Fast Fiber Orientation Estimation in Diffusion MRI from kq-Space Sampling and Anatomical Priors
Abstract
1. Introduction
2. Materials and Methods
2.1. Background
2.1.1. k and q Spaces Overview
2.1.2. FOD Recovery via SD Framework
2.2. Inverse Problem Formulation
2.2.1. Proposed Measurement Model
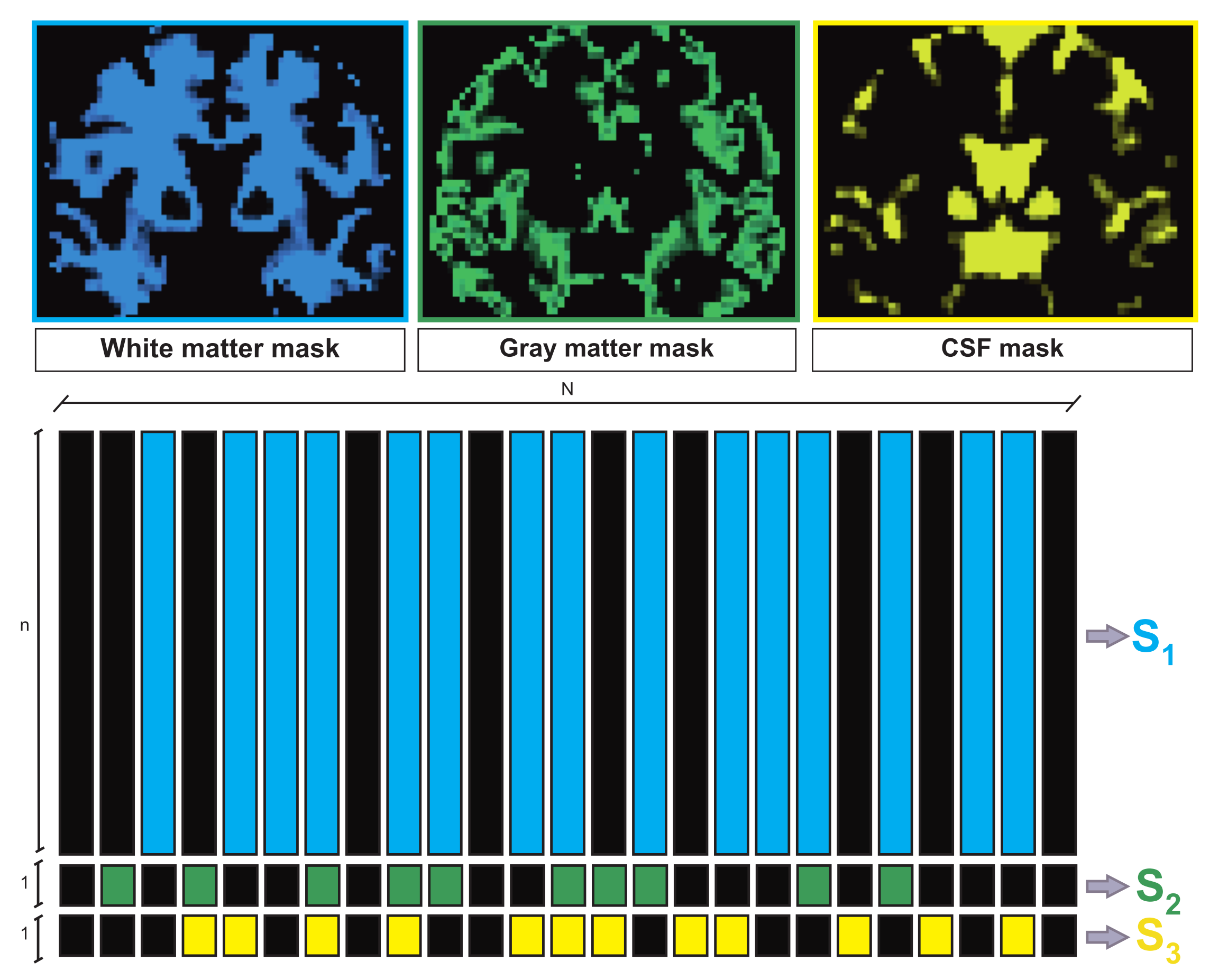
2.2.2. Tissue Segmentation Constraints
2.3. Minimization Problem and Algorithm
2.3.1. Constrained Weighted- Minimization Problem
2.3.2. Algorithm for Constrained Weighted Minimization
| Algorithm 1 Stochastic FB to solve (7) |
|
2.3.3. Weights Computation and Reweighting Procedure
| Algorithm 2 Reweighting procedure |
|
2.3.4. Data Post-Processing
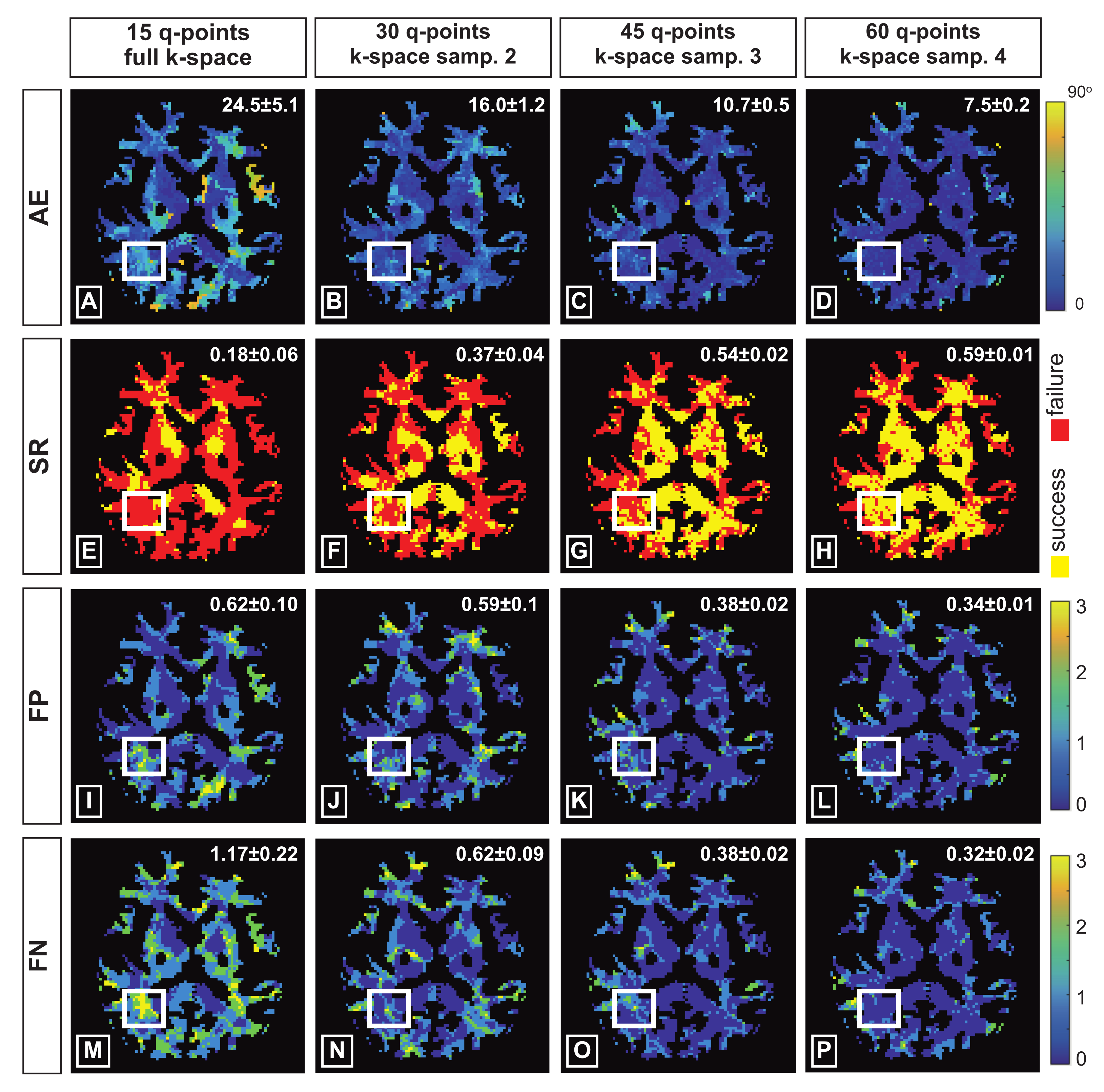
3. Experimental Setting
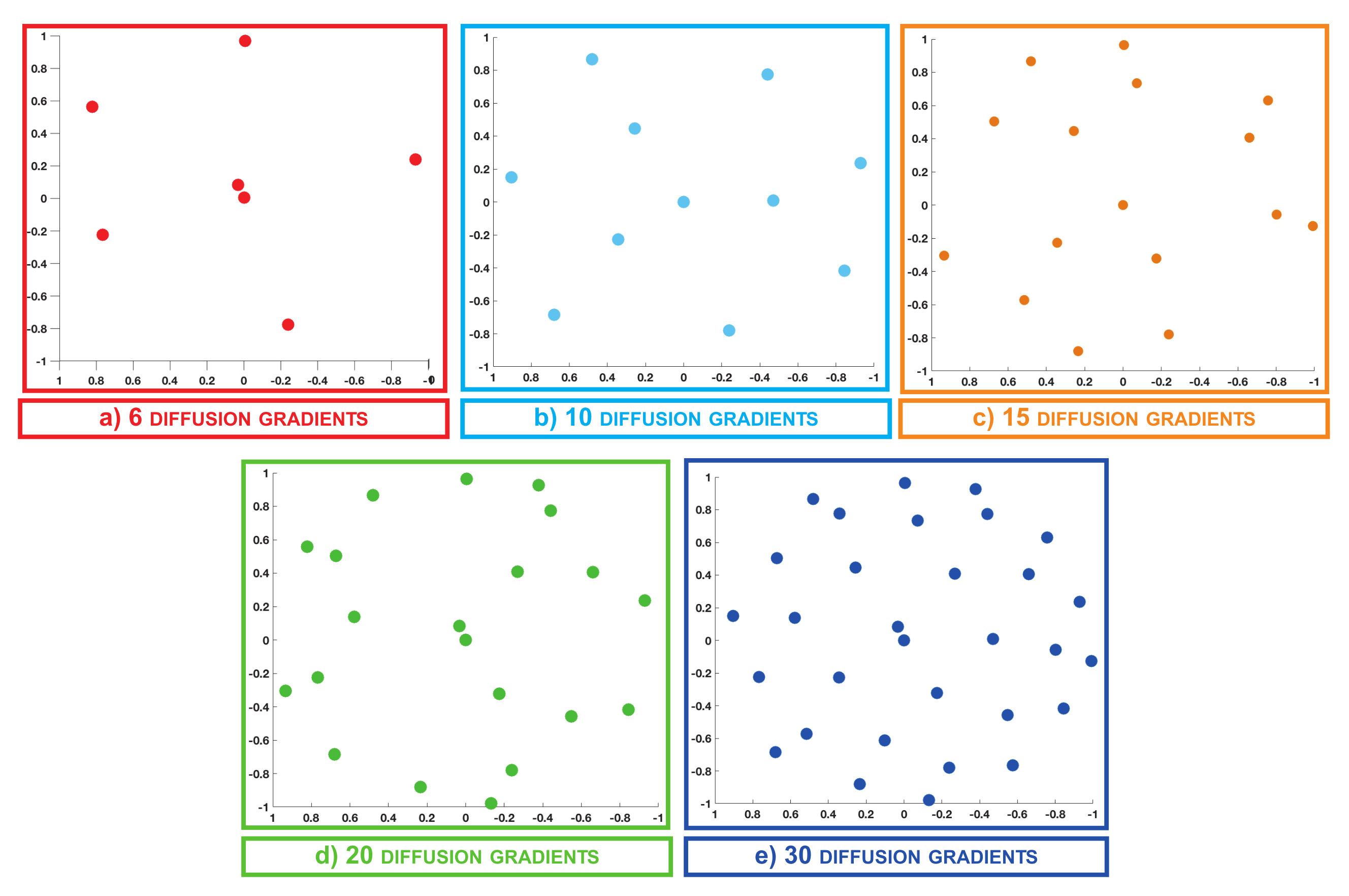
3.1. q-Space Under-Sampling
3.2. k-Space Under-Sampling
3.3. Dictionary Generation
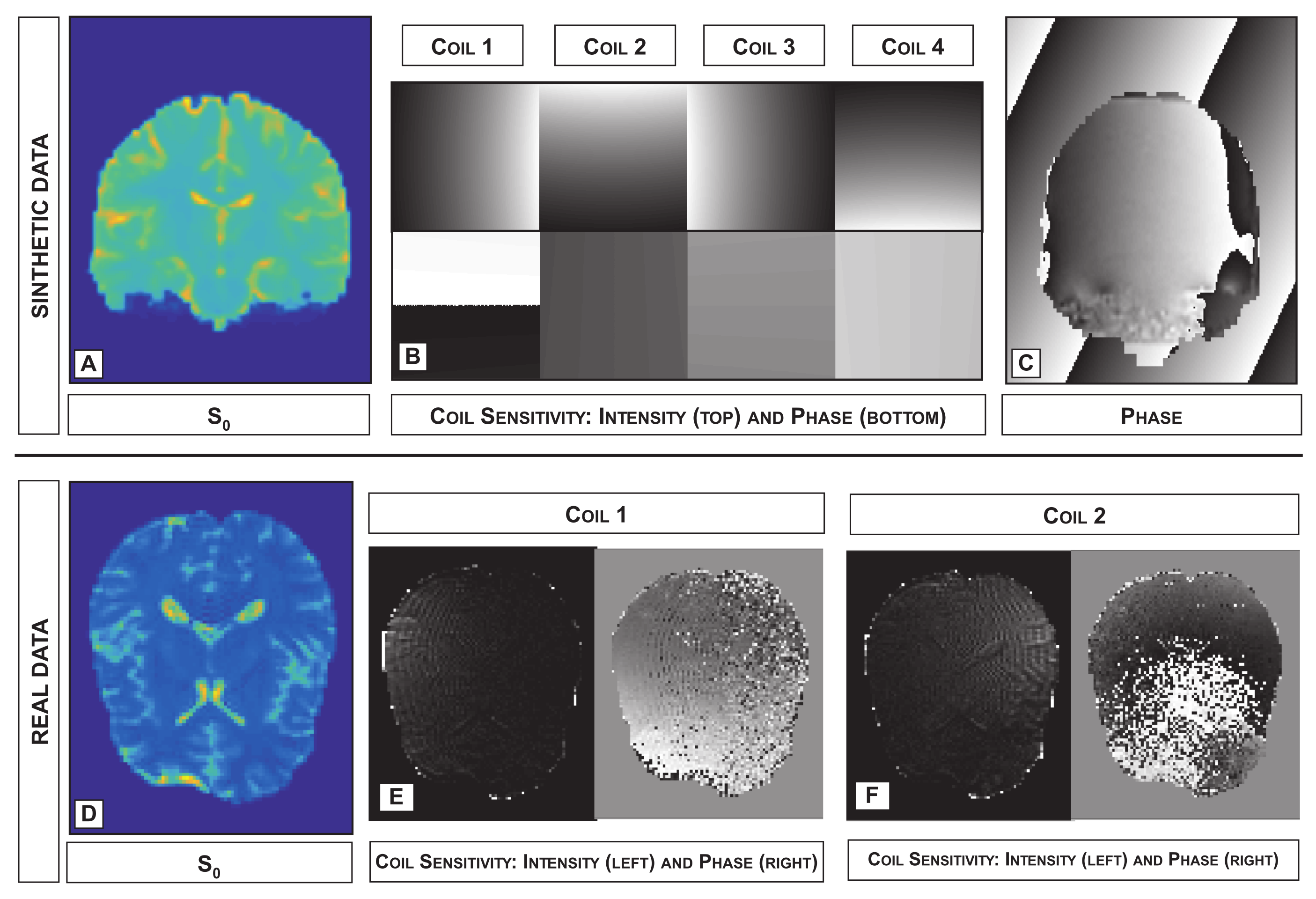
3.4. Phase Estimation
3.5. Coil Sensitivity Estimation
3.6. Evaluation Criteria
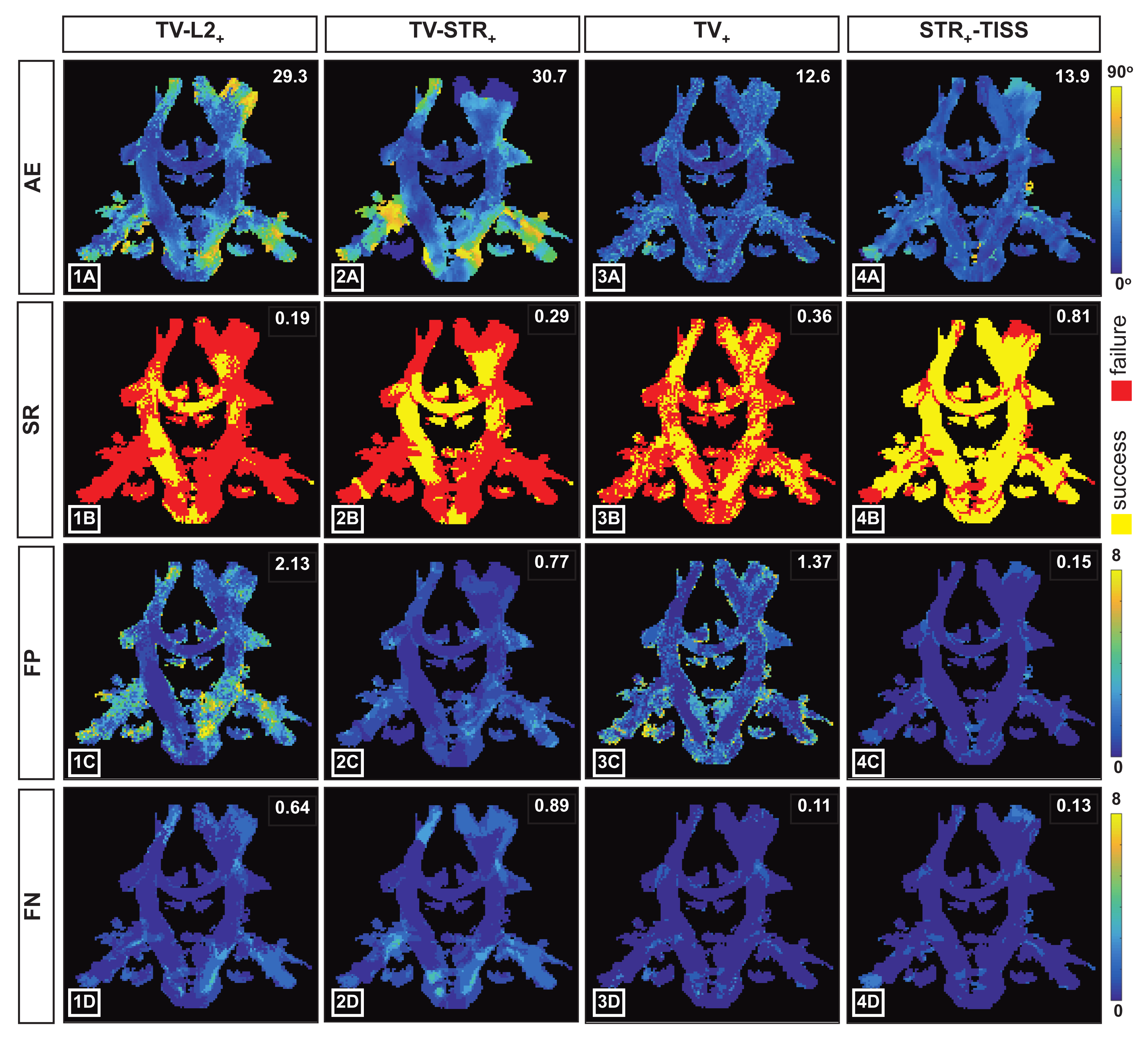
4. Results
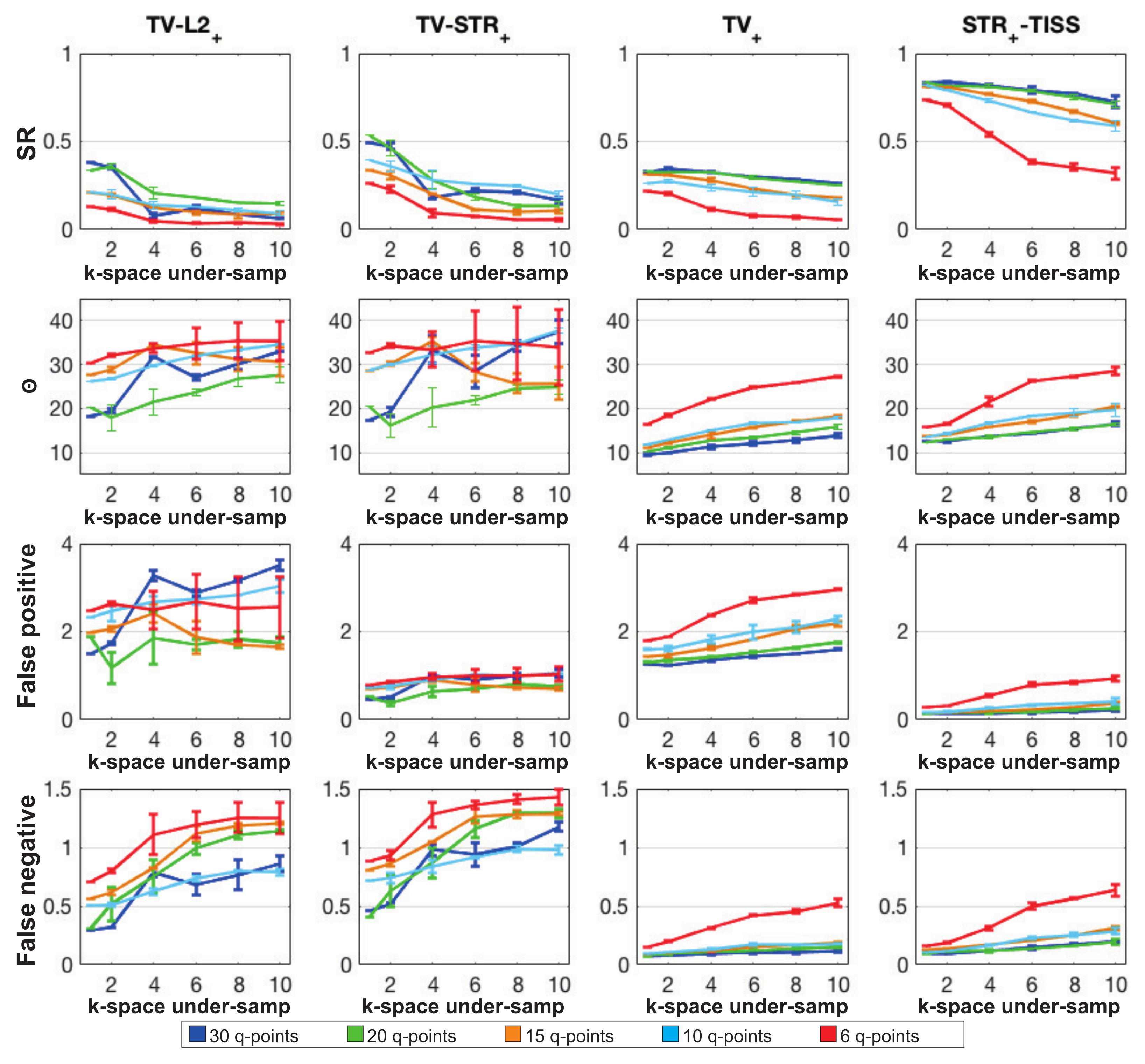
4.1. Synthetic Data
- 1.
- is a two-step approach consisting in a first step where the DW volumes are recovered by using the regularization and in a second step where FOD coefficients are recovered by relying on the non-negative least squares problem.where contains the signal in kq-space. Complex DW volumes are recovered in from the TV regularized problem, where , with for and . To exclude the remaining phase contamination, the magnitude of the complex images is considered for the FOD recovery (i.e., ).where for all . Positivity of the FOD coefficients is imposed in this step.
- 2.
- is a two-step approach consisting in the recovery of both DW volumes and FOD coefficients from regularized problems. In the first step, the prior is considered while, in the second step, the structured sparsity prior proposed in [24] is exploited.
- 3.
- is a one-step approach consisting in the recovery of the FOD coefficients from the kq-space signal applying the prior to the images in order to implicitly promote a smooth variation of the FOD coefficients within neighbor voxels.
