Explorative Imaging and Its Implementation at the FleX-ray Laboratory
Abstract
1. Introduction
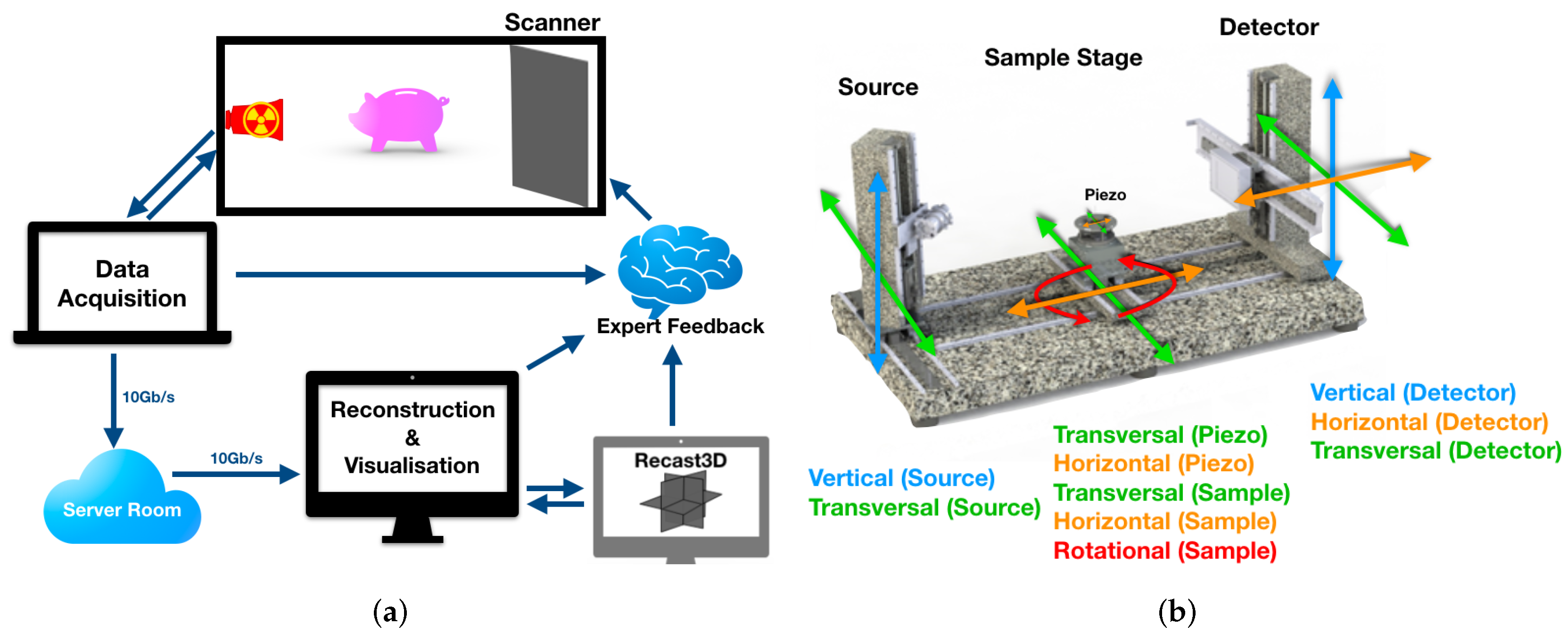
2. FleX-ray Laboratory
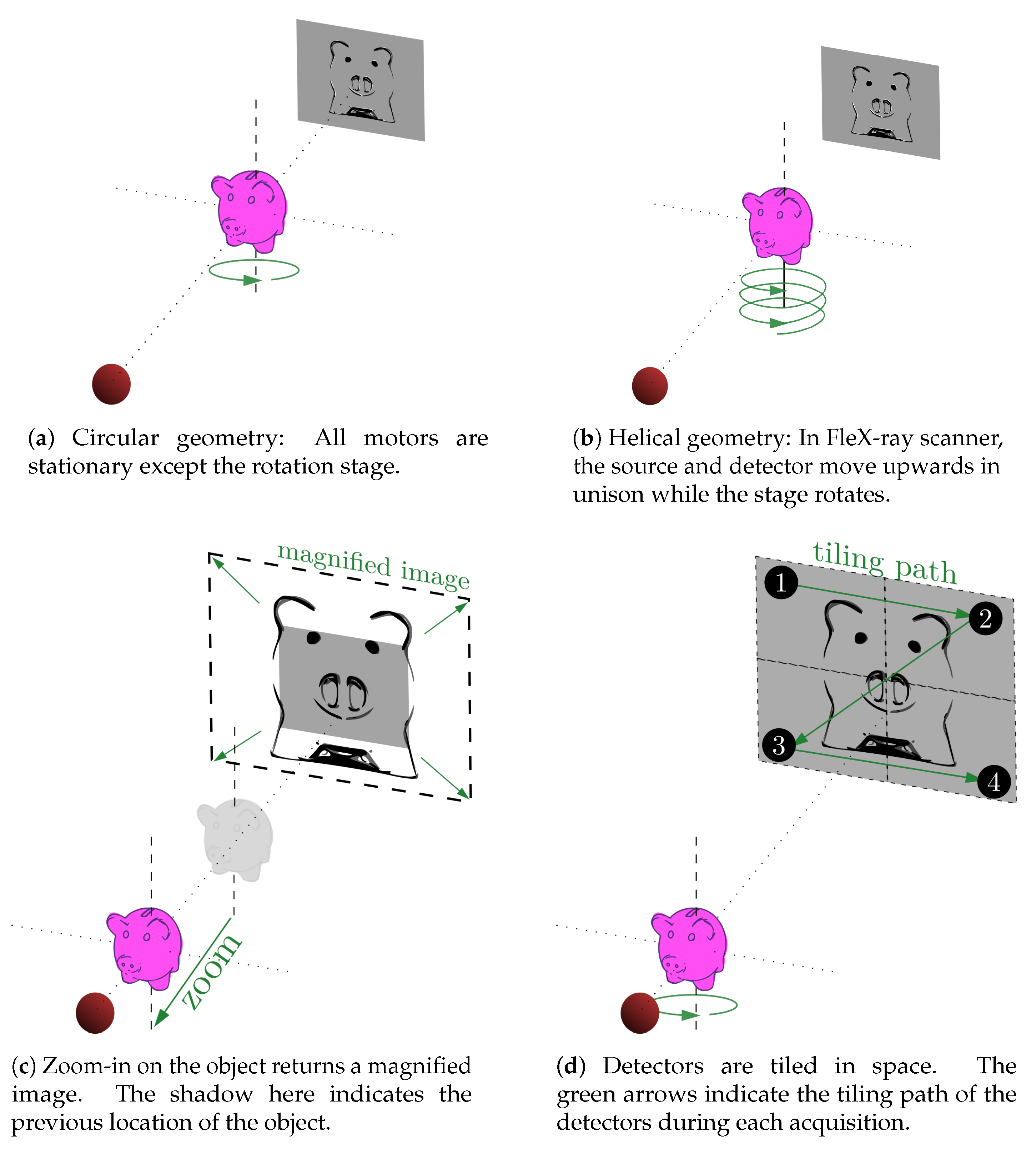
2.1. A Custom Built Scanner
2.2. Software Details
3. Explorative Imaging in Action
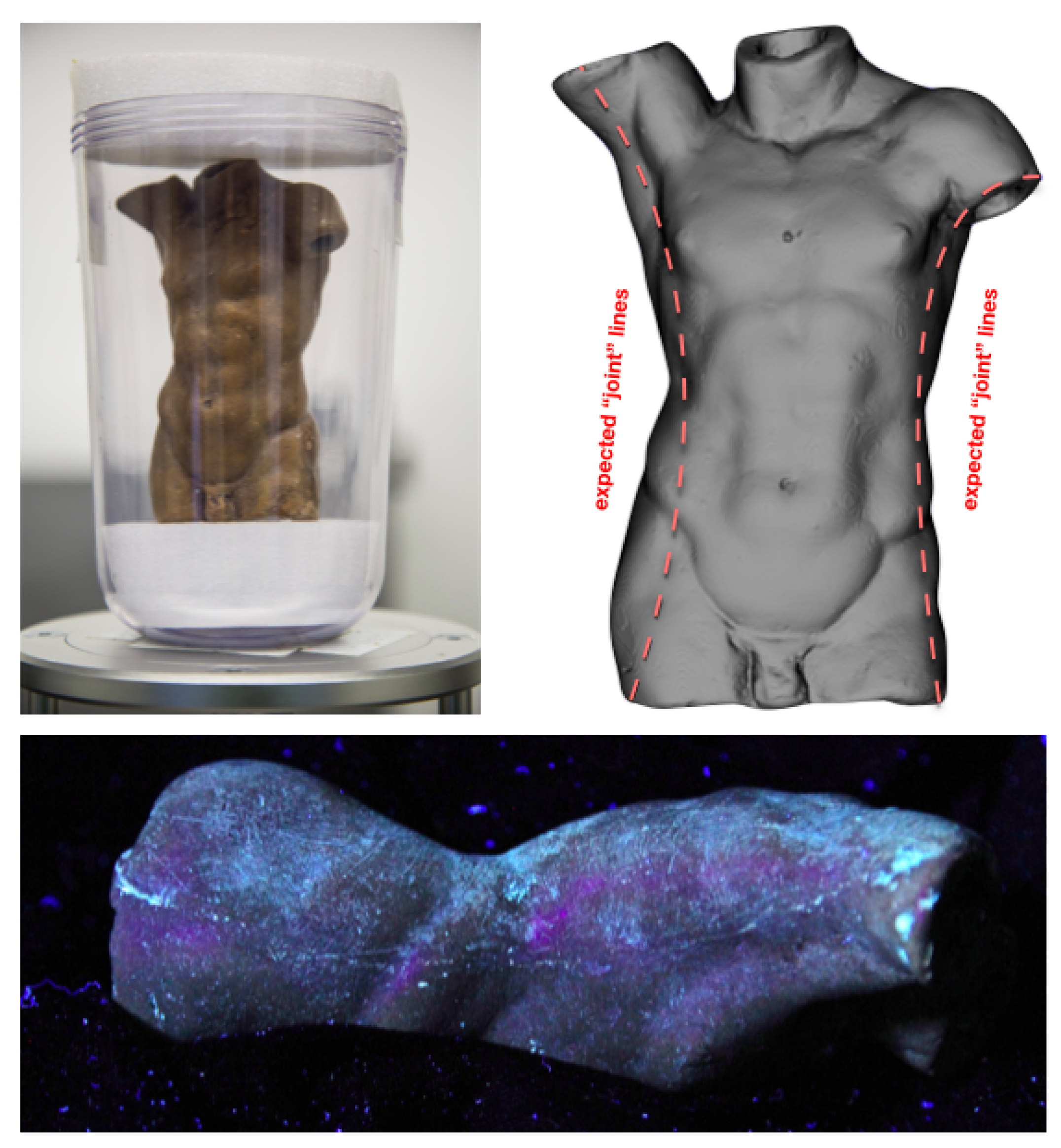
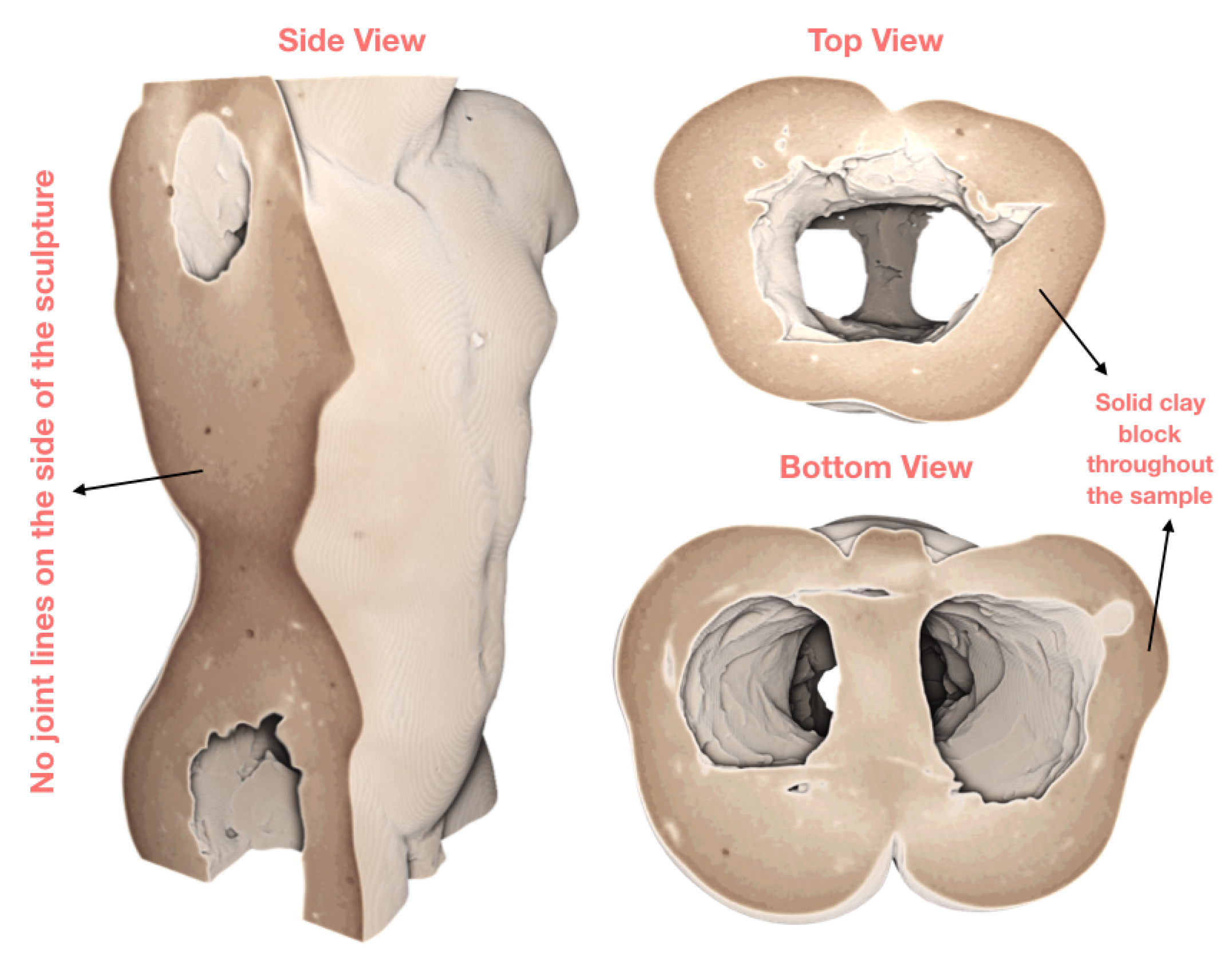
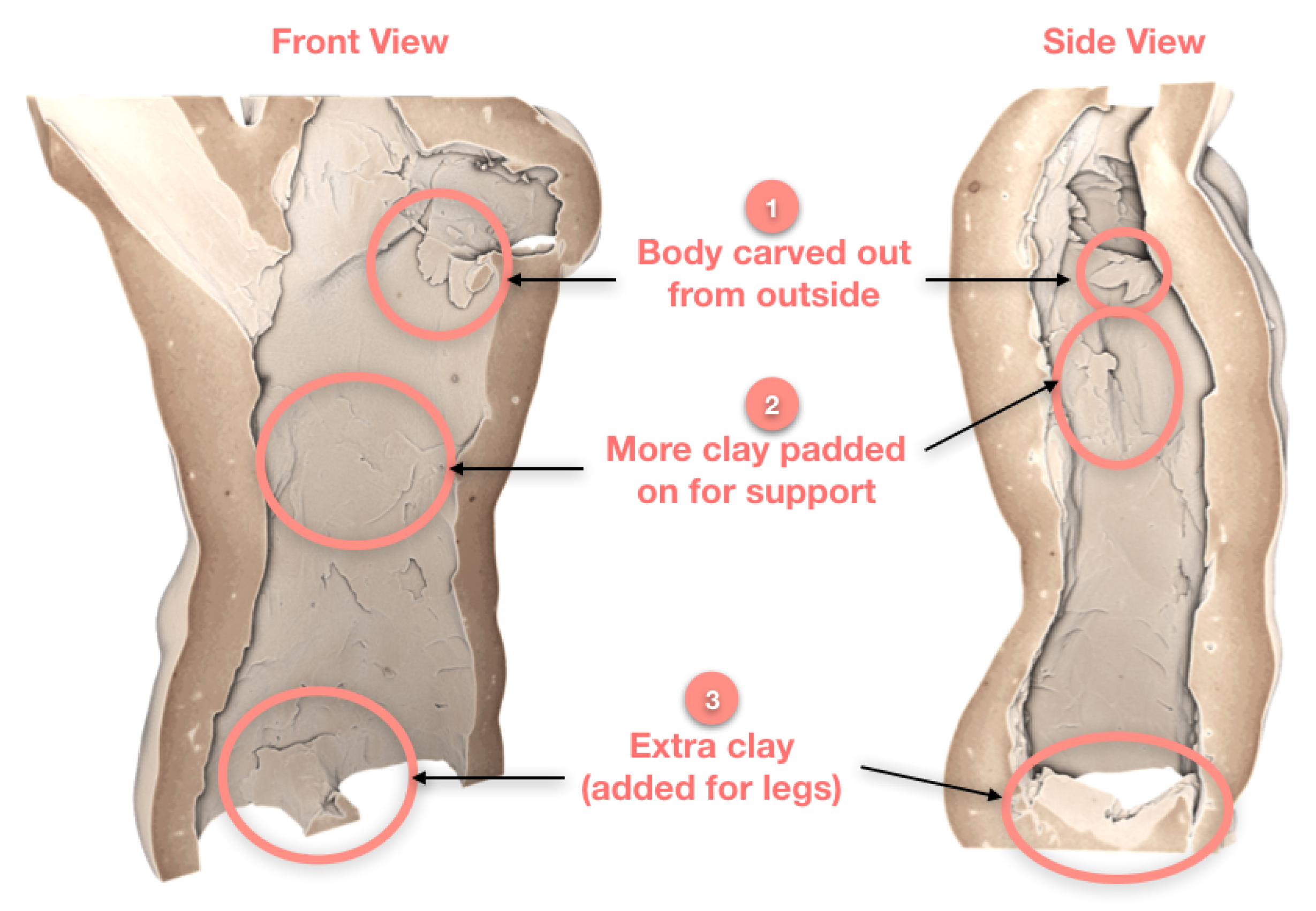
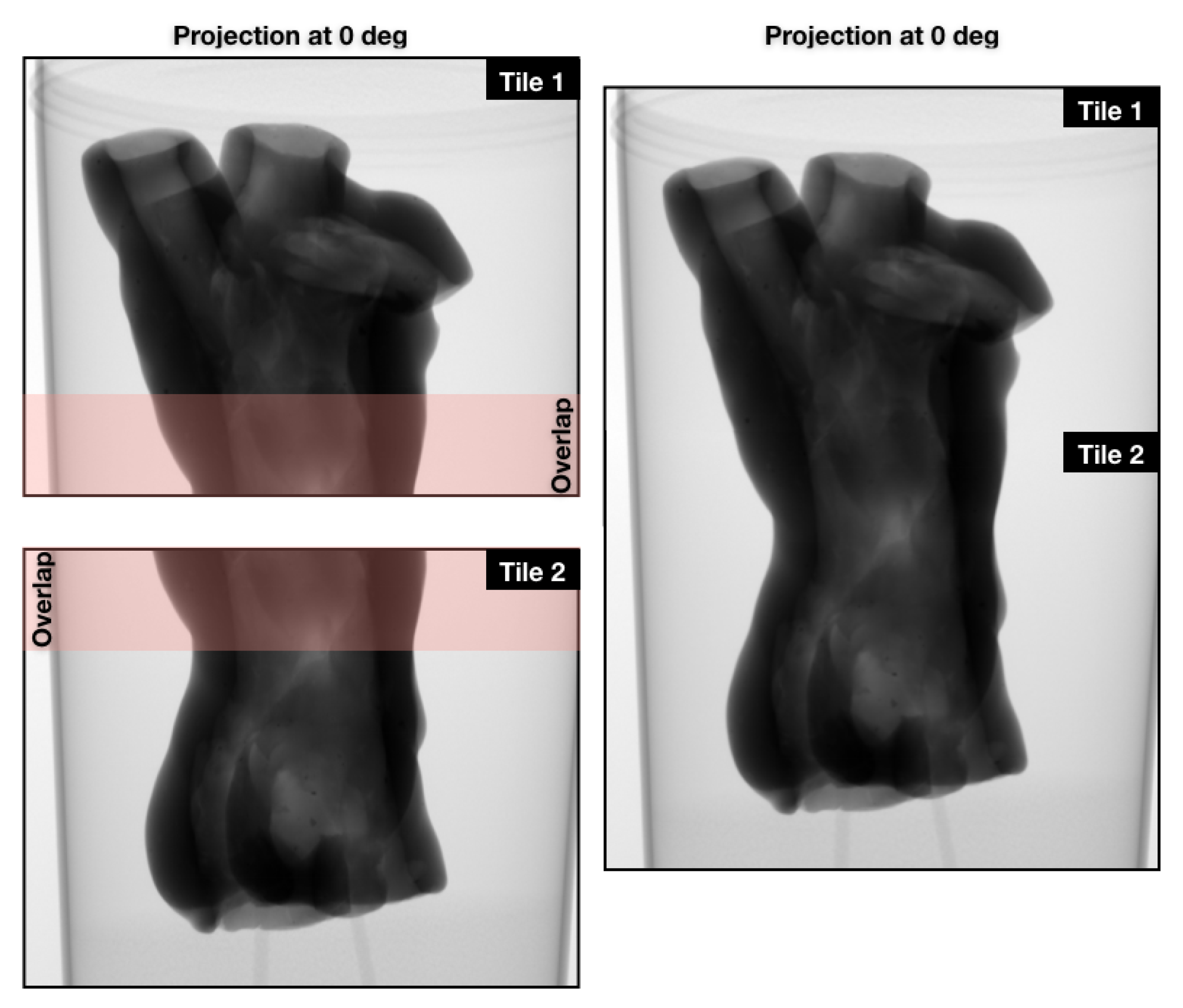
