRef] [PubMed]
- Velazco-garcia, J.D.; Shah, D.J.; Leiss, E.L.; Tsekos, N.V. A modular and scalable computational framework for interactive immersion into imaging data with a holographic augmented reality interface. Comput. Methods Programs Biomed. 2021, 198, 105779. [Google Scholar] [CrossRef] [PubMed]
- Collins, M.K.; Ding, V.Y.; Ball, R.L.; Dolce, D.L.; Henderson, J.M.; Halpern, C.H. Novel application of virtual reality in patient engagement for deep brain stimulation: A pilot study. Brain Stimul. 2018, 11, 935–937. [Google Scholar] [CrossRef] [PubMed]
- Wacker, F.K.; Vogt, S.; Khamene, A.; Jesberger, J.A.; Nour, S.G.; Elgort, D.R.; Sauer, F.; Duerk, J.L.; Lewin, J.S. An augmented reality system for MR image–guided needle biopsy: Initial results in a swine model. Radiology 2006, 238, 497–504. [Google Scholar] [CrossRef] [PubMed]
- Perkins, S.L.; Lin, M.A.; Hargreaves, B.A.; Daniel, B.L.; Wheeler, A.J. A mixed-reality system for breast surgical planning. In Proceedings of the 2017 IEEE International Symposium on Mixed and Augmented Reality (ISMAR-Adjunct), Nantes, France, 9–13 October 2017; pp. 269–274. [Google Scholar] [CrossRef]
- Mewes, A.; Heinrich, F.; Kägebein, U.; Hensen, B.; Wacker, F.; Hansen, C. Projector-based augmented reality system for interventional visualization inside MRI scanners. Int. J. Med. Robot. Comput. Assist. Surg. 2019, 15, e1950. [Google Scholar] [CrossRef] [Green Version]
- Franson, D.; Dupuis, A.; Gulani, V.; Griswold, M.; Seiberlich, N. Real-time acquisition, reconstruction, and mixed-reality display system for 2D and 3D cardiac MRI. In Proceedings of the ISMRM, Paris, France, 16–21 June 2018; p. 598. [Google Scholar]
- Franson, D.; Ahad, J.; Hamilton, J.; Lo, W.C.; Jiang, Y.; Chen, Y.; Seiberlich, N. Real-time 3D cardiac MRI using through-time radial GRAPPA and GPU-enabled reconstruction pipelines in the Gadgetron framework. In Proceedings of the ISMRM, Honolulu, HI, USA, 22–27 April 2017; p. 0448. [Google Scholar]
- Sorensen, T.S.; Schaeffter, T.; Noe, K.O.; Hansen, M.S. Accelerating the nonequispaced fast Fourier transform on commodity graphics hardware. IEEE Trans. Med. Imaging 2008, 27, 538–547. [Google Scholar] [CrossRef] [PubMed]
- Dupuis, A.; Franson, D.; Jiang, Y.; Mlakar, J.; Eastman, H.; Gulani, V.; Seiberlich, N.; Griswold, M. Collaborative volumetric magnetic resonance image rendering on consumer-grade devices. In Proceedings of the ISMRM, Paris, France, 16–21 June 2018; p. 3417. [Google Scholar]
- Chen, Y.; Lo, W.C.; Hamilton, J.I.; Barkauskas, K.; Saybasili, H.; Wright, K.L.; Batesole, J.; Griswold, M.A.; Gulani, V.; Seiberlich, N. Single breath-hold 3D cardiac T1mapping using through-time spiral GRAPPA. NMR Biomed. 2018, 31, e3923. [Google Scholar] [CrossRef] [PubMed]
- Jaubert, O.; Montalt-Tordera, J.; Knight, D.; Coghlan, G.J.; Arridge, S.; Steeden, J.A.; Muthurangu, V. Real-time deep artifact suppression using recurrent U-Nets for low-latency cardiac MRI. Magn. Reson. Med. 2021, 86, 1904–1916. [Google Scholar] [CrossRef]
