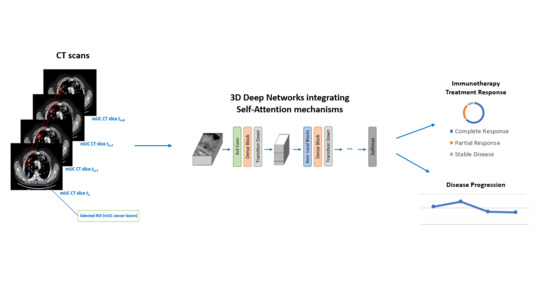
3D Non-Local Neural Network: A Non-Invasive Biomarker for Immunotherapy Treatment Outcome Prediction. Case-Study: Metastatic Urothelial Carcinoma
Abstract
1. Introduction
2. Related Works
- We present a generalizable deep model that combines 3D densely connected convolutional layers empowered with self-attention mechanisms for estimating automatically the efficacy of bladder cancer immunotherapy treatment, purely based on CT imaging analysis.
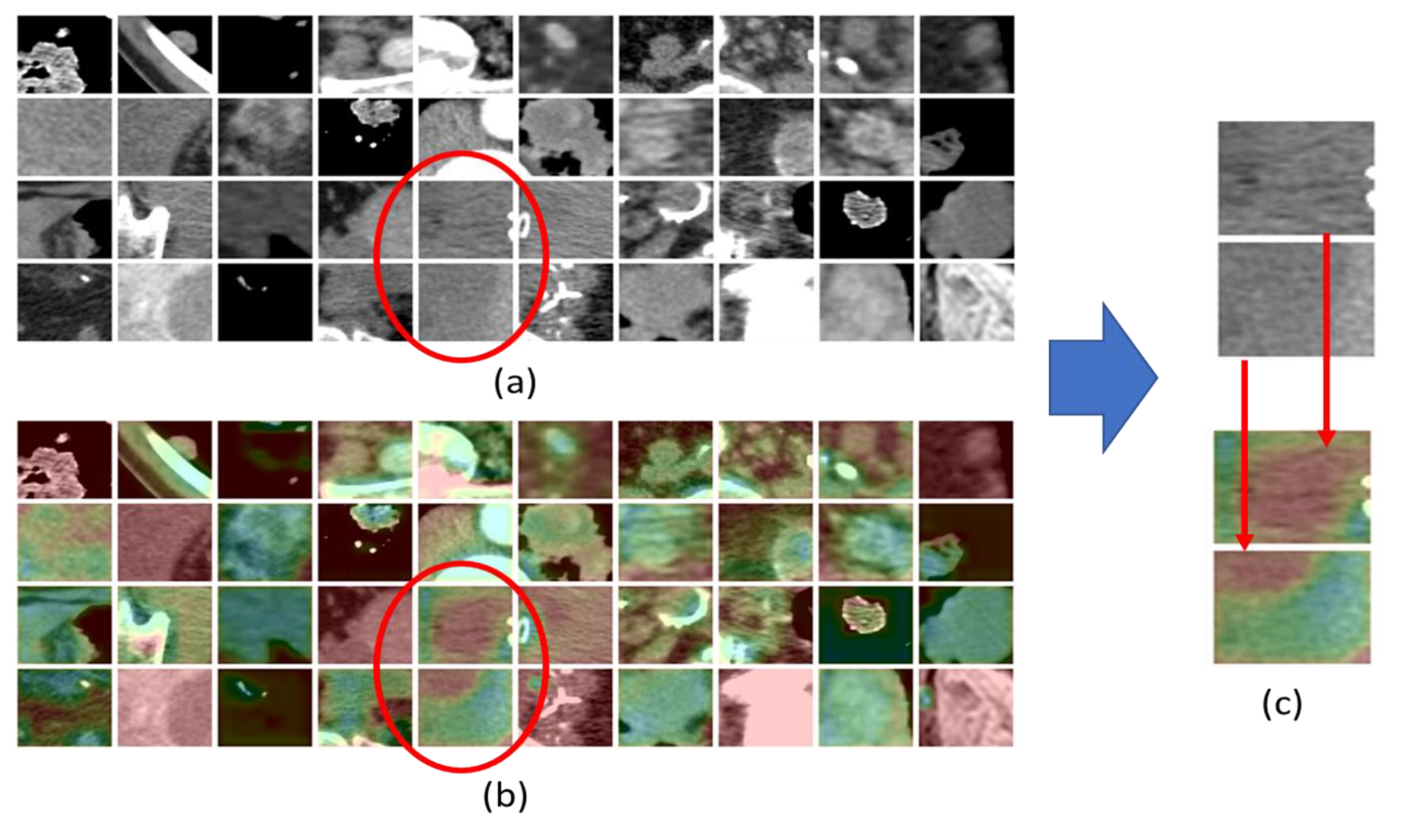
- We investigate, through interpretability methods, such as Grad-CAM [40], what are the radiological CT visual features that most likely act as biomarkers for immunotherapy treatment outcome, thus, providing a potentially invaluable support to medical staff in evaluating the progress of bladder cancer. To the best of our knowledge, to date, no method has tackled the task herein proposed, from both the automated treatment outcome prediction and interpretability perspectives.