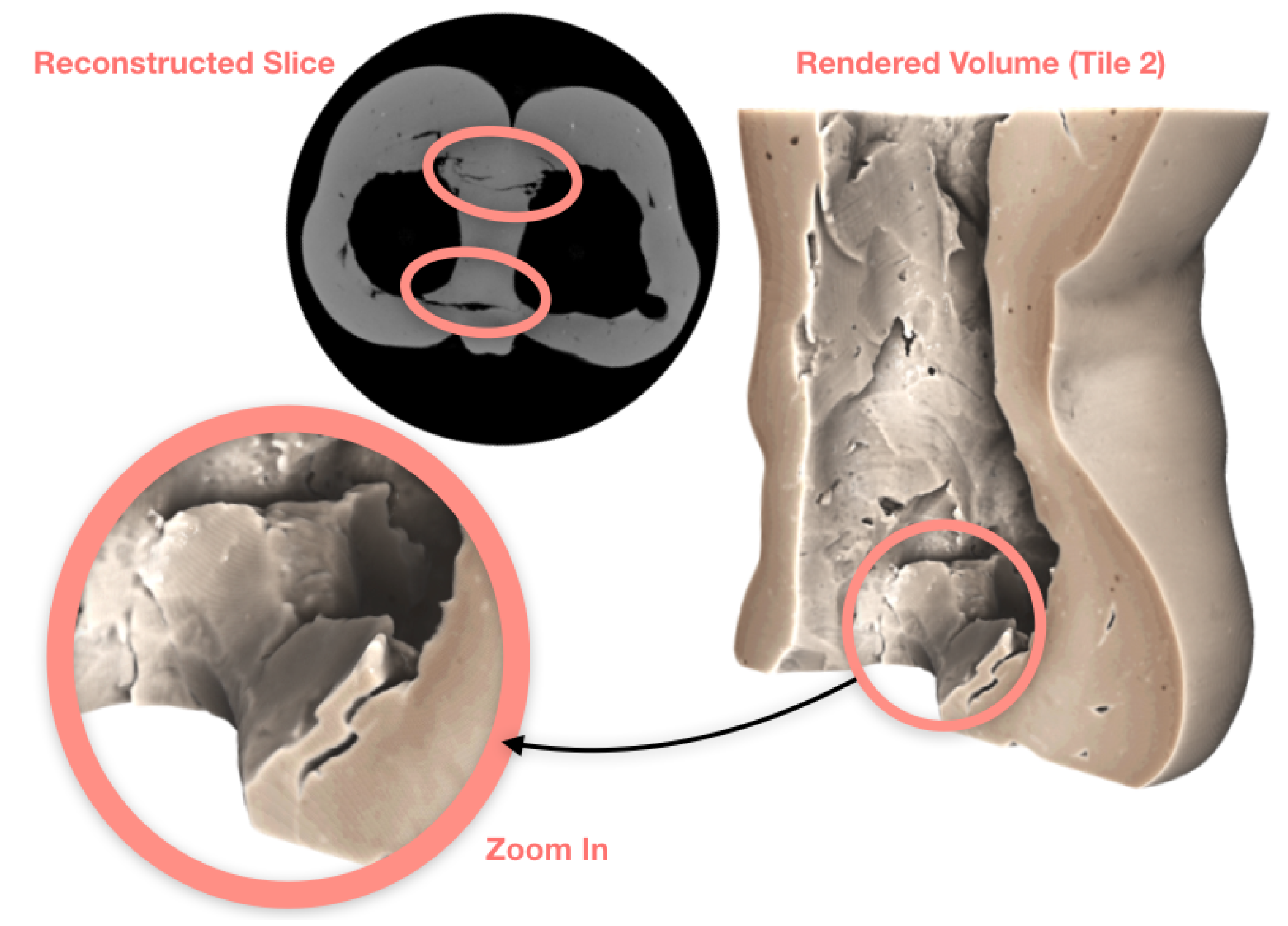
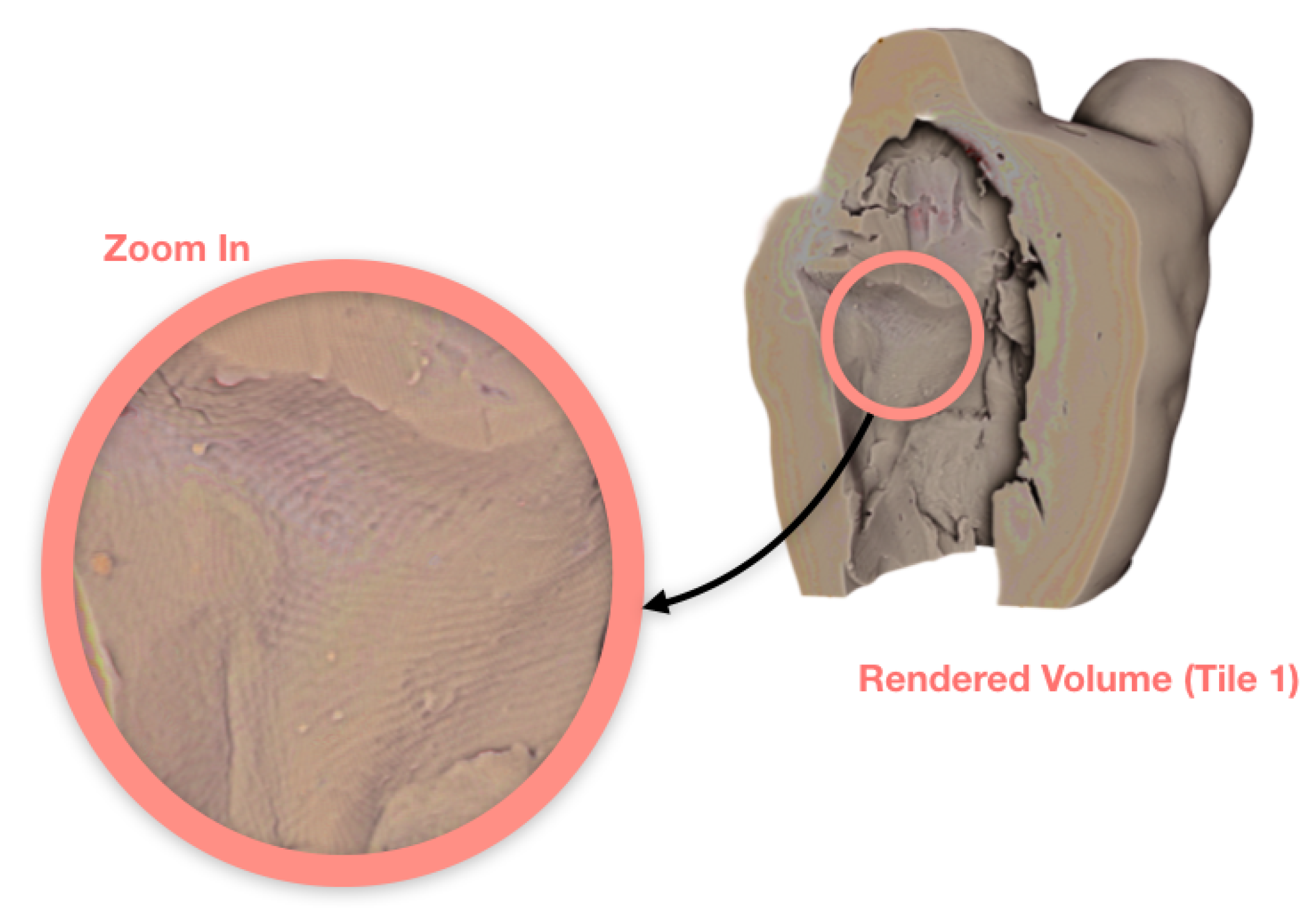
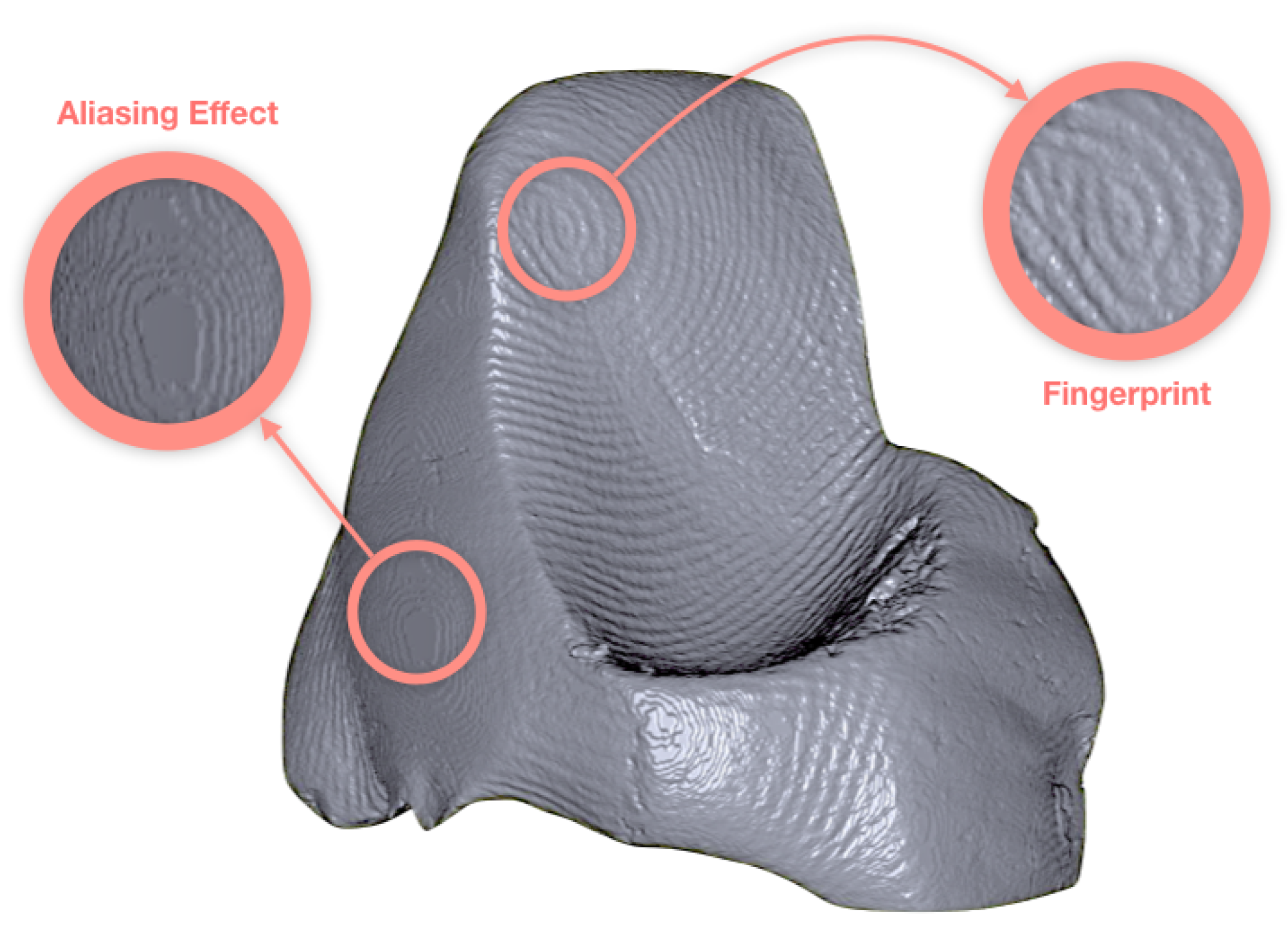
3.1. Case Study I: Cultural Heritage
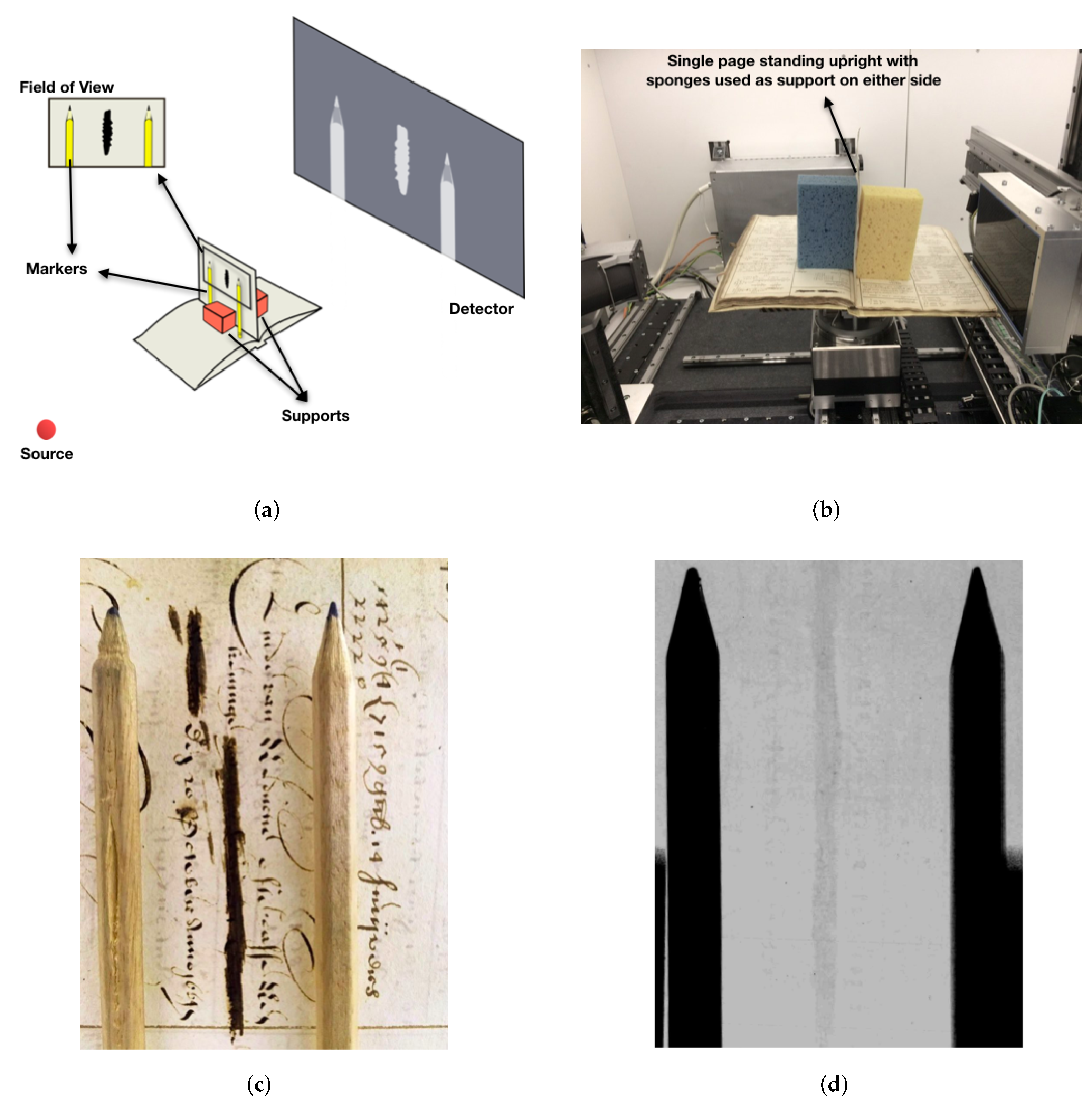
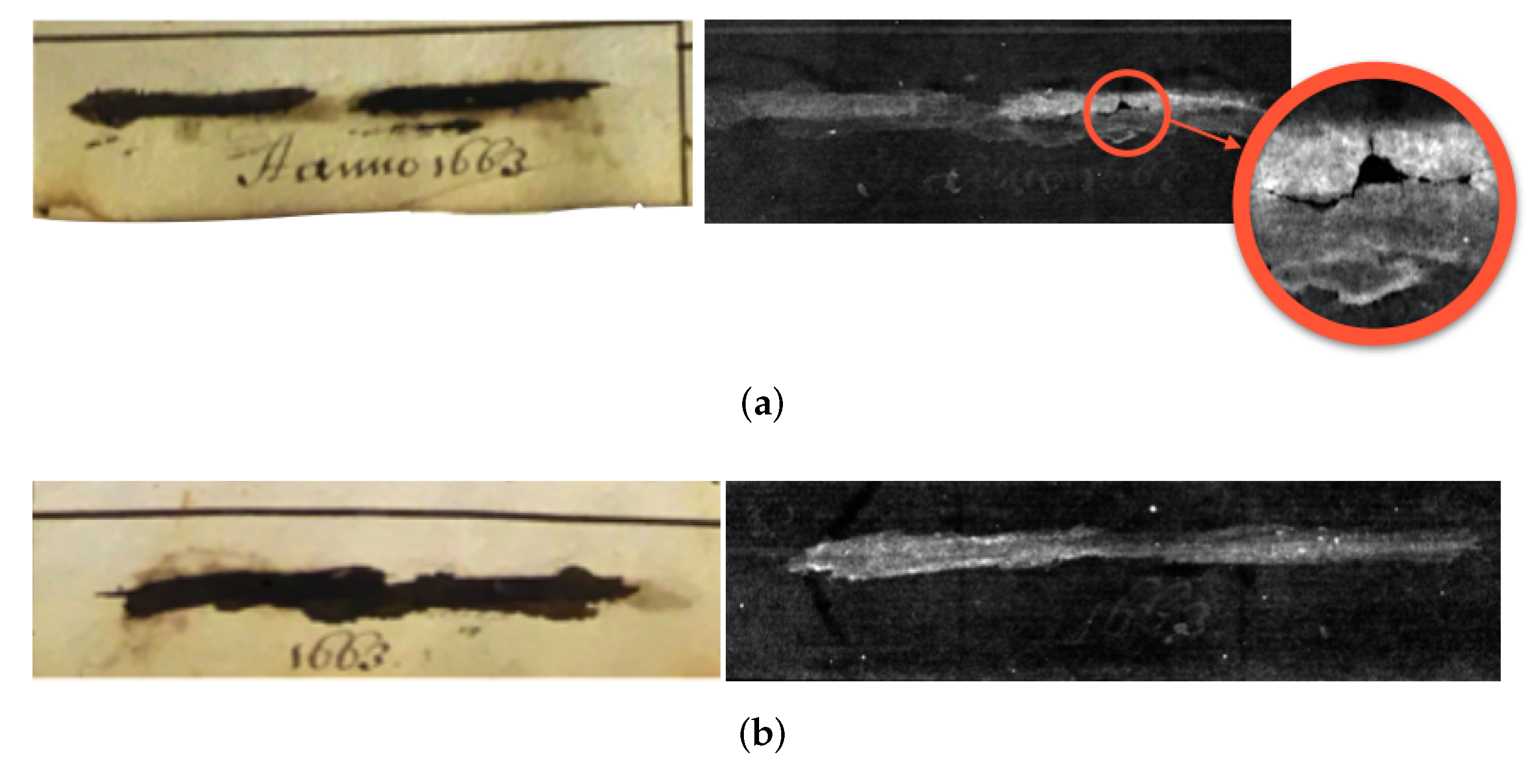
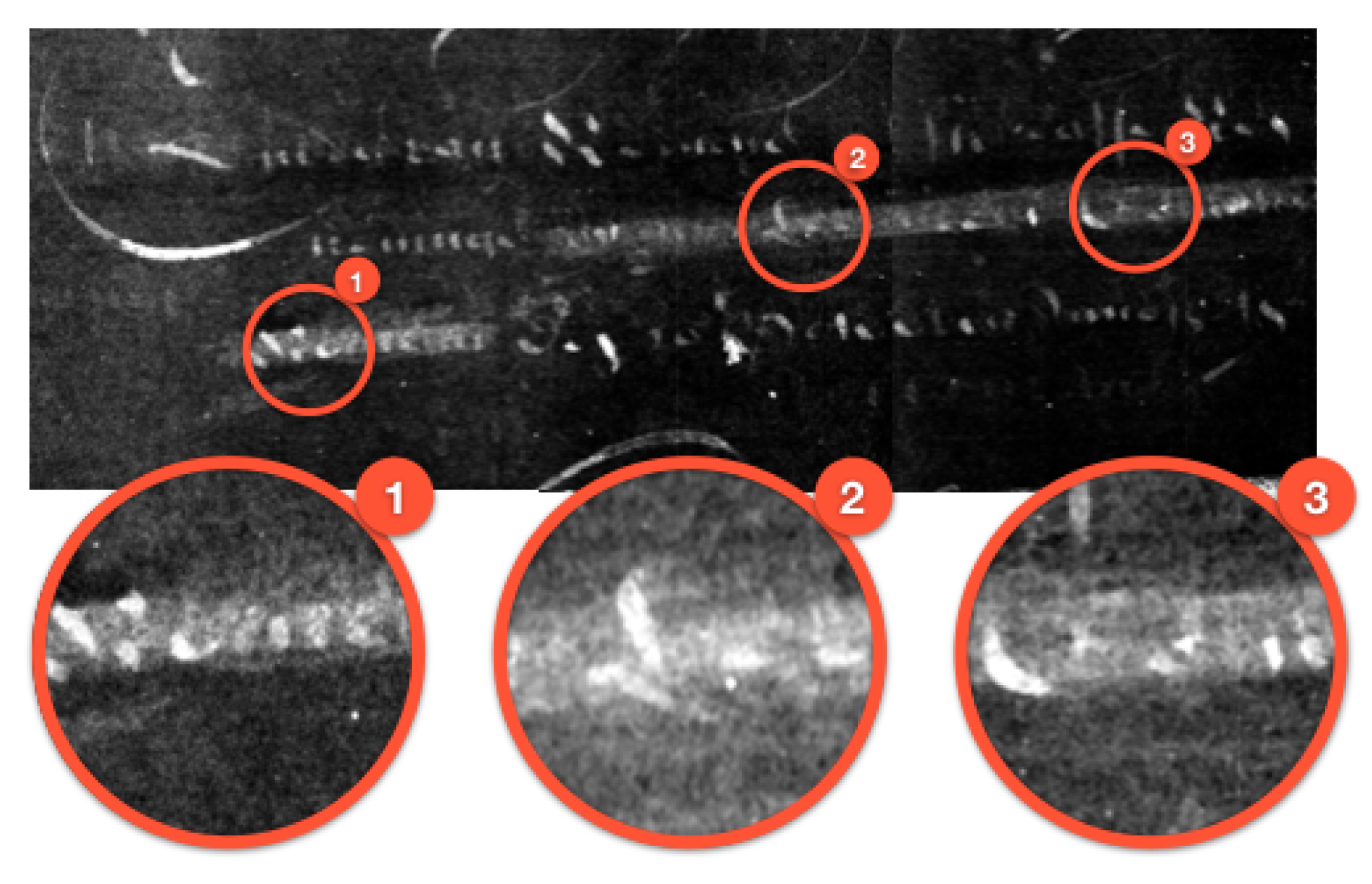
3.2. Case Study II: Ink Layers
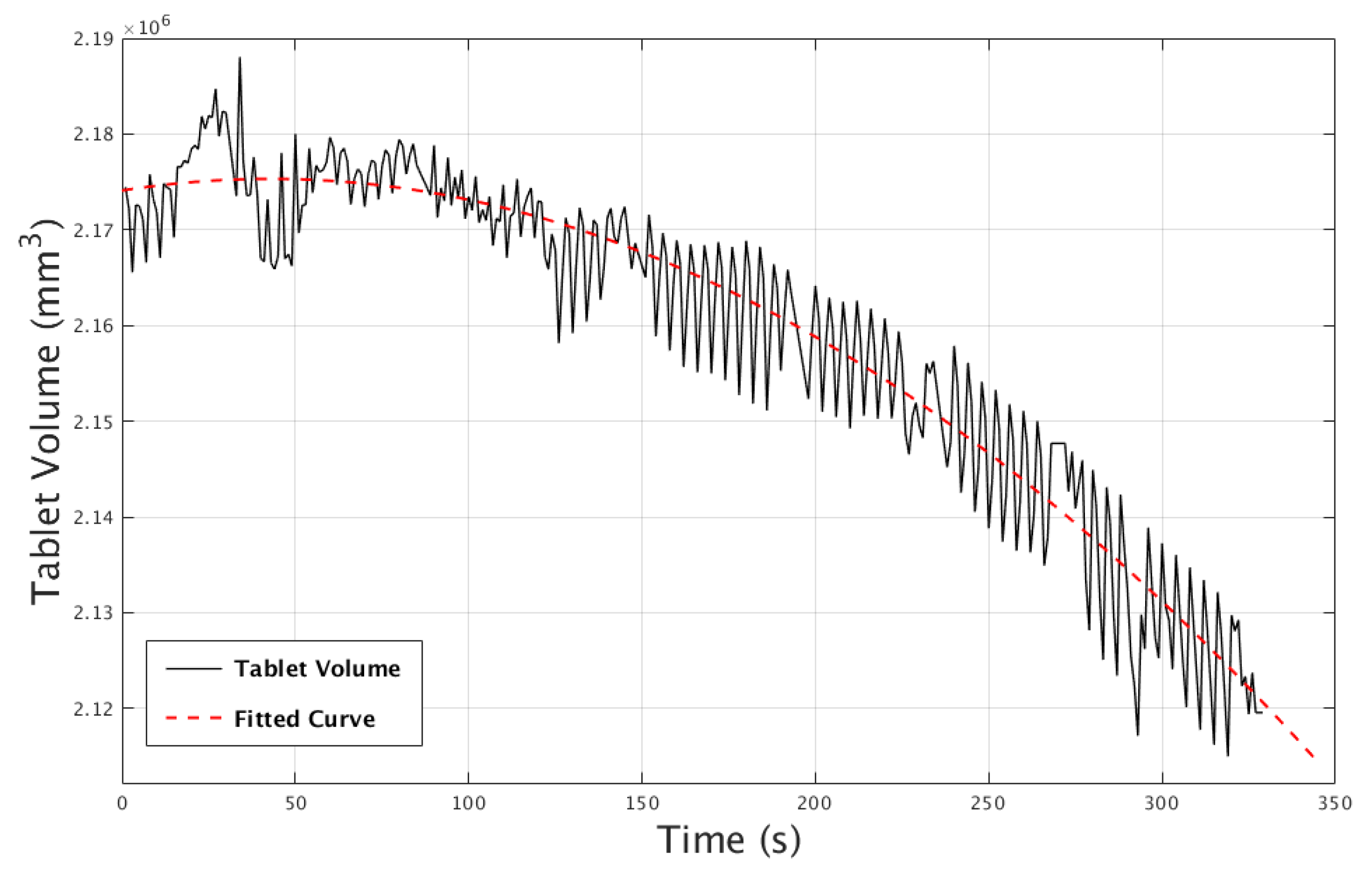
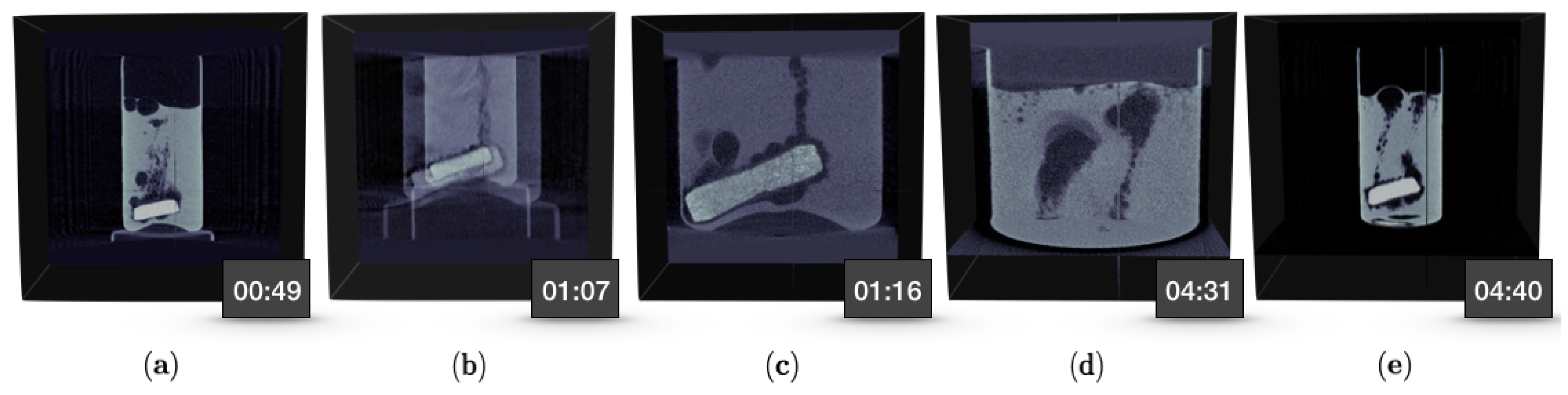
3.3. Case Study III: Gas Bubble Travel Through Liquid