- Valsangiacomo Buechel, E.; Grosse-Wortmann, L.; Fratz, S.; Eichhorn, J.; Sarikouch, S.; Greil, G.; Beerbaum, P.; Bucciarelli-Ducci, C.; Bonello, B.; Sieverding, L.; et al. Indications for cardiovascular magnetic resonance in children with congenital and acquired heart disease: An expert consensus paper of the Imaging Working Group of the AEPC and the Cardiovascular Magnetic Resonance Section of the EACVI. Eur. Hear. J.-Cardiovasc. Imaging 2015, 16, 281–297. [Google Scholar] [CrossRef]
- Southworth, M.K.; Silva, J.R.; Silva, J.N. Use of extended realities in cardiology. Trends Cardiovasc. Med. 2020, 30, 143–148. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Poirot, N.L.; Franson, D.; Seiberlich, N.; Griswold, M.A.; Cavusoglu, M.C. Modeling and validation of the three-dimensional deflection of an MRI-compatible magnetically actuated steerable catheter. IEEE Trans. Biomed. Eng. 2016, 63, 2142–2154. [Google Scholar] [CrossRef]
- Linder, N.; Schaudinn, A.; Petersen, T.O.; Bailis, N.; Stumpp, P.; Horn, L.C.; Stolzenburg, J.U.; Kahn, T.; Moche, M.; Busse, H. In-bore biopsies of the prostate assisted by a remote-controlled manipulator at 1.5 T. Magn. Reson. Mater. Physics, Biol. Med. 2019, 32, 599–605. [Google Scholar] [CrossRef]
- Patel, N.A.; Li, G.; Shang, W.; Wartenberg, M.; Heffter, T.; Burdette, E.C.; Iordachita, I.; Tokuda, J.; Hata, N.; Tempany, C.M.; et al. System integration and preliminary clinical evaluation of a robotic system for MRI-guided transperineal prostate biopsy. J. Med. Robot. Res. 2019, 4, 1950001. [Google Scholar] [CrossRef] [PubMed]
- Velazco Garcia, J.D.; Navkar, N.V.; Gui, D.; Morales, C.M.; Christoforou, E.G.; Ozcan, A.; Abinahed, J.; Al-Ansari, A.; Webb, A.; Seimenis, I.; et al. A platform integrating acquisition, reconstruction, visualization, and manipulator control modules for MRI-guided interventions. J. Digit. Imaging 2019, 32, 420–432. [Google Scholar] [CrossRef]
- Codella, N.C.F.; Spincemaille, P.; Prince, M.; Wang, Y. A radial self-calibrated (RASCAL) generalized autocalibrating partially parallel acquisition (GRAPPA) method using weight interpolation. NMR Biomed. 2011, 24, 844–854. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sayin, O.; Saybasili, H.; Zviman, M.M.; Griswold, M.; Halperin, H.; Seiberlich, N.; Herzka, D.A. Real-time free-breathing cardiac imaging with self-calibrated through-time radial GRAPPA. Magn. Reson. Med. 2017, 77, 250–264. [Google Scholar] [CrossRef] [Green Version]
- Franson, D.; Hamilton, J.; Griswold, M.; Seiberlich, N. Self-calibrating through-time spiral GRAPPA for flexible real-time imaging. In Proceedings of the ISMRM, Montréal, QC, Canada, 11–16 May 2019; p. 1184. [Google Scholar]
- Chieh, S.W.; Kaveh, M.; Akçakaya, M.; Moeller, S. Self-calibrated interpolation of non-Cartesian data with GRAPPA in parallel imaging. Magn. Reson. Med. 2020, 83, 1837–1850. [Google Scholar] [CrossRef] [PubMed]