- 4.
- is the proposed one-step approach consisting in the recovery of the FOD coefficients by solving the problem proposed in (7) where the structured sparsity prior and the spatial distribution of the different tissues is taken into account.
4.2. Real Data
5. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jara, H. Theory of Quantitative Magnetic Resonance Imaging; World Scientific: Singapore, 2013. [Google Scholar]
- Le Bihan, D. Looking into the functional architecture of the brain with diffusion MRI. Nat. Rev. Neurosci. 2003, 4, 469–480. [Google Scholar] [CrossRef] [PubMed]
- Sporns, O.; Tononi, G.; Kötter, R. The human connectome: A structural description of the human brain. PLoS Comput. Biol. 2005, 1, 0245–0251. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Schuff, N.; Du, A.T.; Rosen, H.J.; Kramer, J.H.; Gorno-Tempini, M.L.; Miller, B.L.; Weiner, M.W. White matter damage in frontotemporal dementia and Alzheimer’s disease measured by diffusion MRI. Brain 2009, 132, 2579–2592. [Google Scholar] [CrossRef] [PubMed]
- Park, H.J.; Westin, C.F.; Kubicki, M.; Maier, S.E.; Niznikiewicz, M.; Baer, A.; Frumin, M.; Kikinis, R.; Jolesz, F.A.; McCarley, R.W.; et al. White matter hemisphere asymmetries in healthy subjects and in schizophrenia: A diffusion tensor MRI study. NeuroImage 2004, 23, 213–223. [Google Scholar] [CrossRef]
- Callaghan, P.T.; Eccles, C.D.; Xia, Y. NMR microscopy of dynamic displacements: K-space and q-space imaging. J. Phys. E Sci. Instrum. 2000, 21, 820–822. [Google Scholar] [CrossRef]
- Basser, P.J.; Mattiello, J.; Le Bihan, D. MR Diffusion Tensor Spectroscopy and Imaging. Biophys. J. 1994, 66, 259–267. [Google Scholar] [CrossRef]
- Tuch, D.S.; Reese, T.G.; Wiegell, M.R.; Makris, N.; Belliveau, J.W.; Wedeen, V.J. High angular resolution diffusion imaging reveals intravoxel white matter fiber heterogeneity. Magn. Reson. Med. 2002, 48, 577–582. [Google Scholar] [CrossRef] [PubMed]