4. Discussions
Our Vision for the Future
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Software Information
Data Availability
References
- Zhang, X.; Shan, X.; Withers, P.J.; Zhao, X.; Xiao, P. Tracking the calcium-magnesium-alumino-silicate (CMAS) infiltration into an air-plasma spray thermal barrier coating using X-ray imaging. Scr. Mater. 2020, 176, 94–98. [Google Scholar] [CrossRef]
- Erdem, S.; Gürbüz, E.; Uysal, M. Micro-mechanical analysis and X-ray computed tomography quantification of damage in concrete with industrial by-products and construction waste. J. Clean. Prod. 2018, 189, 933–940. [Google Scholar] [CrossRef]
- Schoeman, L.; Williams, P.; du Plessis, A.; Manley, M. X-ray micro-computed tomography (microCT) for non-destructive characterisation of food microstructure. Trends Food Sci. Technol. 2016, 47, 10–24. [Google Scholar] [CrossRef]
- Takahashi, M.; Kato, M.; Lin, W.; Sato, M. Three-dimensional pore geometry and permeability anisotropy of Berea sandstone under hydrostatic pressure: Connecting path and tortuosity data obtained by microfocus X-ray CT. Geol. Soc. Lond. Eng. Geol. Spec. Publ. 2016, 27, 207–215. [Google Scholar] [CrossRef]
- Yajvinder; Gulati, V. Implementation of Micro CT in CAD/CAM dentistry for image processing and soft computing: A review. J. Phys. Conf. Ser. 2020, 1432, 012079. [Google Scholar] [CrossRef]
- Wadeson, N.; Basham, M. Savu: A Python-based, MPI Framework for Simultaneous Processing of Multiple, N-dimensional, Large Tomography Datasets. arXiv 2016, arXiv:1610.08015. [Google Scholar]
- Dexela Limited. DEXELA 1512 CMOS X-ray Detector Product Specifications. Available online: http://file.yizimg.com/344621/2010061015232418.pdf (accessed on 12 April 2019).