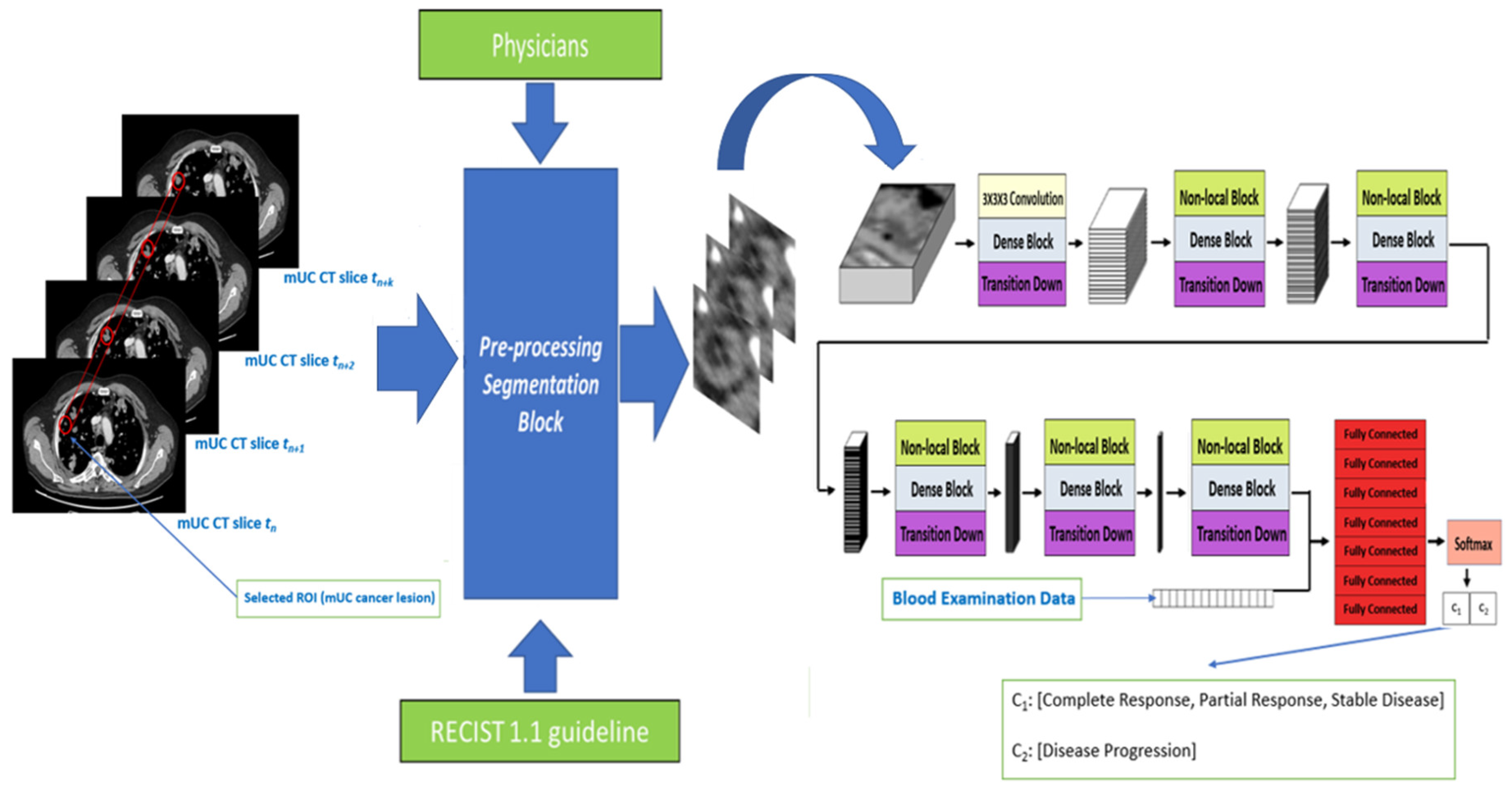
3. Materials and Methods
3.1. Dense Blocks
3.2. Self-Attention through Non-Local Blocks
3.3. Classification Layer: The Stack of Fully Connected
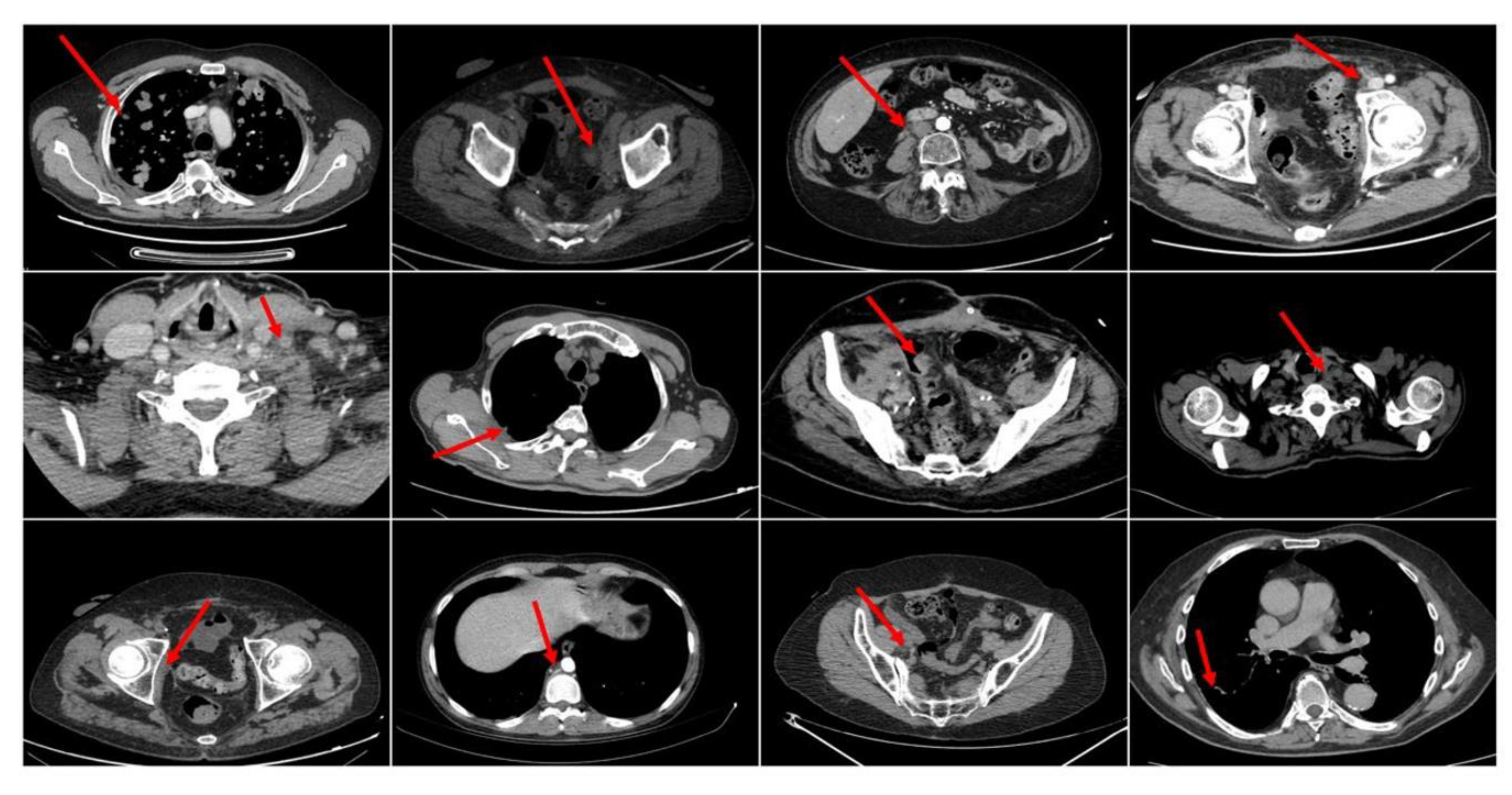
3.4. Dataset: Recruitment and Data Pre-Processing
- A patient shows a complete response (CR) to the medical treatment if all identified target lesions (LD sum) disappear at the end-treatment CT imaging.
- A patient shows a partial response (PR) to drug treatment if the target lesions (LD sum) are reduced by at least 30%.
- A patient shows a progressive disease (PD) if the Longest Diameter(LD) sum increases by, at least, 20% of the LD (LD sum, in case of multiple target lesions).
- A patient instead reports stable disease (SD) if no significant increase or decrease is observed on the target lesions.