- Wedeen, V.J.; Hagmann, P.; Tseng, W.Y.I.; Reese, T.G.; Weisskoff, R.M. Mapping complex tissue architecture with diffusion spectrum magnetic resonance imaging. Magn. Reson. Med. 2005, 54, 1377–1386. [Google Scholar] [CrossRef] [PubMed]
- Tuch, D.S. Q-ball imaging. Magn. Reson. Med. 2004, 52, 1358–1372. [Google Scholar] [CrossRef]
- Zhang, H.; Hubbard, P.L.; Parker, G.J.M.; Alexander, D.C. Axon diameter mapping in the presence of orientation dispersion with diffusion MRI. NeuroImage 2011, 56, 1301–1315. [Google Scholar] [CrossRef]
- Zhang, H.; Schneider, T.; Wheeler-Kingshott, C.A.; Alexander, D.C. NODDI: Practical in vivo neurite orientation dispersion and density imaging of the human brain. NeuroImage 2012, 61, 1000–1016. [Google Scholar] [CrossRef]
- Daducci, A.; Canales-Rodriguez, E.J.; Descoteaux, M.; Garyfallidis, E.; Gur, Y.; Lin, Y.C.; Mani, M.; Merlet, S.; Paquette, M.; Ramirez-Manzanares, A.; et al. Quantitative comparison of reconstruction methods for intra-voxel fiber recovery from diffusion MRI. IEEE Trans. Med. Imaging 2014, 33, 384–399. [Google Scholar] [CrossRef]
- Ning, L.; Laun, F.; Gur, Y.; DiBella, E.V.; Deslauriers-Gauthier, S.; Megherbi, T.; Ghosh, A.; Zucchelli, M.; Menegaz, G.; Fick, R.; et al. Sparse Reconstruction Challenge for diffusion MRI: Validation on a physical phantom to determine which acquisition scheme and analysis method to use? Med. Image Anal. 2015, 26, 316–331. [Google Scholar] [CrossRef] [PubMed]
- Tournier, J.D.; Calamante, F.; Gadian, D.G.; Connelly, A. Direct estimation of the fiber orientation density function from diffusion-weighted MRI data using spherical deconvolution. NeuroImage 2004, 23, 1176–1185. [Google Scholar] [CrossRef]
- Alexander, D.C. Maximum entropy spherical deconvolution for diffusion MRI. Inf. Process. Med. Imaging 2005, 19, 76–87. [Google Scholar]
- Acqua, F.D.; Rizzo, G.; Scifo, P.; Clarke, R.A.; Scotti, G.; Fazio, F. A Model-Based Deconvolution Approach to Solve Fiber Crossing in Diffusion-Weighted MR Imaging. IEEE Trans. Biomed. Eng. 2007, 54, 462–472. [Google Scholar] [CrossRef] [PubMed]