- Buurlage, J.W.; Kohr, H.; Palenstijn, W.J.; Batenburg, K.J. Real-time quasi-3D tomographic reconstruction. Meas. Sci. Technol. 2018, 29, 064005. [Google Scholar] [CrossRef]
- van Aarle, W.; Palenstijn, W.J.; Beenhouwer, J.D.; Altantzis, T.; Bals, S.; Batenburg, K.J.; Sijbers, J. The ASTRA Toolbox: A platform for advanced algorithm development in electron tomography. Ultramicroscopy 2015, 157, 35–47. [Google Scholar] [CrossRef]
- Kostenko, A.; Palenstijn, W.J.; Coban, S.B.; Hendriksen, A.A.; Batenburg, K.J.; van Liere, R. Prototyping X-ray tomographic reconstruction pipelines with FleXbox. SoftwareX 2020, 11, 100364. [Google Scholar] [CrossRef]
- Vermeulen, M.; Leona, M. Evidence of early amorphous arsenic sulfide production and use in Edo period Japanese woodblock prints by Hokusai and Kunisada. Herit. Sci. 2019, 7, 73. [Google Scholar] [CrossRef]
- Beaugnon, F.; Gariani, G.; Gouillart, E.; Bouquillon, A.; Bormand, M.; Wallez, G. Microstructure imaging of Florentine stuccoes through X-ray tomography: A new insight on ancient plaster-making techniques. J. Cult. Herit. 2019, 40, 17–24. [Google Scholar] [CrossRef]
- Li, H.; Zuo, Z.; Cui, J.; Tian, J.; Yang, Y.; Yi, L.; Zhou, Z.; Fan, J. Bronze production in the Ancient Chengdu Plains: A diachronic metallurgical perspective on a separate cultural region. J. Cult. Heritage 2019. [Google Scholar] [CrossRef]
- Rijksmuseum Collection. Study Models of Parts of the Body, Johan Gregor van der Schardt “the Torso”(BK-2016-44-4). Available online: https://www.rijksmuseum.nl/en/collection/BK-2016-44-4 (accessed on 17 April 2019).
- Scholten, F. Acquisitions: Sculpture. Rijksmus. Bull. 2014, 62, 288–327. [Google Scholar]
- Boswell, R. Canadian-Owned Sculptures Found Not to be Michelangelo’s But Expected to Sell for Tidy Sum This Month. Available online: https://o.canada.com/news/canadian-owned-sculptures-found-not-to-be-michelangelos-but-expected-to-sell-for-tidy-sum-this-month (accessed on 17 April 2019).
- Baines, D. Blockbuster Donation of ’Michelangelo’ Sculptures Turns into Multi-Million-Dollar Bust. Available online: http://www.vancouversun.com/news/Blockbuster+donation+Michelangelo+sculptures+turns+into+multi+million+dollar+bust/7875342/story.html (accessed on 17 April 2019).
- Coban, S. A single- and two-tile tomographic micro-CT data of the terracotta sculpture “the Torso”. Available online: https://doi.org/10.5281/zenodo.3630710 (accessed on 25 September 2017).
- Conservatory, Y.U.L.S.C. Medieval Manuscripts: Some ink and Pigment Recipes. Available online: https://travelingscriptorium.files.wordpress.com/2012/03/scopa-recipes-booklet_web.pdf (accessed on 26 April 2019).
- Feller, R.L. Artists’ Pigments: A Handbook of Their History and Characteristics, Volume 1; Cambridge University Press: Cambridge, UK, 1986. [Google Scholar]
- Baker, A. Common Medieval Pigments; Technical Report; The Cochineal, University of Texas: Austin, TX, USA, 2004. [Google Scholar]
- Mocella, V.; Brun, E.; Ferrero, C.; Delattre, D. Revealing letters in rolled Herculaneum papyri by X-ray phase-contrast imaging. Nat. Commun. 2015, 6, 5895. [Google Scholar] [CrossRef] [PubMed]
- Rosin, P.L.; Lai, Y.K.; Liu, C.; Davis, G.R.; Mills, D.; Tuson, G.; Russell, Y. Virtual Recovery of Content from X-Ray Micro-Tomography Scans of Damaged Historic Scrolls. Sci. Rep. 2018, 8, 11901. [Google Scholar] [CrossRef] [PubMed]
- Bergmann, U. Archimedes brought to light. Phys. World 2007, 20, 39–42. [Google Scholar] [CrossRef]
- Bergmann, U.; Manning, P.L.; Wogelius, R.A. Chemical Mapping of Paleontological and Archeological Artifacts with Synchrotron X-Rays. Annu. Rev. Anal. Chem. 2012, 5, 361–389. [Google Scholar] [CrossRef]
- Albertin, F.; Astolfo, A.; Stampanoni, M.; Peccenini, E.; Hwu, Y.; Kaplan, F.; Margaritondo, G. Ancient administrative handwritten documents: X-ray analysis and imaging. J. Synchrotron Radiat. 2015, 22, 446–451. [Google Scholar] [CrossRef]
- D’Almeida, M.L.O.; de Souza Medeiros Barbosa, P.; Boaratti, M.F.G.; Borrely, S.I. Radiation effects on the integrity of paper. Radiat. Phys. Chem. 2009, 78, 489–492. [Google Scholar] [CrossRef]
- Gonzalez, M.; Calvo, A.; Kairiyama, E. Gamma radiation for preservation of biologically damaged paper. Radiat. Phys. Chem. 2002, 63, 263–265. [Google Scholar] [CrossRef]
- Maire, E.; Withers, P. Acquisitions: Sculpture. Quant. X-Ray Tomogr. 2014, 59, 1–43. [Google Scholar]
- Bultreys, T.; Boone, M.A.; Boone, M.N.; Schryver, T.D.; Masschaele, B.; Hoorebeke, L.V.; Cnudde, V. Fast laboratory-based micro-computed tomography for pore-scale research: Illustrative experiments and perspectives on the future. Adv. Water Resour. 2016, 95, 341–351. [Google Scholar] [CrossRef]
- Dobson, K.J.; Coban, S.B.; McDonald, S.A.; Walsh, J.N.; Atwood, R.C.; Withers, P.J. 4-D imaging of sub-second dynamics in pore-scale processes using real-time synchrotron X-ray tomography. Solid Earth 2016, 7, 1059–1073. [Google Scholar] [CrossRef]
- Maire, E.; Le Bourlot, C.; Adrien, J.; Mortensen, A.; Mokso, R. 20 Hz X-ray tomography during an in situ tensile test. Int. J. Fract. 2016, 200, 3–12. [Google Scholar] [CrossRef]
- Polacci, M.; Arzilli, F.; La Spina, G.; Le Gall, N.; Cai, B.; Hartley, M.E.; Di Genova, D.; Vo, N.T.; Nonni, S.; Atwood, R.C.; et al. Crystallisation in basaltic magmas revealed via in situ 4D synchrotron X-ray microtomography. Sci. Rep. 2018, 8, 8377. [Google Scholar] [CrossRef]
- Tikhonov, A. Solutions of Ill-Posed Problems; Wiley: New York, NY, USA, 1977. [Google Scholar]
- Calvetti, D.; Lewis, B.; Reichel, L. A hybrid GMRES and TV-norm-based method for image restoration. In Advanced Signal Processing Algorithms, Architectures, and Implementations XII; Luk, F.T., Ed.; International Society for Optics and Photonics, SPIE: Seattle, WA, USA, 2002; Volume 4791, pp. 192–200. [Google Scholar]
- Jensen, T.; Jørgensen, J.; Hansen, P.; Jensen, S. Implementation of an optimal first-order method for strongly convex total variation regularization. BIT Numer. Math. 2011, 1–28. [Google Scholar] [CrossRef]