3.5. Data Annotation, Training Procedure, and Evaluation Metrics
4. Results and Discussion
Performance Analysis
5. Conclusions
6. Patents
Author Contributions
Funding
Conflicts of Interest
References
- Ferlay, J.; Soerjomataram, I.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.M.; Forman, D.; Bray, F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in Globocan 2012. Int. J. Cancer 2015, 136, E359–E386. [Google Scholar] [CrossRef] [PubMed]
- Vaidya, A.; Soloway, M.S.; Hawke, C.; Tiguert, R.; Civantos, F. De Novo Muscle Invasive Bladder Cancer: Is There a Change in Trend? J. Urol. 2001, 165, 47–50. [Google Scholar] [CrossRef] [PubMed]
- De Santis, M.; Bellmunt, J.; Mead, G.; Kerst, J.M.; Leahy, M.; Maroto, P.; Gil, T.; Marreaud, S.; Daugaard, G.; Skoneczna, I.; et al. Randomized Phase II/III Trial Assessing Gemcitabine/Carboplatin and Methotrexate/Carboplatin/Vinblastine in Patients With Advanced Urothelial Cancer Who Are Unfit for Cisplatin-Based Chemotherapy: EORTC Study 30986. J. Clin. Oncol. 2012, 30, 191–199. [Google Scholar] [CrossRef] [PubMed]
- Seront, E.; Machiels, J.-P. Molecular biology and targeted therapies for urothelial carcinoma. Cancer Treat. Rev. 2015, 41, 341–353. [Google Scholar] [CrossRef] [PubMed]
- Powles, T.; Durán, I.; van der Heijden, M.S.; Loriot, Y.; Vogelzang, N.J.; De Giorgi, U.; Oudard, S.; Retz, M.M.; Castellano, D.; Bamias, A. Atezolizumab versus chemotherapy in patients with platinum-treated locally advanced or metastatic urothelial carcinoma (IMvigor211): A multicentre, open-label, phase 3 randomised controlled trial. Lancet 2018, 391, 748–757. [Google Scholar] [CrossRef]
- Bellmunt, J.; De Wit, R.; Vaughn, D.J.; Fradet, Y.; Lee, J.-L.; Fong, L.; Vogelzang, N.J.; Climent, M.A.; Petrylak, D.P.; Choueiri, T.K.; et al. Pembrolizumab as Second-Line Therapy for Advanced Urothelial Carcinoma. N. Engl. J. Med. 2017, 376, 1015–1026. [Google Scholar] [CrossRef]
- Sternberg, C.N.; Loriot, Y.; James, N.; Choy, E.; Castellano, D.; Lopez-Rios, F.; Banna, G.L.; De Giorgi, U.; Masini, C.; Bamias, A.; et al. Primary Results from SAUL, a Multinational Single-arm Safety Study of Atezolizumab Therapy for Locally Advanced or Metastatic Urothelial or Nonurothelial Carcinoma of the Urinary Tract. Eur. Urol. 2019, 76, 73–81. [Google Scholar] [CrossRef]
- Balar, A.V.; Galsky, M.D.; E Rosenberg, J.; Powles, T.; Petrylak, D.P.; Bellmunt, J.; Loriot, Y.; Necchi, A.; Hoffman-Censits, J.; Perez-Gracia, J.L.; et al. Atezolizumab as first-line treatment in cisplatin-ineligible patients with locally advanced and metastatic urothelial carcinoma: A single-arm, multicentre, phase 2 trial. Lancet 2017, 389, 67–76. [Google Scholar] [CrossRef]
- Balar, A.V.; Dreicer, R.; Loriot, Y.; Perez-Gracia, J.L.; Hoffman-Censits, J.H.; Petrylak, D.P.; Van Der Heijden, M.S.; Ding, B.; Shen, X.; Rosenberg, J.E. Atezolizumab (atezo) in first-line cisplatin-ineligible or platinum-treated locally advanced or metastatic urothelial cancer (mUC): Long-term efficacy from phase 2 study IMvigor210. J. Clin. Oncol. 2018, 36, 4523. [Google Scholar] [CrossRef]
- Balar, A.V.; Castellano, D.; O’Donnell, P.H.; Grivas, P.; Vuky, J.; Powles, T.; Plimack, E.R.; Hahn, N.M.; De Wit, R.; Pang, L.; et al. First-line pembrolizumab in cisplatin-ineligible patients with locally advanced and unresectable or metastatic urothelial cancer (Keynote-052): A multicentre, single-arm, phase 2 study. Lancet Oncol. 2017, 18, 1483–1492. [Google Scholar] [CrossRef]
- Vuky, J.; Balar, A.V.; Castellano, D.E.; O’Donnell, P.H.; Grivas, P.; Bellmunt, J. (Joaquim); Powles, T.; Bajorin, D.F.; Hahn, N.M.; De Wit, R.; et al. Updated efficacy and safety of KEYNOTE-052: A single-arm phase 2 study investigating first-line pembrolizumab (pembro) in cisplatin-ineligible advanced urothelial cancer (UC). J. Clin. Oncol. 2018, 36, 4524. [Google Scholar] [CrossRef]