- Jian, B.; Vemuri, B.C. A unified computational framework for deconvolution to reconstruct multiple fibers from diffusion weighted MRI. IEEE Trans. Med. Imaging 2007, 26, 1464–1471. [Google Scholar] [CrossRef]
- Ramirez-Manzanares, A.; Rivera, M.; Vemuri, B.C.; Carney, P.; Mareei, T. Diffusion basis functions decomposition for estimating white matter intravoxel fiber geometry. IEEE Trans. Med. Imaging 2007, 26, 1091–1102. [Google Scholar] [CrossRef]
- Tristán-Vega, A.; Westin, C.F. Probabilistic ODF estimation from reduced HARDI data with sparse regularization. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Berlin/Heidelberg, Germany, 2011; Volume 6892 LNCS, pp. 182–190. [Google Scholar]
- Tournier, J.D.; Calamante, F.; Connelly, A. Robust determination of the fibre orientation distribution in diffusion MRI: Non-negativity constrained super-resolved spherical deconvolution. NeuroImage 2007, 35, 1459–1472. [Google Scholar] [CrossRef] [PubMed]
- Daducci, A.; Van De Ville, D.; Thiran, J.P.; Wiaux, Y. Sparse regularization for fiber ODF reconstruction: From the suboptimality of ℓ2 and ℓ1 priors to ℓ0. Med. Image Anal. 2014, 18, 820–833. [Google Scholar] [CrossRef]
- Landman, B.A.; Bogovic, J.A.; Wan, H.; ElShahaby, F.E.Z.; Bazin, P.L.; Prince, J.L. Resolution of crossing fibers with constrained compressed sensing using diffusion tensor MRI. NeuroImage 2012, 59, 2175–2186. [Google Scholar] [CrossRef] [PubMed]
- Auría, A.; Daducci, A.; Thiran, J.P.; Wiaux, Y. Structured sparsity for spatially coherent fibre orientation estimation in diffusion MRI. NeuroImage 2015, 115, 245–255. [Google Scholar] [CrossRef][Green Version]
- Michailovich, O.; Rathi, Y.; Dolui, S. Spatially Regularized Compressed Sensing for High Angular Resolution Diffusion Imaging. IEEE Trans. Med. Imaging 2011, 30, 1100–1115. [Google Scholar] [CrossRef]
- Mani, M.; Jacob, M.; Guidon, A.; Magnotta, V.; Zhong, J. Acceleration of high angular and spatial resolution diffusion imaging using compressed sensing with multichannel spiral data. Magn. Reson. Med. 2015, 73, 126–138. [Google Scholar] [CrossRef] [PubMed]