- Dupuis, A.; Ma, D.; Griswold, M.A. Scanner control and realtime visualization via wireless augmented reality. In Proceedings of the ISMRM, Online, 4–8 October 2020; p. 4135. [Google Scholar]
- Dupuis, A.; Franson, D.; Seiberlich, N.; Griswold, M.A. Interactive hand gestures for HoloLens rendering control of real-time MR images. In Proceedings of the ISMRM, Montréal, QC, Canada, 11–16 May 2019; p. 1581. [Google Scholar]



| Planar (Slices) | Volumetric (Partitions) | ||||
|---|---|---|---|---|---|
| 1 | 2 | 3 | 8 | 12 | |
| Remove Readout Oversampling | 8.94 (±1.28) | 8.74 (±1.17) | 8.74 (±1.21) | 95.3 (±7.35) | 112 (±7.64) |
| Coil Compression | 0.478 (±0.149) | 0.486 (±0.109) | 0.480 (±0.110) | 5.08 (±0.889) | 6.08 (±1.14) |
| GRAPPA | 10.7 (±1.49) | 10.7 (±1.51) | 10.7 (±1.49) | 119 (±3.49) | 159 (±4.23) |
| NUFFT | 15.3 (±1.76) | 15.3 (±1.79) | 15.3 (±1.81) | 46.3 (±2.73) | 54.8 (±2.52) |
| Export Data to TCP | 0.750 (±0.461) | 0.720 (±0.429) | 0.727 (±0.555) | 3.69 (±1.16) | 3.47 (±0.908) |
| Overhead | 1.60 (±0.203) | 1.60 (±0.236) | 1.60 (±0.178) | 5.79 (±0.956) | 6.86 (±0.881) |
| Total Reconstruction Latency (ms) | 29.1 (±2.42) | 29.1 (±2.37) | 29.1 (±2.42) | 181 (±5.36) | 230 (±5.54) |
| Parse TCP Data | 2.24 (±2.68) | 2.28 (±2.30) | 2.35 (±1.95) | 32.0 (±12.5) | 46.7 (±14.3) |
| Process Position and Texture Data | 1.53 (±2.76) | 1.60 (±1.04) | 1.33 (±6.71) | - | - |
| Wait for Frame Update (vSync) | 5.25 (±4.66) | 5.65 (±5.90) | 6.09 (±5.74) | 6.23 (±5.27) | 5.88 (±4.50) |
| Render and Blit to Screen | 3.02 (±0.789) | 2.90 (±0.737) | 2.93 (±0.756) | 3.10 (±1.05) | 3.04 (±1.22) |
| Total Rendering Display Latency (ms) | 12.2 (±7.42) | 12.5 (±8.20) | 13.0 (±7.71) | 41.3 (±17.2) | 55.6 (±18.3) |
| Net Display Latency (ms) | 41.3 (±10.2) | 41.6 (±10.8) | 42.1 (±10.1) | 222 (±22.6) | 286 (±24.0) |
| Acquisition (ms) | 46 | 92 | 138 | 467 | 588 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Franson, D.; Dupuis, A.; Gulani, V.; Griswold, M.; Seiberlich, N. A System for Real-Time, Online Mixed-Reality Visualization of Cardiac Magnetic Resonance Images. J. Imaging 2021, 7, 274. https://doi.org/10.3390/jimaging7120274
Franson D, Dupuis A, Gulani V, Griswold M, Seiberlich N. A System for Real-Time, Online Mixed-Reality Visualization of Cardiac Magnetic Resonance Images. Journal of Imaging. 2021; 7(12):274. https://doi.org/10.3390/jimaging7120274
Chicago/Turabian StyleFranson, Dominique, Andrew Dupuis, Vikas Gulani, Mark Griswold, and Nicole Seiberlich. 2021. "A System for Real-Time, Online Mixed-Reality Visualization of Cardiac Magnetic Resonance Images" Journal of Imaging 7, no. 12: 274. https://doi.org/10.3390/jimaging7120274
APA StyleFranson, D., Dupuis, A., Gulani, V., Griswold, M., & Seiberlich, N. (2021). A System for Real-Time, Online Mixed-Reality Visualization of Cardiac Magnetic Resonance Images. Journal of Imaging, 7(12), 274. https://doi.org/10.3390/jimaging7120274