- Seliger, B.; Bono, P.; Kim, J.W.; Spiliopoulou, P.; Calvo, E.; Pillai, R.N.; Ott, P.A.; De Braud, F.G.; Morse, M.A.; Le, D.T.; et al. Efficacy and safety of nivolumab monotherapy in metastatic urothelial cancer (mUC): Results from the phase I/II CheckMate 032 study. J. Clin. Oncol. 2016, 34, 4501. [Google Scholar] [CrossRef]
- Massard, C.; Gordon, M.S.; Sharma, S.; Rafii, S.; Wainberg, Z.A.; Luke, J.; Curiel, T.J.; Colon-Otero, G.; Hamid, O.; Sanborn, R.E.; et al. Safety and Efficacy of Durvalumab (MEDI4736), an Anti–Programmed Cell Death Ligand-1 Immune Checkpoint Inhibitor, in Patients With Advanced Urothelial Bladder Cancer. J. Clin. Oncol. 2016, 34, 3119–3125. [Google Scholar] [CrossRef] [PubMed]
- Apolo, A.B.; Infante, J.R.; Balmanoukian, A.; Patel, M.R.; Wang, D.; Kelly, K.; Mega, A.E.; Britten, C.D.; Ravaud, A.; Mita, A.C.; et al. Avelumab, an Anti–Programmed Death-Ligand 1 Antibody, In Patients With Refractory Metastatic Urothelial Carcinoma: Results From a Multicenter, Phase Ib Study. J. Clin. Oncol. 2017, 35, 2117–2124. [Google Scholar] [CrossRef] [PubMed]
- Aggen, D.H.; Drake, C.G. Biomarkers for immunotherapy in bladder cancer: A moving target. J. Immunother. Cancer 2017, 5, 94. [Google Scholar] [CrossRef] [PubMed]
- Paratore, S.; Banna, G.; D’Arrigo, M.; Saita, S.; Iemmolo, R.; Lucenti, L.; Bellia, D.; Lipari, H.; Buscarino, C.I.; Cunsolo, R.; et al. CXCR4 and CXCL12 immunoreactivities differentiate primary non-small-cell lung cancer with or without brain metastases. Cancer Biomark. 2012, 10, 79–89. [Google Scholar] [CrossRef]
- Rundo, F.; Libertino, S.; Banna, G.L.; Ortis, A.; Stanco, F.; Battiato, S. Evaluation of Levenberg–Marquardt neural networks and stacked autoencoders clustering for skin lesion analysis, screening and follow-up. IET Comput. Vis. 2018, 12, 957–962. [Google Scholar] [CrossRef]
- Addeo, A.; Banna, G.L.; Weiss, G.J. Tumor Mutation Burden—From Hopes to Doubts. JAMA Oncol. 2019, 5, 934–935. [Google Scholar] [CrossRef]
- Banna, G.; Olivier, T.; Rundo, F.; Malapelle, U.; Fraggetta, F.; Libra, M.; Addeo, A. The Promise of Digital Biopsy for the Prediction of Tumor Molecular Features and Clinical Outcomes Associated with Immunotherapy. Front. Med. 2019, 6, 172. [Google Scholar] [CrossRef]
- Lambin, P.; Rios-Velazquez, E.; Leijenaar, R.; Carvalho, S.; Van Stiphout, R.G.P.M.; Granton, P.; Zegers, C.M.L.; Gillies, R.; Boellard, R.; Dekker, A.; et al. Radiomics: Extracting more information from medical images using advanced feature analysis. Eur. J. Cancer 2012, 48, 441–446. [Google Scholar] [CrossRef]
- Wang, X.; Girshick, R.; Gupta, A.; He, K. Non-local Neural Networks. In Proceedings of the 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 7794–7803. [Google Scholar]
- Williams, H.; Cattani, L.; Li, W.; Tabassian, M.; Vercauteren, T.; Deprest, J.; D’Hooge, J. 3D Convolutional Neural Network for Segmentation of the Urethra in Volumetric Ultrasound of the Pelvic Floor. In Proceedings of the 2019 IEEE International Ultrasonics Symposium (IUS), Glasgow, UK, 6–9 October 2019; pp. 1473–1476. [Google Scholar]
- Moradi, P.; Jamzad, M. Detecting Lung Cancer Lesions in CT Images using 3D Convolutional Neural Networks. In Proceedings of the 2019 4th International Conference on Pattern Recognition and Image Analysis (IPRIA), Tehran, Iran, 6–7 March 2019; pp. 114–118. [Google Scholar]
- Wang, G.; Lam, K.-M.; Deng, Z.; Choi, K.-S. Prediction of mortality after radical cystectomy for bladder cancer by machine learning techniques. Comput. Biol. Med. 2015, 63, 124–132. [Google Scholar] [CrossRef] [PubMed]
- Garapati, S.S.; Hadjiiski, L.M.; Cha, K.H.; Chan, H.-P.; Caoili, E.M.; Cohan, R.H.; Weizer, A.; Alva, A.; Paramagul, C.; Wei, J.; et al. Urinary bladder cancer staging in CT urography using machine learning. Med. Phys. 2017, 44, 5814–5823. [Google Scholar] [CrossRef] [PubMed]