- Donoho, D.L. Compressed sensing. IEEE Trans. Inf. Theory 2006, 52, 1289–1306. [Google Scholar] [CrossRef]
- Candès, E.J. Compressive sampling. In Proceedings of the International Congress of Mathematicians, Madrid, Spain, 22–30 August 2006; pp. 1433–1452. [Google Scholar]
- Candes, E.J.; Wakin, M.B.; Boyd, S.P. Enhancing sparsity by reweighted L1 minimisation. J. Fourier Anal. Appl. 2008, 14, 877–905. [Google Scholar] [CrossRef]
- Calabrese, E.; Badea, A.; Coe, C.L.; Lubach, G.R.; Styner, M.A.; Johnson, G.A. Investigating the tradeoffs between spatial resolution and diffusion sampling for brain mapping with diffusion tractography: Time well spent? Hum. Brain Mapp. 2014, 35, 5667–5685. [Google Scholar] [CrossRef]
- Zhan, L.; Jahanshad, N.; Ennis, D.B.; Jin, Y.; Bernstein, M.A.; Borowski, B.J.; Jack, C.R.; Toga, A.W.; Leow, A.D.; Thompson, P.M. Angular Versus Spatial Resolution Trade-Offs for Diffusion Imaging Under Time Constraints. Hum. Brain Mapp. 2013, 32, 2688–2706. [Google Scholar] [CrossRef][Green Version]
- Vos, S.B.; Aksoy, M.; Han, Z.; Holdsworth, S.J.; Maclaren, J.; Viergever, M.A.; Leemans, A.; Bammer, R. NeuroImage Trade-off between angular and spatial resolutions in vivo fiber tractography. NeuroImage 2016, 129, 117–132. [Google Scholar] [CrossRef] [PubMed]
- Scherrer, B.; Afacan, O.; Taquet, M.; Prabhu, S.P.; Gholipour, A.; Warfield, K. Super-resolution reconstruction to increase the spatial resolution of diffusion weighted images from orthogonal anisotropic acquisition. HHS Pubblic Access 2012, 16, 1465–1476. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Li, L.; Zhang, K.; Zhou, W.; Hu, X. PCLR: Phase-constrained low-rank model for compressive diffusion-weighted MRI. Magn. Reson. Med. 2014, 72, 1330–1341. [Google Scholar] [CrossRef] [PubMed]
- Haldar, J.P.; Wedeen, V.J.; Nezamzadeh, M.; Dai, G.; Weiner, M.W.; Schuff, N.; Liang, Z.P. Improved diffusion imaging through SNR-enhancing joint reconstruction. Magn. Reson. Med. 2013, 69, 277–289. [Google Scholar] [CrossRef]