- Coban, S.; Lionheart, W. Regularised GMRES-type Methods for X-Ray Computed Tomography. In Proceedings of the Third International Conference on Image Formation in X-Ray Computed Tomography. Utah Center For Advanced Imaging Research (UCAIR), Salt Lake City, UT, USA, 22–25 June 2014. [Google Scholar]
- Kazantsev, D.; Ourselin, S.; Hutton, B.; Dobson, K.; Kaestner, A.; Lionheart, W.; Withers, P.; Lee, P.; Arridge, S. A novel technique to incorporate structural prior information into multi-modal tomographic reconstruction. Inverse Probl. 2014, 30. [Google Scholar] [CrossRef]
- Batenburg, K.; Sijbers, J. DART: A practical reconstruction algorithm for discrete tomography. IEEE Trans Image Process 2011, 20, 2542–2553. [Google Scholar] [CrossRef]
- Choi, K.; Wang, J.; Zhu, L.; Suh, T.S.; Boyd, S.; Xing, L. Compressed sensing based cone-beam computed tomography reconstruction with a first-order method. Med. Phys. 2010, 37, 5113–5125. [Google Scholar] [CrossRef]
- Li, X.; Luo, S. A compressed sensing-based iterative algorithm for CT reconstruction and its possible application to phase contrast imaging. BioMedical Eng. OnLine 2011, 10, 73. [Google Scholar] [CrossRef]
- Jørgensen, J.; Coban, S.; Lionheart, W.; McDonald, S.; Withers, P. SparseBeads data: Benchmarking sparsity-regularized computed tomography. Meas. Sci. Technol. 2017, 28. [Google Scholar] [CrossRef]
- Pelt, D.M.; Batenburg, K.J. Improving Filtered Backprojection Reconstruction by Data-Dependent Filtering. IEEE Trans. Image Process. 2014, 23, 4750–4762. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ravishankar, S.; Ye, J.C.; Fessler, J.A. Image Reconstruction: From Sparsity to Data-Adaptive Methods and Machine Learning. Proc. IEEE 2019, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Yoshimura, S.; Kuritani, T.; Matsumoto, A.; Nakagawa, M. Fingerprint of silicic magma degassing visualised through chlorine microscopy. Sci. Rep. 2019, 9, 786. [Google Scholar] [CrossRef]
- Sparks, R.S.J. Dynamics of magma degassing. Geol. Soc. Lond. Spec. Publ. 2003, 213, 5–22. [Google Scholar] [CrossRef]
- Martel, C.; Bureau, H. In situ high-pressure and high-temperature bubble growth in silicic melts. Earth Planet. Sci. Lett. 2001, 191, 115–127. [Google Scholar] [CrossRef]
- Liger-Belair, G.; Polidori, G.; Jeandet, P. Recent advances in the science of champagne bubbles. Chem. Soc. Rev. 2008, 37, 2490–2511. [Google Scholar] [CrossRef]
- Liger-Belair, G.; Beaumont, F.; Vialatte, M.A.; Jegou, S.; Jeandet, P.; Polidori, G. Kinetics and stability of the mixing flow patterns found in champagne glasses as determined by laser tomography techniques: Likely impact on champagne tasting. Anal Chim Acta 2008, 621, 30–37. [Google Scholar] [CrossRef]
- Babin, P.; Valle, G.D.; Chiron, H.; Cloetens, P.; Hoszowska, J.; Pernot, P.; Réguerre, A.; Salvo, L.; Dendievel, R. Fast X-ray tomography analysis of bubble growth and foam setting during breadmaking. J. Cereal Sci. 2006, 43, 393–397. [Google Scholar] [CrossRef]
- Narsimhan, G. Model for growth of bubbles during proofing of viscoelastic dough. Bubble Sci. Eng. Technol. 2012, 4, 63–71. [Google Scholar] [CrossRef]
- Plank, B.; Helmus, R.; Gschwandtner, M.; Hinterhölzl, R.; Kastner, J. In-Situ observation of bubble formation in neat resin during the curing process by means of X-ray computed tomography. In Proceedings of the 19th World Conference on Non-Destructive Testing, Munich, Germany, 13–17 June 2016. [Google Scholar]
- Obreschkow, D.; Kobel, P.; Dorsaz, N.; de Bosset, A.; Nicollier, C.; Farhat, M. Cavitation bubble dynamics inside liquid drops in microgravity. Phys Rev Lett 2006, 97. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Atlar, M.; Sampson, R. An experimental investigation on cavitation, noise, and slipstream characteristics of ocean stream turbines. Proc. Inst. Mech. Eng. Part A J. Power Energy 2007, 221, 219–231. [Google Scholar] [CrossRef]
- Kumar, P.; Saini, R. Study of cavitation in hydro turbines—A review. Renew. Sustain. Energy Rev. 2010, 14, 374–383. [Google Scholar] [CrossRef]
- Iben, U.; Wolf, F.; Freudigmann, H.E.A. Optical measurements of gas bubbles in oil behind a cavitating micro-orifice flow. Exp Fluids 2015, 56, 114. [Google Scholar] [CrossRef]
- Coban, S.; Lucka, F. Dynamic 3D X-ray Micro-CT Data of A Tablet Dissolution in A Water-Based Gel. Available online: https://doi.org/10.5281/zenodo.3610187 (accessed on 18 October 2019).
- Feldkamp, L.; Davis, L.; Kress, J. Practical cone-beam algorithm. JOSA A 1984, 1, 612–619. [Google Scholar] [CrossRef]
- Curve Fitting Toolbox: For Use with MATLAB®: User’s Guide; Version 1; MathWorks: Natick, MA, USA, 2001.
- Coban, S.; Lucka, F.; Palenstijn, W. Dynamic 3D X-ray Micro-CT Data of A Tablet Dissolution in A Water-Based Gel With Dynamic Changes in the Scanning Geometry. Available online: https://doi.org/10.5281/zenodo.3675371 (accessed on 17 February 2020).
- Liu, S.; Cao, R.; Huang, Y.; Ouypornkochagorn, T.; Jia, J. Time Sequence Learning for Electrical Impedance Tomography Using Bayesian Spatiotemporal Priors. IEEE Trans. Instrum. Meas. 2020. [Google Scholar] [CrossRef]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Coban, S.B.; Lucka, F.; Palenstijn, W.J.; Van Loo, D.; Batenburg, K.J. Explorative Imaging and Its Implementation at the FleX-ray Laboratory. J. Imaging 2020, 6, 18. https://doi.org/10.3390/jimaging6040018
Coban SB, Lucka F, Palenstijn WJ, Van Loo D, Batenburg KJ. Explorative Imaging and Its Implementation at the FleX-ray Laboratory. Journal of Imaging. 2020; 6(4):18. https://doi.org/10.3390/jimaging6040018
Chicago/Turabian StyleCoban, Sophia Bethany, Felix Lucka, Willem Jan Palenstijn, Denis Van Loo, and Kees Joost Batenburg. 2020. "Explorative Imaging and Its Implementation at the FleX-ray Laboratory" Journal of Imaging 6, no. 4: 18. https://doi.org/10.3390/jimaging6040018
APA StyleCoban, S. B., Lucka, F., Palenstijn, W. J., Van Loo, D., & Batenburg, K. J. (2020). Explorative Imaging and Its Implementation at the FleX-ray Laboratory. Journal of Imaging, 6(4), 18. https://doi.org/10.3390/jimaging6040018