- Hasnain, Z.; Mason, J.; Gill, K.; Miranda, G.; Gill, I.S.; Kuhn, P.; Newton, P.K. Machine learning models for predicting post-cystectomy recurrence and survival in bladder cancer patients. PLoS ONE 2019, 14, e0210976. [Google Scholar] [CrossRef] [PubMed]
- Cha, K.H.; Hadjiiski, L.; Samala, R.K.; Chan, H.-P.; Caoili, E.M.; Cohan, R.H. Urinary bladder segmentation in CT urography using deep-learning convolutional neural network and level sets. Med. Phys. 2016, 43, 1882–1896. [Google Scholar] [CrossRef] [PubMed]
- Gordon, M.; Hadjiiski, L.; Cha, K.; Chan, H.-P.; Samala, R.; Cohan, R.H.; Caoili, E.M. Segmentation of inner and outer bladder wall using deep-learning convolutional neural network in CT urography. In Medical Imaging 2017: Computer-Aided Diagnosis, Proceedings of the SPIE Medical Imaging, Orlando, FL, USA, 13–16 February 2017; International Society for Optics and Photonics: Orlando, FL, USA, 2017. [Google Scholar]
- Ma, X.; Hadjiiski, L.M.; Wei, J.; Chan, H.-P.; Cohan, R.H.; Caoili, E.M.; Samala, R.K.; Zhou, C.; Lu, Y.; Cha, K.H. 2D and 3D bladder segmentation using U-Net-based deep-learning. In Medical Imaging 2019: Computer-Aided Diagnosis, Proceedings of the SPIE Medical Imaging, San Diego, CA, USA, 13 March 2019; International Society for Optics and Photonics: San Diego, CA, USA, 2019. [Google Scholar]
- Shkolyar, E.; Jia, X.; Chang, T.C.; Trivedi, D.; Mach, K.E.; Meng, M.Q.-H.; Xing, L.; Liao, J.C. Augmented Bladder Tumor Detection Using Deep Learning. Eur. Urol. 2019, 76, 714–718. [Google Scholar] [CrossRef] [PubMed]
- Wu, E.; Hadjiiski, L.M.; Samala, R.K.; Chan, H.-P.; Cha, K.H.; Richter, C.; Cohan, R.H.; Caoili, E.M.; Paramagul, C.; Alva, A.; et al. Deep Learning Approach for Assessment of Bladder Cancer Treatment Response. Tomography 2019, 5, 201–208. [Google Scholar] [CrossRef] [PubMed]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. Commun. ACM 2017, 60, 84–90. [Google Scholar] [CrossRef]
- Cha, K.H.; Hadjiiski, L.M.; Chan, H.-P.; Weizer, A.Z.; Alva, A.; Cohan, R.H.; Caoili, E.M.; Paramagul, C.; Samala, R.K. Bladder Cancer Treatment Response Assessment in CT using Radiomics with Deep-Learning. Sci. Rep. 2017, 7, 1–12. [Google Scholar] [CrossRef]
- Rundo, F.; Spampinato, C.; Banna, G.; Conoci, S. Advanced Deep Learning Embedded Motion Radiomics Pipeline for Predicting Anti-PD-1/PD-L1 Immunotherapy Response in the Treatment of Bladder Cancer: Preliminary Results. Electronics 2019, 8, 1134. [Google Scholar] [CrossRef]
- Holzinger, A.; Biemann, C.; Kell, D.; Pattichis, C.S. What do we need to build explainable AI systems for the medical domain? arXiv 2017, arXiv:1712.09923. [Google Scholar]
- Samek, W.; Montavon, G.; Vedaldi, A.; Hansen, L.K.; Müller, K.R. (Eds.) Explainable AI: Interpreting, Explaining and Visualizing Deep Learning; Springer: Berlin/Heidelberg, Germany, 2019. [Google Scholar]
- Lee, H.; Yune, S.; Mansouri, M.; Kim, M.; Tajmir, S.H.; Guerrier, C.E.; Ebert, S.A.; Pomerantz, S.R.; Romero, J.M.; Kamalian, S.; et al. An explainable deep-learning algorithm for the detection of acute intracranial haemorrhage from small datasets. Nat. Biomed. Eng. 2018, 3, 173–182. [Google Scholar] [CrossRef]
- Arrieta, A.B.; Díaz-Rodríguez, N.; Del Ser, J.; Bennetot, A.; Tabik, S.; Barbado, A.; Garcia, S.; Gil-Lopez, S.; Molina, D.; Benjamins, R.; et al. Explainable Artificial Intelligence (XAI): Concepts, taxonomies, opportunities and challenges toward responsible AI. Inf. Fusion 2020, 58, 82–115. [Google Scholar] [CrossRef]