- Jeong, E.K.; Kim, S.E.; Parker, D.L. High-resolution diffusion-weighted 3D MRI, using diffusion-weighted driven-equilibrium (DW-DE) and multishot segmented 3D-SSFP without navigator echoes. Magn. Reson. Med. 2003, 50, 821–829. [Google Scholar] [CrossRef]
- Cheng, J.; Shen, D.; Basser, P.J.; Yap, P.T. Joint 6D k-q Space Compressed Sensing for Accelerated High Angular Resolution Diffusion MRI. In International Conference on Information Processing in Medical Imaging; Springer: Cham, Switzerland, 2015; Volume 9123, pp. 782–793. [Google Scholar]
- Mani, M.M.M.; Jacob, M. qModeL: A plug-and-play model-based reconstruction for highly accelerated multi-shot diffusion MRI using learned priors. Magn. Reson. Med. 2021, 86, 835–851. [Google Scholar] [CrossRef]
- Ramos-Llordén, G.; Ning, L.; Liao, R.M.C.; Michailovich, O.; Setsompop, Y.R.K. High-fidelity, accelerated whole-brain submillimeter in vivo. Magn. Reson. Med. 2020, 84, 1781–1795. [Google Scholar] [CrossRef]
- Sun, J.; Sakhaee, E.; Entezari, A.; Vemuri, B.C. Leveraging EAP-Sparsity for Compressed Sensing of MS-HARDI in (k,q)-Space. In International Conference on Information Processing in Medical Imaging; Springer: Cham, Switzerland, 2015; Volume 24, pp. 375–386. [Google Scholar]
- Awate, S.P.; Dibella, V.R.E. Compressed sensing HARDI via rotation-invariant concise dictionaries, flexible k-space under-sampling, and multiscale spatial regularity. In Proceedings of the 2013 IEEE 10th International Symposium on Biomedical Imaging, San Francisco, CA, USA, 7–11 April 2013; pp. 9–12. [Google Scholar] [CrossRef]
- Combettes, P.L.; Pesquet, J.C. Stochastic forward-backward and primal-dual approximation algorithms with application to online image restoration. In Proceedings of the 2016 24th European Signal Processing Conference (EUSIPCO), Budapest, Hungary, 29 August–2 September 2016; Volume 1, pp. 1813–1817. [Google Scholar]
- Gudbajartsson, S.P.H. The Rician Distribution of Noisy MRI Data. Changes 1995, 29, 997–1003. [Google Scholar] [CrossRef]