- Tonekaboni, S.; Joshi, S.; McCradden, M.; Goldenberg, A. What clinicians want: Contextualizing explainable machine learning for clinical end use. Mach. Learn. Res. 2019, 106, 1–21. [Google Scholar]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-CAM: Visual Explanations from Deep Networks via Gradient-Based Localization. In Proceedings of the 2017 IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017; pp. 618–626. [Google Scholar]
- Eisenhauer, E.; Therasse, P.; Bogaerts, J.; Schwartz, L.; Sargent, D.; Ford, R.; Dancey, J.; Arbuck, S.; Gwyther, S.; Mooney, M.; et al. New response evaluation criteria in solid tumours: Revised RECIST guideline (version 1.1). Eur. J. Cancer 2009, 45, 228–247. [Google Scholar] [CrossRef]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention is all you need. In Advances in Neural Information Processing System, Proceedings of the First 12 Conferences (The MIT Press), 21 November 2001; Jordan, M.I., LeCun, Y., Solla, S.A., Eds.; The MIT Press: Cambridge, MA, USA, 2017. [Google Scholar]
- Huang, G.; Liu, Z.; Pleiss, G.; Van Der Maaten, L.; Weinberger, K. Convolutional Networks with Dense Connectivity. IEEE Trans. Pattern Anal. Mach. Intell. 2019, 1. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Cao, P. Deep Learning Based Data Augmentation and Classification for Limited Medical Data Learning. In Proceedings of the 2019 IEEE International Conference on Power, Intelligent Computing and Systems; Institute of Electrical and Electronics Engineers (IEEE), Shenyang, China, 12–14 July 2019. [Google Scholar]
- Dromain, C.; Beigelman, C.; Pozzessere, C.; Duran, R.; Digklia, A. Imaging of tumour response to immunotherapy. Eur. Radiol. Exp. 2020, 4, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Alsaab, H.O.; Sau, S.; Alzhrani, R.; Tatiparti, K.; Bhise, K.; Kashaw, S.K.; Iyer, A. PD-1 and PD-L1 Checkpoint Signaling Inhibition for Cancer Immunotherapy: Mechanism, Combinations, and Clinical Outcome. Front. Pharmacol. 2017, 8, 561. [Google Scholar] [CrossRef]
- Ding, X.; Chen, Q.; Yang, Z.; Li, J.; Zhan, H.; Lu, N.; Chen, M.; Yang, Y.; Wang, J.; Yang, D. Clinicopathological and prognostic value of PD-L1 in urothelial carcinoma: A meta-analysis. Cancer Manag. Res. 2019, 11, 4171–4184. [Google Scholar] [CrossRef]
- Zhou, T.C.; Sankin, A.I.; Porcelli, S.A.; Perlin, D.S.; Schoenberg, M.P.; Zang, X. A review of the PD-1/PD-L1 checkpoint in bladder cancer: From mediator of immune escape to target for treatment 1 1MPS is an investor in and consultant for Urogen. SAP is consultant and advisor for Vaccinex. The remaining authors have nothing to disclose. Urol. Oncol. Semin. Orig. Investig. 2017, 35, 14–20. [Google Scholar] [CrossRef]
- Spencer, K.R.; Wang, J.; Silk, A.W.; Ganesan, S.; Kaufman, H.L.; Mehnert, J.M. Biomarkers for Immunotherapy: Current Developments and Challenges. Am. Soc. Clin. Oncol. Educ. Book 2016, 36, e493–e503. [Google Scholar] [CrossRef]
- Rundo, F.; Trenta, F.; Di Stallo, A.L.; Battiato, S. Advanced Markov-Based Machine Learning Framework for Making Adaptive Trading System. Computation 2019, 7, 4. [Google Scholar] [CrossRef]
- Rundo, F.; Trenta, F.; Di Stallo, A.L.; Battiato, S. Grid Trading System Robot (GTSbot): A Novel Mathematical Algorithm for trading FX Market. Appl. Sci. 2019, 9, 1796. [Google Scholar] [CrossRef]
- Rundo, F.; Petralia, S.; Fallica, G.; Libertino, S. Lecture Notes in Electrical Engineering; Springer: Berlin/Heidelberg, Germany, 2019; Volume 539, pp. 473–480. [Google Scholar]
| Block | Output Size | Layer(s) Description | Layers Number |
|---|---|---|---|
| Convolution | 32 × 16 × 64 × 64 | 3 × 3 × 3 convolution | 1 |
| Dense Block | 128 × 16 × 64 × 64 | Batch Normalization Rectified Linear Unit (ReLU) 3 × 3 × 3 depth-wise convolution 1 × 1 × 1 point-wise convolution | 6 |
| Transition layer | 128 × 8 × 32 × 32 | 1 × 1 × 1 convolution, 2 × 2 × 2 maxpool | 1 |
| Dense Block | 256 × 8 × 32 × 32 | 8 | |
| Transition layer | 256 × 4 × 16 × 16 | 1 × 1 × 1 convolution, 2 × 2 × 2 maxpool | 1 |
| Dense Block | 384 × 4 × 16 × 16 | 8 | |
| Transition layer | 384 × 2 × 8 × 8 | 1 × 1 × 1 convolution, 2 × 2 × 2 maxpool | 1 |
| Dense Block | 512 × 2 × 8 × 8 | 8 | |
| Transition layer | 512 × 1 × 4 × 4 | 1 × 1 × 1 convolution, 2 × 2 × 2 maxpool | 1 |
| Dense Block | 640 × 1 × 4 × 4 | 8 | |
| Transition layer | 640 × 1 × 2 × 2 | 1 × 1 × 1 convolution, 2 × 2 × 2 maxpool | 1 |
| Dense Block | 736 × 1 × 2 × 2 | 6 | |
| Transition layer | 736 × 1 × 1 × 1 | 1 × 1 × 1 convolution, 2 × 2 × 2 maxpool | 1 |
| Concatenation | 751 | Integrates hematochemical patient’s data | 1 |
| Fully Connected | 375 | FC, ReLU | 1 |
| Fully Connected | 187 | FC, ReLU | 1 |
| Fully Connected | 93 | FC, ReLU | 1 |
| Fully Connected | 46 | FC, ReLU | 1 |
| Fully Connected | 46 | FC, ReLU | 1 |
| Fully Connected | 46 | FC, ReLU | 1 |
| Classification | 2 | FC, Softmax | 1 |
| Dataset Field Description | Number | % |
|---|---|---|
| Age | ||
| ≤60 | 13 | 30 |
| >60 | 30 | 80 |
| Gender | ||
| Male | 39 | 91 |
| Female | 4 | 9 |
| Tobacco Use | ||
| Never | 5 | 12 |
| Current | 16 | 37 |
| Former | 22 | 51 |
| Therapy Line | ||
| 2 | 38 | 88 |
| ≥3 | 5 | 12 |
| Primary Tumor Site | ||
| Upper urinary tract | 4 | 9 |
| Lower urinary tract | 36 | 84 |
| Both | 3 | 7 |
| Metastases Site | ||
| Lymph-nodes only | 14 | 33 |
| Visceral | 29 | 67 |
| Treatment Response | ||
| CR/PR/SD | 16 (43 target lesions) | 37 |
| PD | 27 (63 target lesions) | 63 |
| Follow-up Median -Months- | ||
| CR/PR/SD/PD | 13.4 | 11.1–15.6 |
| Follow-up Imaging | ||
| CT-scan | 41 | |
| MRI (Magnetic Resonance Imaging) | 2 |
| Model | Accuracy | Sensitivity | Specificity | F1-Score |
|---|---|---|---|---|
| 2D ResNet-50 | 0.620 ± 0.052 | 0.604 ± 0.0078 | 0.636 ± 0.061 | 0.613 ± 0.058 |
| 3D DenseNet + H | 0.713 ± 0.047 | 0.711 ± 0.041 | 0.716 ± 0.064 | 0.713 ± 0.043 |
| 3D DenseNet + SepConv + H | 0.733 ± 0.049 | 0.729 ± 0.069 | 0.738 ± 0.047 | 0.731 ± 0.054 |
| 3D DenseNet | 0.640 ± 0.034 | 0.636 ± 0.034 | 0.644 ± 0.048 | 0.638 ± 0.032 |
| 3D DenseNet + NLB + SepConv | 0.878 ± 0.039 | 0.871 ± 0.054 | 0.884 ± 0.075 | 0.877 ± 0.041 |
| Proposed | 0.922 ± 0.037 | 0.929 ± 0.053 | 0.916 ± 0.047 | 0.922 ± 0.038 |
| 2D ResNet-101 | 0.829 ± 0.043 | 0.822 ± 0.054 | 0.836 ± 0.061 | 0.828 ± 0.043 |
| 3D DenseNet-201 | 0.856 ± 0.033 | 0.871 ± 0.047 | 0.840 ± 0.055 | 0.858 ± 0.032 |
| 2D VGG-19 | 0.667 ± 0.033 | 0.662 ± 0.069 | 0.671 ± 0.059 | 0.664 ± 0.041 |
| Previous [34] | 0.861 ± 0.023 | 0.815 ± 0.011 | 0.883 ± 0.048 | 0.810 ± 0.037 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rundo, F.; Banna, G.L.; Prezzavento, L.; Trenta, F.; Conoci, S.; Battiato, S. 3D Non-Local Neural Network: A Non-Invasive Biomarker for Immunotherapy Treatment Outcome Prediction. Case-Study: Metastatic Urothelial Carcinoma. J. Imaging 2020, 6, 133. https://doi.org/10.3390/jimaging6120133
Rundo F, Banna GL, Prezzavento L, Trenta F, Conoci S, Battiato S. 3D Non-Local Neural Network: A Non-Invasive Biomarker for Immunotherapy Treatment Outcome Prediction. Case-Study: Metastatic Urothelial Carcinoma. Journal of Imaging. 2020; 6(12):133. https://doi.org/10.3390/jimaging6120133
Chicago/Turabian StyleRundo, Francesco, Giuseppe Luigi Banna, Luca Prezzavento, Francesca Trenta, Sabrina Conoci, and Sebastiano Battiato. 2020. "3D Non-Local Neural Network: A Non-Invasive Biomarker for Immunotherapy Treatment Outcome Prediction. Case-Study: Metastatic Urothelial Carcinoma" Journal of Imaging 6, no. 12: 133. https://doi.org/10.3390/jimaging6120133
APA StyleRundo, F., Banna, G. L., Prezzavento, L., Trenta, F., Conoci, S., & Battiato, S. (2020). 3D Non-Local Neural Network: A Non-Invasive Biomarker for Immunotherapy Treatment Outcome Prediction. Case-Study: Metastatic Urothelial Carcinoma. Journal of Imaging, 6(12), 133. https://doi.org/10.3390/jimaging6120133