- Henkelman, R.M. Measurement of signal intensities in the presence of noise in MR images. Med. Phys. 1985, 12, 232–233. [Google Scholar] [CrossRef]
- Chen, S.S.; Donoho, D.L.; Saunders, M.A. Atomic Decomposition by Basis Pursuit. SIAM J. Sci. Comput. 2001, 43, 129–159. [Google Scholar] [CrossRef]
- Donoho, D.L. De-Noising by Soft-Thresholding. IEEE Trans. Inf. Theory 1995, 41, 613–627. [Google Scholar] [CrossRef]
- Ochs, P.; Dosovitskiy, A.; Brox, T.; Pock, T. On iteratively reweighted algorithms for non-smooth nonconvex optimization in computer vision. SIAM J. Imaging Sci. 2015, 8, 331–372. [Google Scholar] [CrossRef]
- Geiping, J.; Moeller, M. Composite optimization by nonconvex majorization-minimization. SIAM J. Imaging Sci. 2018, 11, 2494–2598. [Google Scholar] [CrossRef]
- Ochs, P.; Fadili, J.; Brox, T. Non-smooth non-convex Bregman minimization: Unification and new algorithms. J. Optim. Theory Appl. 2019, 181, 244–278. [Google Scholar] [CrossRef]
- Repetti, A.; Wiaux, Y. Variable Metric Forward-Backward Algorithm for Composite Minimization Problems. SIAM J. Optim. 2021, 31, 1215–1241. [Google Scholar] [CrossRef]
- Combettes, P.L.; Wajs, V.R. Signal Recovery by Proximal Forward-Backward Splitting. Multiscale Model. Simul. 2005, 4, 1168–1200. [Google Scholar] [CrossRef]
- Caruyer, E.; Lenglet, C.; Sapiro, G.; Deriche, R. Design of multishell sampling schemes with uniform coverage in diffusion MRI. Magn. Reson. Med. 2013, 69, 1534–1540. [Google Scholar] [CrossRef]
- Miller, K.L.; Pauly, J.M. Nonlinear phase correction for navigated diffusion imaging. Magn. Reson. Med. 2003, 50, 343–353. [Google Scholar] [CrossRef] [PubMed]
- Pipe, J.G.; Farthing, V.G.; Forbes, K.P. Multishot diffusion-weighted FSE using PROPELLER MRI. Magn. Reson. Med. 2002, 47, 42–52. [Google Scholar] [CrossRef] [PubMed]
- Roemer, P.B.; Edelstein, W.A.; Hayes, C.E.; Souza, S.P.; Muller, O.M. The NMR Phase Array. Magn. Reson. Med. 1990, 16, 192–225. [Google Scholar] [CrossRef]
- Maier-Hein, K.H.; Neher, P.F.; Houde, J.-C.; Côté, M.-A.; Garyfallidis, E.; Zhong, J.; Chamberland, M.; Yeh, F.-C.; Lin, Y.-C.; Ji, Q.; et al. The challenge of mapping the human connectome based on diffusion tractography. Nat. Commun. 2017, 8, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Komodakis, N.; Pesquet, J.C. Playing with Duality: An Overview of Recent Primal-Dual Approaches for Solving Large-Scale Optimization Problems. IEEE Signal Process. Mag. 2015, 32, 31–54. [Google Scholar] [CrossRef]
- Beck, A.; Teboulle, M. A Fast Iterative Shrinkage-Thresholding Algorithm for Linear Inverse Problems. SIAM J. Imaging Sci. 2009, 2, 183–202. [Google Scholar] [CrossRef]


| Samp. | 17 Coils (Deterministic) | 12 Coils (Stochastic) | |||
|---|---|---|---|---|---|
| q | k | Time per Iter. (s) | Iter. | Time per Iter. (s) | Iter. |
| 60 | 4 | 21.5 ± 0.13 | 3520 | 16.4 ± 0.15 | 3536 |
| 45 | 3 | 17.9 ± 0.08 | 3603 | 13.5 ± 0.12 | 3618 |
| 30 | 2 | 12.8 ± 0.07 | 3445 | 9.9 ± 0.13 | 3445 |
| 15 | 1 | 7.7 ± 0.25 | 1856 | 6.1 ± 0.21 | 1854 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pesce, M.; Repetti, A.; Auría, A.; Daducci, A.; Thiran, J.-P.; Wiaux, Y. Fast Fiber Orientation Estimation in Diffusion MRI from kq-Space Sampling and Anatomical Priors. J. Imaging 2021, 7, 226. https://doi.org/10.3390/jimaging7110226
Pesce M, Repetti A, Auría A, Daducci A, Thiran J-P, Wiaux Y. Fast Fiber Orientation Estimation in Diffusion MRI from kq-Space Sampling and Anatomical Priors. Journal of Imaging. 2021; 7(11):226. https://doi.org/10.3390/jimaging7110226
Chicago/Turabian StylePesce, Marica, Audrey Repetti, Anna Auría, Alessandro Daducci, Jean-Philippe Thiran, and Yves Wiaux. 2021. "Fast Fiber Orientation Estimation in Diffusion MRI from kq-Space Sampling and Anatomical Priors" Journal of Imaging 7, no. 11: 226. https://doi.org/10.3390/jimaging7110226
APA StylePesce, M., Repetti, A., Auría, A., Daducci, A., Thiran, J.-P., & Wiaux, Y. (2021). Fast Fiber Orientation Estimation in Diffusion MRI from kq-Space Sampling and Anatomical Priors. Journal of Imaging, 7(11), 226. https://doi.org/10.3390/jimaging7110226





